Back to Journals » Journal of Inflammation Research » Volume 15
DNA Repair Mechanisms are Activated in Circulating Lymphocytes of Hospitalized Covid-19 Patients
Authors Olsen MB , Huse C, Sousa MMLD, Murphy SL , Sarno A, Obermann TS, Yang K , Holter JC , Jørgensen MJ, Christensen EE, Wang W, Ji P, Heggelund L , Hoel H, Dyrhol-Riise AM, Gregersen I, Aukrust P, Bjørås M, Halvorsen B , Dahl TB
Received 30 June 2022
Accepted for publication 20 October 2022
Published 7 December 2022 Volume 2022:15 Pages 6629—6644
DOI https://doi.org/10.2147/JIR.S379331
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Maria Belland Olsen,1,2 Camilla Huse,1,2 Mirta Mittelstedt Leal de Sousa,3,4 Sarah Louise Murphy,1,2 Antonio Sarno,3,5 Tobias Sebastian Obermann,3 Kuan Yang,1 Jan Cato Holter,2,6 Marte Jøntvedt Jørgensen,2,7 Erik Egeland Christensen,2,7 Wei Wang,3 Ping Ji,3 Lars Heggelund,8,9 Hedda Hoel,1,10 Anne Margarita Dyrhol-Riise,2,7 Ida Gregersen,1 Pål Aukrust,1,2,11 Magnar Bjørås,3,6 Bente Halvorsen,1,2 Tuva Børresdatter Dahl12
1Research Institute of Internal Medicine, Oslo University Hospital, Oslo, Norway; 2Institute of Clinical Medicine, University of Oslo, Oslo, Norway; 3Department of Clinical and Molecular Medicine, Norwegian University of Science and Technology, Trondheim, Norway; 4Proteomics and Modomics Experimental Core Facility (PROMEC), NTNU, Trondheim, Norway; 5Department of Fisheries and New Biomarine Industry, SINTEF Ocean, Trondheim, Norway; 6Department of Microbiology, Oslo University Hospital and University of Oslo, Oslo, Norway; 7Department of Infectious Diseases, Oslo University Hospital, Oslo, Norway; 8Department of Internal Medicine, Vestre Viken Hospital Trust, Drammen, Norway; 9Department of Clinical Science, Faculty of Medicine, University of Bergen, Bergen, Norway; 10Department of Medicine, Lovisenberg Diaconal Hospital, Oslo, Norway; 11Section of Clinical Immunology and Infectious Diseases, Oslo University Hospital, Oslo, Norway; 12Division of Critical Care and Emergencies, Oslo University Hospital, Oslo, Norway
Correspondence: Tuva Børresdatter Dahl, Division of Critical Care and Emergencies and Research Institute of Internal Medicine, Oslo University Hospital, Sognsvannsveien 20, Oslo, Norway, Tel +4723072786, Email [email protected]
Purpose: Reactive oxygen species (ROS) are an important part of the inflammatory response during infection but can also promote DNA damage. Due to the sustained inflammation in severe Covid-19, we hypothesized that hospitalized Covid-19 patients would be characterized by increased levels of oxidative DNA damage and dysregulation of the DNA repair machinery.
Patients and Methods: Levels of the oxidative DNA lesion 8-oxoG and levels of base excision repair (BER) proteins were measured in peripheral blood mononuclear cells (PBMC) from patients (8-oxoG, n = 22; BER, n = 17) and healthy controls (n = 10) (Cohort 1). Gene expression related to DNA repair was investigated in two independent cohorts of hospitalized Covid-19 patients (Cohort 1; 15 patents and 5 controls, Cohort 2; 15 patients and 6 controls), and by publicly available datasets.
Results: Patients and healthy controls showed comparable amounts of oxidative DNA damage as assessed by 8-oxoG while levels of several BER proteins were increased in Covid-19 patients, indicating enhanced DNA repair in acute Covid-19 disease. Furthermore, gene expression analysis demonstrated regulation of genes involved in BER and double strand break repair (DSBR) in PBMC of Covid-19 patients and expression level of several DSBR genes correlated with the degree of respiratory failure. Finally, by re-analyzing publicly available data, we found that the pathway Hallmark DNA repair was significantly more regulated in circulating immune cells during Covid-19 compared to influenza virus infection, bacterial pneumonia or acute respiratory infection due to seasonal coronavirus.
Conclusion: Although beneficial by protecting against DNA damage, long-term activation of the DNA repair machinery could also contribute to persistent inflammation, potentially through mechanisms such as the induction of cellular senescence. However, further studies that also include measurements of additional markers of DNA damage are required to determine the role and precise molecular mechanisms for DNA repair in SARS-CoV-2 infection.
Keywords: Covid-19, oxidative stress, DNA damage, DNA repair, base excision repair, double strand break repair
Introduction
Reactive oxygen species (ROS) are produced during inflammation to combat pathogens and to stimulate tissue regeneration. However, sustained inflammation and ROS production will also damage DNA.1 The DNA damage response is tightly linked to immune regulation, and persistent DNA damage triggers senescence-associated inflammatory cytokine secretion, creating a positive feedback loop.2–4 Moreover, if inflammation-induced DNA damage is not followed by DNA repair, the result will be DNA damage accumulation and genome instability which over time can have severe pathogenic consequences for the host. Numerous review articles discuss the relevance of oxidative stress and DNA damage linked to inflammation in Covid-19,5–9 but only one study has reported the level of extracellular oxidative RNA/DNA damage to be associated with mortality of Covid-19 patients.10 Furthermore, data on intracellular oxidative DNA damage and in particular DNA repair mechanisms in clinical samples during Covid-19 infection are scarce or lacking.
Covid-19 is caused by SARS-CoV-2, a virus first detected in Wuhan 2019, which rapidly spread across the globe. Most infected individuals experience mild or moderate respiratory tract symptoms, but some develop severe respiratory failure with the need of hospitalization and in the most severe cases treatment at an intensive care unit (ICU).11 To date, several studies have shown that an attenuated anti-viral immune response involving various interferons is associated with disease severity particularly in the initial phase of the disease, accompanied and followed by a marked and sustained inflammation.12 However, the pathogenesis of severe Covid-19 disease is far from completely understood, and the identification of the disease mediators, especially at the molecular level, is a prerequisite for the development of novel treatment options in this complex disorder.
DNA is constantly exposed to damage, and to ensure genome stability the cells contain several DNA repair mechanisms to detect and repair these damages. Among several cellular DNA damage response pathways, base excision repair (BER) is the major pathway for repair of oxidative DNA lesions. The lesions are recognized and excised by DNA glycosylases generating an abasic site that is further processed by short-patch or long-patch BER, leaving the DNA strand repaired and intact.13
Unrepaired, abasic sites may spontaneously yield strand breaks, which if proximal to a strand break on the opposite DNA strand can generate a double-strand break (DSB). The risk of DSB increases during gene transcription, thus activated cells with high transcription rates are more prone to DSBs.14,15 DSBs are a great threat to genome stability by increasing the risk of chromosome rearrangements, and thus disrupting gene structure and function. Therefore, DSB accumulation is detrimental to the cell and DBSs are considered the most severe form of DNA damage. To avoid such harmful events, the cells have developed multiple pathways consisting of separate steps and proteins that if activated can repair DSB.16
Due to the sustained inflammation in patients with moderate to severe Covid-19, we hypothesize that these patients would be characterized by altered oxidative DNA damage and dysregulation of the DNA repair machinery. To test this hypothesis, we examined these parameters in peripheral blood mononuclear cells (PBMC) in a cohort of Norwegian hospitalized Covid-19 patients with various degree of disease severity,17 as well as reanalyzing and extending RNA sequencing data from the same and an additional Norwegian COVID-19 cohort. By using publicly available RNA sequencing data, we investigated Covid-19-related DNA repair gene expression in various types of blood cells. Finally, we also used publicly available RNA sequencing data to compare DNA repair responses in Covid-19 patients to responses in patients with other acute airways infections (ie, seasonal coronavirus, influenza virus and bacterial pneumonia).
Materials and Methods
Study Design and Participants
Study participants in Cohort 1 were recruited from a prospective cohort study of hospitalized adult patients (≥18 years old) with Covid-19 (Norwegian SARS-CoV-2 study; NCT04381819), described elsewhere.17 Briefly, 22 patients with confirmed COVID-19 by a positive SARS-CoV-2 PCR targeting the E-gene on oropharyngeal and nasopharyngeal specimens were consecutively recruited from Oslo University Hospital, Norway, between March 9th and June 12th 2020. Peripheral blood was collected within eight days of admission. PO2/FIO2 (P/F) ratio was measured concurrently (median 41.5, 8–58). Median age at admission was 54 years (30–87), 7 (36%) were females and 11 (68%) Caucasian. Median symptom duration upon sampling was 13 days (range 4–48). Seven (32%) were smokers or former smokers, 6 (28%) had chronic pulmonary disease, 9 (41%) had hypertension, 5 (23%) had chronic heart disease and 1 (5%) had diabetes mellitus. None of the patients were treated with dexamethasone or other specific treatment for Covid-19.
Study participants in Cohort 2 were recruited from an independent add-on study to the WHO Solidarity Trial (The Nor Solidarity Trial, NCT04321616), a multicenter, open-label, adaptive randomized controlled trial evaluating the effect of antiviral agents (hydroxychloroquine and remdesivir) as described elsewhere.18 Eligible patients consisted of adults (≥18 years old), with positive PCR confirmed SARS-CoV-2 infection. In brief, from a subpopulation of the cohort (n = 15), peripheral blood was sampled on the day of inclusion (from June to October 2020), and PBMC was isolated. The median age at admission was 56 years (42–87), and 11 were men (74%). Blood samples were taken before any anti-viral treatment and none were receiving Dexamethasone. In both cohorts, patients with severe co-morbidities such as cancer were not included.
Gender, age and ethnicity matched healthy controls, without any known disease and no current or previous known Covid-19 disease or detectable plasma antibodies for SARS-CoV-2, were recruited from the general population.
PBMC Isolation and Handling
Peripheral blood was collected in BD CPT™ Cell Preparation Tube containing sodium heparin, mixed by inverting the tube 8 times and centrifuged (1650 G,15 minutes, RT). The layer containing PBMC was transferred to falcon tubes and washed twice in PBS followed by centrifugation (300 G, 15 minutes, RT). For Cohort 1, cells were then resuspended in 1 mL RPMI containing 50/ fetal calf serum (FCS) and placed on ice. FCS containing 20% DMSO were added drop-vise (50 μL x 20 in 3 sec intervals). PBMC was placed in a freezing container and stored in DMSO at −150°C until thawed for RNA, DNA and protein isolation. The cells were thawed in a water bath at 37°C, before one volume of pre-warmed complete RPMI 1640 (10% FCS, penicillin, streptomycin, glutamine) was added dropwise. The volume was adjusted to 10 mL, and the cells were spun down and resuspended in complete RPMI media. The cells rested for 2 h in a 37°C incubator with 5% CO2. For Cohort 2, PBMC was pelleted by centrifugation (500 G, 15 min, 4°C). After the supernatant was removed, samples were snap frozen and stored at −80°C until RNA extraction.
Isolation of RNA, DNA and Protein for Cohort 1
The thawed PBMC were spun down, and total RNA and DNA was isolated under RNase-free conditions using Allprep DNA/RNA/Protein Mini Kit (Qiagen) following the manufacturer’s instruction. This kit allows simultaneous purification of genomic DNA, total RNA and protein from the same cell pellet. Briefly, the lysed samples are loaded on DNA columns for column-based DNA isolation. The flow through is used for column-based RNA isolation, and the remaining flow through depleted of RNA and DNA is used for protein isolation with the APP protein precipitation buffer. RNA and DNA concentration and purity based on the 260/230 and 260/280 ratios were determined by spectrophotometer absorbance (NanoDrop ND-1000, Thermo Fisher Scientific). Isolated RNA and DNA were stored at −80°C for further analyses. The protein fraction was stored at −80°C for later isolation before the samples were thawed on ice and protein was precipitated according to manufacturer’s instructions, except for keeping the protein as a dry pellet for targeted mass spectrometry analysis.
Isolation of RNA for Cohort 2
Total RNA was isolated from PBMC with miRNeasy Kit (Qiagen, Cat. No. / ID: 217004), according to manufactures instruction. To increase yield, isopropanol was added to the RLT lysis buffer, per the manufacturer’s recommendation. In brief, RNA was separated from the DNA and protein fraction by phase separation. The aqueous phase (top layer) contains RNA loaded on RNA columns for column-based RNA isolation. Meanwhile, the DNA interphase and organic phase (protein and lipids) were stored for later use. After isolation, RNA concentration and purity were determined by spectrophotometer absorbance (NanoDrop ND-1000, Thermo Fisher Scientific), based on their 260/230 and 260/280 ratios. Isolated RNA was then stored at −80°C until further analysis.
Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) Analysis of 8-Oxoguanine in Genomic DNA
DNA 8-oxoG levels were measured as previously described.19 Briefly, genomic DNA was enzymatically hydrolyzed to deoxyribonucleotides (using benzonase, nuclease P1, and alkaline phosphatase), acetonitrile precipitated, and measured by reverse phase LC-MS/MS (Agilent 1260 HPLC coupled to a 4670 triple quadruple).
Targeted Mass Spectrometry of BER Proteins
Protein pellets obtained using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen) were resuspended in 8M urea, 30 mM Tris pH 8.0, 4% CHAPS containing 5 mM TCEP and incubated for 1 hour at room temperature. 20 µg protein was alkylated with iodoacetamide (1 µmol/mg protein) for 30 min in the dark, followed by precipitation via methanol-chloroform20 and overnight Trypsin (Thermo Scientific) digestion in 50 mM NH4HCO3 at 1:50 ratio (w/w, enzyme:protein) at 37 ◦C in a shaker. Samples were dried, resuspended in 40 µL formic acid and centrifuged for 10 minutes at max. speed (16000 g). 20 µL samples were transferred to HPLC vials and a pool of synthetic peptides (4 fmol/µL) containing heavy labeled Lysine (+8) or Arginine (+10) (PEPotec SRM Grade 2 Peptides, Thermo scientific) was spiked into the samples.
Parallel reaction monitoring (PRM)-based targeted mass spectrometry methods were designed, analyzed, and processed using Skyline software version 21.1.0.278.21 Peptide standards and samples were separated using an EASY-nLC 1200 UHPLC system (Thermo Scientific) using columns and set up described in (3). The following 180 min method was used at 300 nl/min flow rate: starting with 2% buffer B (80% Acetonitrile, 0.1% Formic acid) with an increase to 30% buffer B in 150 min, followed by an increase to 100% Buffer B over 15 min, where it was subsequently held for 15 min. Buffer A consisted of 0.1% Formic acid. The peptides eluting from the column were analyzed on a Thermo Scientific Q Exactive HF mass spectrometer operating in PRM mode using the instrument settings described in.22
Peptide areas for multiple peptides of the same protein were summed to assign relative protein abundance. Endogenous β-actin levels were used for data normalization. Values of relative abundance of each protein in samples belonging to the same group (Control or Patient) were averaged. The error bars represent standard deviation of biological replicates. P-values were calculated based on Student’s t-tests driven by mean difference of the log2 space values. Highly significant values representing p-value <0.01 when compared with Control group are indicated by asterisks (**), and (*) indicates p-value <0.05. Statistics and graphs where prepared by GraphPad Prism 8.3.0. A minimum of 2 peptides per protein was used for quantification, except for MUTYH, where only one peptide was detectable due to low levels of this protein. UNG/UNG2, TDG, OGG1 and NEIL1 were targeted, but not detected. A list of peptides used for quantification is provided in Supplemental File 1. Five patient samples were excluded due to lack of material or sample quality.
RNA Sequencing and Analysis of Data
For Cohort 1, the RNA was sequenced with 2×150 bp flow cell on the Illumina-NovaSeq 6000 platform (OGC, Oxford, UK) at the Norwegian Sequencing Centre, Oslo, Norway. For Cohort 2 the isolated RNA was sent to Novogene (UK) Company Limited, for stranded library preparation and sequencing on the Illumina platform. The fastp software (v0.20.1) was used to remove contaminated adapters and low-quality reads with Phred score below 30 in the pair-end 150 bp raw sequencing files.23 Mapping of filtered reads to the human transcriptome (Gencode Human Release H37) was performed with Salmon (v1.5.2) with 200 bootstrap iterations.24,25 To obtain the differentially expressed transcripts (DETs), the Salmon outputs were imported into DESeq2 (v1.32.0) via tximport (v.1.20.0).26,27 Only the protein coding transcripts (p ≤ 0.05) were selected and uploaded to Metascape for gene functional enrichment analysis.28
To examine the correlation between the gene expression and P/F ratio, the Pearson correlation coefficient was calculated between natural log transformed normalized transcript counts and p/f ratio by using Hmisc R package (v4.6). Significantly correlated protein coding transcripts (p < 0.02, correlation coefficient > ±0.6) were uploaded to Metascape for gene functional enrichment analysis. Network plot was generated by Cytoscape. GraphPad Prism 8.3.0 was used for additional statistics and presentation of genes involved in mismatch repair (MMR), nucleotide excision repair (NER), BER and DSBR.
Expression of DNA Repair Genes in Publicly Available Datasets
To examine the DNA repair gene expression profile of different cell types in Covid patients and healthy controls, we download the single cell sequencing dataset GSE174072.29 All genes in each single cell were normalized to CPM (counts per million mapped reads) and the average CPM of interested genes in each person was calculated. Then individual’s CPM were calculated into average CPM for the Covid patient group and for the healthy control group. Finally, the averaged CPM were natural log transformed and presented in heatmaps as z-score (pheatmap R package (v1.0.12)).
RNA sequencing data from CD4 T cells were downloaded (GSE1794478)30 and from raw counts differently expressed genes (DEGs) were obtained by DESeq2 (v1.32.0).26 Enrichment analysis including DEGs were performed by Metascape. DEGs belonging to the pathway DNA repair (GO:0006281) and/or Reactome pathway DNA repair (R-HSA- 5693532) were merged in separate heatmaps representing gene expression in conventional T cells (CD4+CD25−) and regulatory T cells (CD4+CD25highCD127low) as defined by the authors of the original data.30 To examine DNA repair gene expression profiles in the periphery between COVID-19 and other lower lung infections, the public whole-blood RNA sequencing dataset GSE161731 was downloaded.31 From this dataset, Covid-19 patients (6 female, 9 male, median age 33 [20–89]) with symptom duration ≤10 days (5 hospitalized and 10 non-hospitalized) were compared to patients hospitalized for acute respiratory infection due to seasonal coronavirus (24 female, 16 male, 15 unknown sex, mean age 18 [18–59]), influenza (8 female, 9 male, median age 18 [0–51]), or bacterial pneumonia (14 female, 10 male, mean age 64 [33–85]). Normalized gene expression was obtained from the raw counts by DESeq2 (v1.36.0). Gene Enrichment Analysis (GSEA32) was performed with the Molecular signatures database (MSigDB, v.7.5.1) on the curated Hallmark (H) gene set.33 In contrast to in house data, all publicly available datasets were analysed at gene level.
Results
Base Excision Repair is Upregulated in PBMC from Hospitalized Covid-19 Patients
Enhanced inflammation, a well-known feature of severe Covid-19, may induce accumulation of DNA damage through increased production of ROS. Therefore, we first examined the levels of in vivo oxidative DNA damage in hospitalized Covid-19 patients (Cohort 1). 8-Oxoguanine (8-oxoG) was measured by liquid chromatography tandem mass spectrometry (LC-MS/MS) in DNA extracted from immune cells (PBMC) collected from hospitalized Covid-19 patients (n = 22, 14 sampled day 1 and 7 sampled day 6–8 after hospitalization) and non-infected healthy controls (n = 10). Notably, 8-oxoG levels from these Covid-19 patients did not differ from levels measured in healthy controls (Figure 1A). Moreover, there was no correlation between 8-oxoG levels and the degree of respiratory failure as assessed by P/F-ratio where low P/F ratio characterize those with the most severe degree of respiratory failure (Figure 1B). Moreover, no significant correlations were found between 8-oxoG levels and other clinical (ie, time since symptom onset, age and co-morbidities as assessed by the Charlson Comorbidity Index34) and inflammatory (ie, C-reactive protein (CRP), ferritin and number of leukocytes, neutrophils, monocytes and lymphocytes) markers (Table S1).
 |
Figure 1 Base excision repair proteins are upregulated in PBMC from hospitalized Covid-19 patients. (A) Levels of 8-oxoG in DNA extracted from PBMC obtained from hospitalized Covid-19 patients (n = 22) and healthy controls (n = 11). 8-oxoG was measured by liquid chromatography with tandem mass spectrometry (LC-MS/MS). (B) Relationship between 8-oxoG levels and the degree of respiratory failure as assessed by pO2/FiO2-(P/F-ratio) where low P/F ratio characterize those with the most severe degree of respiratory failure. Correlation analysis by Pearson correlation coefficient. (C) Comparison of relative BER protein abundances measured by targeted MS. The error bar shows the standard deviation of measured peak area in samples from each group (control or patient). Endogenous β-actin levels were used for data normalization. A list of peptides used for quantification is provided in Supplemental File 1. Student’s t test, *p < 0.05, **p < 0.001. Values are shown as mean ± SEM. PBMC; peripheral blood mononuclear cells, BER; base excision repair. Illustration created with BioRender.com. Related to Figure S2. |
To examine how the DNA repair machinery responded in hospitalized Covid-19, we performed a targeted proteomic analysis of central BER proteins in PBMC isolated from the same cohort of hospitalized Covid-19 patients (Cohort 1; n = 18, 12 day 1–3 and 6 day 6–8 after hospitalization) and non-infected healthy controls (n = 10). Higher levels of several BER proteins were detected in hospitalized Covid-19 patients, but not all components to a significant level as compared with controls (Figure 1C). Among the DNA glycosylases, the levels of Nth like DNA glycosylase 1 (NTHL1), N-methylpurine DNA glycosylase (MPG), Single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1) and Methyl-CpG binding domain 4, DNA glycosylase (MBD4) were significantly higher in Covid-19 patients compared to controls. AP endonuclease 1 (APE1) and X-ray repair cross complementing 1 (XRCC1), which promote short patch repair, and DNA polymerase delta 1 (POLD1), flap structure-specific endonuclease 1 (FEN1) and DNA ligase 1 (LIG1), involved in long patch repair, were also significantly increased in hospitalized Covid-19 patients compared to non-infected healthy controls. The 8-oxoG DNA glycosylase, OGG1, was below detection level in all patient samples. UNG/UNG2, TDG, and NEIL1, all DNA glycosylases of the BER pathway, were also targeted, but not detected. Again, no correlation was found between BER proteins MPG, APE1 or LIG1 levels and clinical (ie, time since symptom onset, age and co-morbidities) and inflammatory (ie, CRP, ferritin and number of leukocytes, neutrophils, monocytes and lymphocytes) markers (Table S2).
Enrichment Analyses Support Regulation of DNA Repair in PBMC from Hospitalized Covid-19 Patients
To explore the regulation of other DNA repair pathways during SARS-CoV-2 infection, we reanalyzed published RNA sequencing data from the same study cohort (Cohort 1).17 Protein coding transcripts regulated in hospitalized Covid-19 patients compared to non-infected controls (2150 transcripts) were included in enrichment analysis. The top 15 enriched gene ontology (GO) terms and Reactome pathways (R-HSA) are presented in Figure 2A. In addition to “Cellular response to DNA damage stimulus”, several of the top 15 terms and pathways are closely linked to DNA repair and DNA damage responses, for example “Mitotic cell cycle process” and “Transcriptional regulation by RUNX1”. Each of these 15 terms/pathways represent groups of enriched terms/pathways sharing many common genes (Supplemental File 2) and Figure 2B presents selected terms and pathways found in our material that relates to stress-induced DNA damage responses. Among these were “DNA repair” (GO) and “DNA Repair” (R-HSA), and their regulated transcripts are presented in Figure 2C. As illustrated in the figure, some of the DNA repair genes could further be linked to BER while the majority were linked to double strand break repair (DSBR) or both. These results were further replicated in another cohort of 15 hospitalized Covid-19 patients compared to 6 healthy controls (Cohort 2; Figure S1). Similar as for Cohort 1, both the GO term and Reactome pathway “DNA Repair” were significantly enriched when including protein coding transcripts regulated in hospitalized Covid-19 patients in the analysis. Many of these DNA repair genes could, also here, be linked to BER and DSBR. Regulation of BER protein coding transcripts in Cohort 1 was comparable to BER protein levels found for Cohort 2 (Figures 1C and S2).
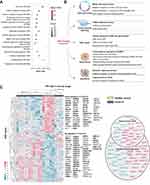 |
Figure 2 Enrichment analyses confirm regulation of DNA repair in PBMC from hospitalized Covid-19 patients. (A) Top 15 regulated groups of Gene Ontology (GO) terms and Reactome (R-HSA) pathways found by enrichment analysis including regulated protein coding transcripts in PBMC from Covid-19 patients (n = 15) vs healthy controls (n = 5). (B) Illustration of cellular processes affected by DNA damage response proteins linked to selected significantly regulated GO terms and Reactome pathways found in the enrichment analysis (all terms and pathways listed in. Supplemental File 2). (C) Heatmap showing the protein coding transcripts enriched in the GO term DNA repair (GO:0006281) and Reactome pathway DNA repair (R-HSA- 5693532). For genes with multiple transcripts, the number of DETs corresponding to each gene is marked by the number brackets. Genes linked to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). PBMC; peripheral blood mononuclear cells. Illustration created with BioRender.com. Related to Figure S1. |
DNA Repair Gene Expression Correlates to Disease Severity
As seen in Figure 2C (Cohort 1), gene expression levels in four patient samples clustered together with healthy controls. Within this cohort, these patients had the highest P/F-ratio values (all above 50 kPa), indicating normal respiratory function. Next, we therefore sought to explore if expression of DNA repair genes was associated with P/F-ratio within the Covid-19 cohort. Pearson correlation analysis was performed to detect association between protein coding transcripts and the degree of respiratory failure as assessed by P/F ratio. By this, we detected 2065 protein coding transcripts significantly correlating to P/F ratio (p < 0.02, r>0.6). Gene enrichment analysis showed enrichment of several terms and pathways closely linked to DNA repair and DNA damage responses as “Cellular response to DNA damage stimulus” and “Oxidative stress induced senescence” (Supplemental File 3). “DNA repair” (GO) was also significantly enriched, and as the Network plot illustrates (Figure 3A), the majority of the transcripts were positively correlated to P/F-ratio (eg, Nei like DNA Glycosylase 2 (NEIL2), ATM serine/threonine kinase (ATM), RPA-related protein RADX and Sirtuin 1 (SIRT1)), ie, low RNA expression in patients with the most severe degree of respiratory failure. Twenty out of 69 were negatively correlated to P/F ratio (eg, DNA polymerase epsilon 2, accessory subunit (POLE2), Poly(ADP-ribose) polymerase 2 (PARP2), and Nudix hydrolase 1 (NUDT1)), thus high RNA levels in those with the most severe degree of respiratory failure. Although several of these genes have roles in DSBR, several DNA repair pathways were represented among the correlated transcripts. As examples, correlation plots are shown for MutL Homolog 3 (MLH3), MMR; General transcription factor IIH subunit 5 (GTF2H5), NER; NEIL2, BER and ATM, DSBR in Figure 3B.
 |
Figure 3 DNA repair gene expression correlates to disease severity. (A) Network plot showing transcripts correlating to P/F-ratio and enriched in DNA repair (GO:0006281). Pearson correlation analysis (p < 0.02, r>±0.6). Genes linked to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). (B) Representative correlation plots are shown. MMR; mismatch repair, NER; nucleotide excision repair, BER; base excision repair and DSBR; double strand break repair. Figure is related to Supplemental File 3. |
Cell Type Specific Regulation of DNA Repair Genes
To exhibit the regulation of DNA repair genes in different immune cell types, we extracted gene expression profiles from previous published single cell RNA sequencing data analyzing PBMC from hospitalized Covid-19 patients (n = 7) and healthy controls (n = 5) (GSE174072).29 From these data we generated heatmaps presenting genes involved in BER (Figure 4A) and DSBR (Figure 4B). These data further suggest regulation of DNA repair genes in Covid-19 patients compared with healthy controls. These data also show that the regulation of DNA repair genes in Covid-19 patients to some degree depend on cell type, where the highest degree of gene regulation was observed in CD4 +T-cells, γδ T-cells, class switched B-cells and IgA and IgG plasma cells. Conveniently, RNA sequencing of CD4+T cells from Covid-19 patients has previously been performed by Galván-Peña and colleagues.30 Reanalyzing of their publicly available dataset (GSE179448) show indeed that DNA repair genes are enriched in both CD4+CD25− conventional T cells (R-HSA-73894 DNA Repair; Log q-value= −2395, 33/335 terms/list) and CD4+CD25highCD127low regulatory T cells (GO:00062 DNA Repair; Log q-value = −11.003, 52/469 terms/list) (Supplemental File 4). Confirming cell type differences in regulation of these genes, DNA repair gene regulation for the two CD4+T cell populations is shown in Figure 5.
 |
Figure 5 CD4 T cell expression of DNA repair genes in hospitalized Covid-19. Heatmaps generated from publicly available RNA sequencing data (GSE1794478) showing DNA repair genes significantly regulated in CD4 T cells from hospitalized Covid-19 patients compared to control. CD4+CD25− cells were defined as (A) conventional T cells and CD4+CD25highCD127low cells were defined as (B) regulatory T cells.30 DNA repair genes were defined as genes included in gene ontology term DNA repair (GO:0006281) and/or Reactome pathway DNA repair (R-HSA- 5693532). Genes affiliated to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). Gene enrichment analysis results are listed in Supplemental File 4). |
Gene Regulation of DNA Repair Genes is Significantly Upregulated in Hospitalized Covid-19 Compared to Patients Hospitalized with Other Lower Airway Infections
In theory, the altered gene regulation of DNA repair genes seen here could be a hallmark of airway infection in general, and not a Covid-19 specific trait. Therefore, we finally sought to examine the regulation of DNA repair genes during infection caused by Sars-CoV-2 compared to other causes of lower airway infections. Wang and colleagues have made a blood atlas that includes RNA-sequencing data of whole blood from healthy controls and patients with Covid-19 (n = 15), acute respiratory infection due to seasonal coronavirus (n = 51), influenza virus (n = 17) or bacterial pneumonia (n = 24),31,35 (Dataset GSE161731). By Gene Set Enrichment Analysis (GSEA), we performed three comparisons of gene expression; (1) Covid-19 vs bacterial pneumonia, (2) Covid-19 vs acute respiratory infection due to seasonal coronavirus and (3) Covid-19 vs Influenza. As shown in Figure 6A, the pathway Hallmark DNA repair was significantly more activated in immune cells during Covid-19 compared to other infections. Figure 6B shows expression levels (ie, fold change) of a selection of DNA repair genes in Covid-19 compared to these other infectious diseases. Several of these genes show the same expression pattern as when comparing Covid-19 with healthy controls (although not all were regulated at a significant level), for example the BER genes MPG, POLD1, LIG1 and FEN1. These data, however, must be interpreted with some caution. Thus, it seems that the Covid-19 patients had in general less severe disease (5 of 15 hospitalized) than other patient groups (all hospitalized). Moreover, although age may influence the levels of DNA repair mechanisms,36 Covid-19 patients were both older (seasonal corona and influenza) and younger (bacterial pneumonia) than the other disease categories (see Methods).
Discussion
In the present study, we show that although severe Covid-19 are known to be characterized by enhanced inflammation and oxidative stress, hospitalized Covid-19 patients did not accumulate 8-oxoG compared to non-infected healthy controls. We did, however, observe a marked regulation of the DNA repair system in these patients with increased levels of several BER enzymes. In addition to BER genes, genes involved in DSBR were also significantly regulated in hospitalized Covid-19, as shown by gene expression analysis. Furthermore, several BER and DSBR-genes correlated with the degree of respiratory failure. Overall, our data show activation of the DNA repair machinery in immune cells of hospitalized Covid-19 patients, and this activation seems to be more prominent for Covid-19 than for other common lower airway infections. Although beneficial by protecting against DNA damage, these responses could also potentially contribute to persistent inflammation through mechanisms such as the induction of cellular senescence. Our data, however, should be interpreted with caution and confirmed in other larger studies that also include measurements of additional markers for DNA damage.
One in vitro study in primary mouse embryonic fibroblasts showed that hydroxychloroquine, a medication administrated to Covid-19 patients in some countries during 2020, but of none in the present study, induced increased 8-oxoG levels in cellular DNA.37 It has also been reported that RNA vaccine against SARS-CoV-2 induced transient increase in DNA damage in PBMC.38 However, to the best of our knowledge, this is the first report on genomic 8-oxoG levels and DNA repair capacity in hospitalized Covid-19 patients. Increased levels of serum oxidative RNA/DNA damage were recently shown to be associated with mortality of Covid-19 patients,10 implying higher ROS production in the severely ill patients. Surprisingly, we found that 8-oxoG levels in immune cells from hospitalized Covid-19 patients were comparable to levels in healthy controls, even in patients with the most severe disease. This could reflect increased DNA repair capacity as the Covid-19 patients showed increased protein levels of several DNA glycosylases initiating BER, including the glycosylase; MPG (recognizing alkylation lesions), SMUG1 (uracil residues) and MBD4 (G:T mismatches). In line with an increased repair capacity, expression levels of NUDT1 were increased in Covid-19 patients compared to healthy controls and NUDT1 expression correlated to the degree of respiratory failure. The protein encoded by this gene is an enzyme that neutralizes oxidized nucleotides and thereby prevents incorporation of damaged bases into DNA.39 Thus, high levels of NUDT1 are indicative of ROS-induced activation of the DNA repair machinery in hospitalized Covid-19 patients.
These findings contrast with our earlier studies on other infectious diseases involving cytomegalovirus (CMV) or HIV infection.40,41 Here, CMV-infected fibroblasts showed declined DNA glycosylase activity in removal of oxidized and alkylated bases.41 Moreover, CD4+ T cells from HIV-infected patients showed increased levels of 8-oxoG accompanied by marked decline in DNA glycosylase activity for repair of oxidative lesions.40 On the other hand, the DNA damage response and DNA repair signaling were shown to be activated by the coronavirus infectious bronchitis virus [15]. It is possible that difference could reflect different regulation of these pathways in chronic infection such as HIV and CMV infection as compared with acute infection such as seen in hospitalized Covid-19 patients. Thus, data on these variables in patients suffering from long-Covid symptoms would be of major interest.
Our data may seem in contrast to the study by Mihaljevic et al, who found enhanced levels of DNA damage, correlated to the degree of inflammation in hospitalized Covid-19 patients.42 The reasons for these apparent discrepancies are not clear but could at least partly reflect differences in the methodology. Thus, Mihaljevic et al used the comet assay with denaturation gel to measure DNA damage, detecting both single- and double strand breaks, and notably, also the present study shows dysregulation of genes involved in DSBR. There are also some differences in the characteristics of the two study populations such as the use of corticosteroid that was not used of any patients in the present study, whereas Mihaljevic et al found a positive correlation between the use of such medications and the degree of DNA damage.42
Transcriptome regulation in immune cells is a dynamic process in order to adapt the immune response to alterations of infection state, and RNA sequencing data reflect a snapshot at the specific time of cell harvest. Therefore, rather than looking at regulation of specific transcripts, it is valuable to take into account the alteration of the biological pathways. We do see a consistent enrichment of DNA repair pathways in cohort 1, cohort 2 and in CD4 T-cells, although the regulated genes vary between cohorts. In both PBMC and CD4 T cells, several of the differentially expressed DNA repair genes had roles in DSBR, as assessed by their affiliation to the GO term and/or Reactome pathway DSBR. Although many of these genes may be involved in repair of multiple forms of DNA damage (eg, poly(ADP-ribose) polymerase 1 PARP1),43 the level of strand breaks (single and DBSs) and the activation of DNA damage responses would be valuable to have investigated further.
Whereas enhanced DNA repair mechanism may protect against DNA damage, increased long-term activation of DNA repair responses could, if not resolved, induce cellular senescence and persistent inflammation.2,4 Interestingly, Lee and colleagues have showed that SARS-Cov-2 evokes cellular senescence as a primary stress response in infected cells, accompanied by a senescence-associated secretory phenotype.44 We find in our RNA datasets that several pathways related to cellular senescence are significantly enriched. In line with this, we have for Cohort 2 included in this study, recently reported an association for the senescence markers chitotriosidase and stathmin 1 (plasma) and cyclin-dependent kinase inhibitor 2A (PBMC) with pulmonary pathology in these patients three months after hospitalization with COVID-19,45 illustrating the potential for these pathways in long-term pathology in Covid-19 disease.
At present, the mechanisms for SARS-CoV-2-related dysregulation of DNA damage and repair are not clear. DNA repair proteins, including proteins of BER and DSBR, are involved in multiple cellular processes beyond canonical DNA repair, including cell cycle regulation,46 inflammation,2,47 immune class switching47 and chromatin remodeling.48 In the present study, we found that the term “Cell cycle” gets the highest score in gene enrichment analyses when included protein coding transcripts regulated in Covid-19 patients. A recent review suggested that SARS-CoV2 induces S-phase cell cycle arrest by promoting replication fork stress, possibly through its virus protein nsp13 which is known to interfere with DNA polymerase δ (POLD).49 We do not know how POLD functions in Covid-19 patients based, but we do however observe an increase of POLD1 protein levels in PBMC and upregulation of POLD3 in regulatory T cells. However, cell cycle regulators do also have important immunomodulatory roles in, for example, interferon signaling and T cell activation.50 In line with this, we have recently suggested that persistent activation of check-point inhibitors may predispose to cellular exhaustion in Covid-19 patients.51
The present study has several limitations such as low number of patients and lack of follow-up data after the first week of hospitalization. Moreover, measurements of other forms of DNA damage, including detection of DSBs are lacking and hampered the conclusion of the study. Future studies should include measurements of both DSBs, additional targets of BER (eg, 8-hydroxy-2’-deoxyguanosine (8-OHdG), 8-hydroxyadenine (8-OHA) or 5-hydroxycytocine (5-OHC)), and other forms of DNA-damage. For example, phosphorylation of serine 139 of the histone variant H2AX, referred to as γH2AX, an early cellular response to DBSs and levels of DBSs could have been estimated by measuring γH2AX levels.52 Further, it would have been valuable to include data on DNA repair capacity for isolated Covid-19 PBMC, and for a selection of the regulated transcripts, protein levels should have been measured. Finally, caution is needed when performing several correlation analyses in the relatively small study group and both positive and negative findings should be interpreted cautiously.
Conclusions
We report that hospitalized Covid-19 patients have normal levels of oxidative stress induced DNA damage in their PBMC as assessed by 8-oxoG, potentially reflecting an up-regulation of several pathways and proteins involved in DNA repair. Further studies are needed to verify and extend these data by including measurements of other forms of DNA damage and posttranslational modifications of DNA repair proteins. How the balance between DNA damage and DNA repair changes over time in relation to long-Covid symptomatology should also be examined.
Abbreviations
BER, base excision repair; DSBR, double strand break repair; DSB, double-strand break; MMR, mismatch repair; NER, nucleotide excision repair; PBMC, peripheral blood mononuclear cells; ROS, Reactive oxygen species.
Data Sharing Statement
Regarding PBMC RNA sequencing data, an institutional data transfer agreement can be established, and data can be shared if the aims of data use are covered by ethical approval and patient consent. The procedure will involve an update to the ethical approval as well as review by legal departments at both institutions, and the process will typically take 2 to 4 months from initial contact.
Ethics Approval and Informed Consent
Informed consent was obtained from all patients or from next-of-kin if the patient was incapacitated and therefore unable to give consent. The studies were approved by the Committee for Medical Research Ethics, Region Southeast Norway (Cohort 1, 106624; Cohort 2, 118684) and the Norwegian Medicines Agency (Cohort 1 20/04950-23, Cohort 2, 20/04950-23). Regional Committee for Medical and Health Research Ethics in South-Eastern Norway (reference numbers 106624 and 2019/306). The study complies with the Declaration of Helsinki.
Acknowledgments
We thank the Norwegian SARS-Cov-2 Study Group and the patients for their contribution of clinical materials. Three publicly available datasets were downloaded and used for this study and we thank the authors of both original papers.29–31,35
Funding
This study has received funding from the Research Council of Norway (grant no. 312780), a private donation from Vivaldi Invest A/S owned by Jon Stephenson von Tetzchner and South-Eastern Norway Regional Health Authority (Helse Sør-Øst RHF, abbreviated HSØ) funding no. 2021071.
Disclosure
The authors declare no conflict of interest.
References
1. Kay J, Thadhani E, Samson L, Engelward B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair (Amst). 2019;83:102673. doi:10.1016/j.dnarep.2019.102673
2. Rodier F, Coppe JP, Patil CK, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi:10.1038/ncb1909
3. Cavanagh MM, Weyand CM, Goronzy JJ. Chronic inflammation and aging: DNA damage tips the balance. Curr Opin Immunol. 2012;24(4):488–493. doi:10.1016/j.coi.2012.04.003
4. Kang C, Xu Q, Martin TD, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349(6255):aaa5612. doi:10.1126/science.aaa5612
5. Chernyak BV, Popova EN, Prikhodko AS, Grebenchikov OA, Zinovkina LA, Zinovkin RA. COVID-19 and Oxidative Stress. Biochemistry (Mosc). 2020;85(12):1543–1553. doi:10.1134/S0006297920120068
6. Cecchini R, Cecchini AL. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med Hypotheses. 2020;143:110102. doi:10.1016/j.mehy.2020.110102
7. Suhail S, Zajac J, Fossum C, et al. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: a Review. Protein J. 2020;39(6):644–656. doi:10.1007/s10930-020-09935-8
8. Laforge M, Elbim C, Frere C, et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol. 2020;20(9):515–516. doi:10.1038/s41577-020-0407-1
9. Shabrish S, Mittra I. Cytokine Storm as a Cellular Response to dsDNA Breaks: a New Proposal. Front Immunol. 2021;12:622738. doi:10.3389/fimmu.2021.622738
10. Lorente L, Martin MM, Gonzalez-Rivero AF, et al. DNA and RNA Oxidative Damage and Mortality of Patients With COVID-19. Am J Med Sci. 2021;361(5):585–590. doi:10.1016/j.amjms.2021.02.012
11. Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi:10.1056/NEJMoa2001017
12. Osuchowski MF, Winkler MS, Skirecki T, et al. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9(6):622–642. doi:10.1016/S2213-2600(21
13. Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012583. doi:10.1101/cshperspect.a012583
14. Schwer B, Wei PC, Chang AN, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc Natl Acad Sci U S A. 2016;113(8):2258–2263. doi:10.1073/pnas.1525564113
15. Puget N, Miller KM, Legube G. Non-canonical DNA/RNA structures during Transcription-Coupled Double-Strand Break Repair: roadblocks or Bona fide repair intermediates? DNA Repair (Amst). 2019;81:102661. doi:10.1016/j.dnarep.2019.102661
16. Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20(11):698–714. doi:10.1038/s41580-019-0152-0
17. Christensen EE, Jorgensen MJ, Nore KG, et al. Critical COVID-19 is associated with distinct leukocyte phenotypes and transcriptome patterns. J Intern Med. 2021;290(3):677–692. doi:10.1111/joim.13310
18. Barratt-Due A, Olsen IC, Nezvalova-Henriksen K, et al. Evaluation of the Effects of Remdesivir and Hydroxychloroquine on Viral Clearance in COVID-19: a Randomized Trial. Ann Intern Med. 2021;174(9):1261–1269. doi:10.7326/M21-0653
19. Olsen MB, Hildrestrand GA, Scheffler K, et al. NEIL3-Dependent Regulation of Cardiac Fibroblast Proliferation Prevents Myocardial Rupture. Cell Rep. 2017;18(1):82–92. doi:10.1016/j.celrep.2016.12.009
20. Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–143. doi:10.1016/0003-2697(84)
21. MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi:10.1093/bioinformatics/btq054
22. Quiles-Jimenez A, Gregersen I, Mittelstedt Leal de Sousa M, et al. N6-methyladenosine in RNA of atherosclerotic plaques: an epitranscriptomic signature of human carotid atherosclerosis. Biochem Biophys Res Commun. 2020;533(4):631–637. doi:10.1016/j.bbrc.2020.09.057
23. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi:10.1093/bioinformatics/bty560
24. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi:10.1038/nmeth.4197
25. Frankish A, Diekhans M, Ferreira AM, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi:10.1093/nar/gky955
26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
27. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi:10.12688/f1000research.7563.2
28. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
29. Wilk AJ, Lee MJ, Wei B, et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J Exp Med. 2021;218(8):548. doi:10.1084/jem.20210582
30. Galvan-Pena S, Leon J, Chowdhary K, et al. Profound Treg perturbations correlate with COVID-19 severity. Proc Natl Acad Sci U S A. 2021;118(37):865. doi:10.1073/pnas.2111315118
31. McClain MT, Constantine FJ, Henao R, et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat Commun. 2021;12(1):1079. doi:10.1038/s41467-021-21289-y
32. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
33. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi:10.1016/j.cels.2015.12.004
34. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi:10.1016/0021-9681(87)
35. Wang L, Balmat TJ, Antonia AL, et al. An atlas connecting shared genetic architecture of human diseases and molecular phenotypes provides insight into COVID-19 susceptibility. Genome Med. 2021;13(1):83. doi:10.1186/s13073-021-00904-z
36. Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35(22):7466–7474. doi:10.1093/nar/gkm756
37. Besaratinia A, Caliri AW, Tommasi S. Hydroxychloroquine induces oxidative DNA damage and mutation in mammalian cells. DNA Repair (Amst). 2021;106:103180. doi:10.1016/j.dnarep.2021.103180
38. Ntouros PA, Vlachogiannis NI, Pappa M, et al. Effective DNA damage response after acute but not chronic immune challenge: SARS-CoV-2 vaccine versus Systemic Lupus Erythematosus. Clin Immunol. 2021;229:108765. doi:10.1016/j.clim.2021.108765
39. Rai P, Sobol RW. Mechanisms of MTH1 inhibition-induced DNA strand breaks: the slippery slope from the oxidized nucleotide pool to genotoxic damage. DNA Repair (Amst). 2019;77:18–26. doi:10.1016/j.dnarep.2019.03.001
40. Aukrust P, Luna L, Ueland T, et al. Impaired base excision repair and accumulation of oxidative base lesions in CD4+ T cells of HIV-infected patients. Blood. 2005;105(12):4730–4735. doi:10.1182/blood-2004-11-4272
41. Ranneberg-Nilsen T, Bjoras M, Luna L, et al. Human cytomegalovirus infection modulates DNA base excision repair in fibroblast cells. Virology. 2006;348(2):389–397. doi:10.1016/j.virol.2006.01.001
42. Mihaljevic O, Zivancevic-Simonovic S, Cupurdija V, et al. DNA damage in peripheral blood lymphocytes of severely ill COVID-19 patients in relation to inflammatory markers and parameters of hemostasis. Mutagenesis. 2022. doi:10.1093/mutage/geac011
43. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–621. doi:10.1038/nrm.2017.53
44. Lee S, Yu Y, Trimpert J, et al. Virus-induced senescence is driver and therapeutic target in COVID-19. Nature. 2021. doi:10.1038/s41586-021-03995-1
45. Lekva T, Ueland T, Halvorsen B, et al. Markers of cellular senescence is associated with persistent pulmonary pathology after COVID-19 infection. Infect Dis. 2022:1–6. doi:10.1080/23744235.2022.2113135
46. Murray JM, Carr AM. Integrating DNA damage repair with the cell cycle. Curr Opin Cell Biol. 2018;52:120–125. doi:10.1016/j.ceb.2018.03.006
47. Bednarski JJ, Sleckman BP. At the intersection of DNA damage and immune responses. Nat Rev Immunol. 2019;19(4):231–242. doi:10.1038/s41577-019-0135-6
48. Azzouz D, Khan MA, Palaniyar N. ROS induces NETosis by oxidizing DNA and initiating DNA repair. Cell Death Discov. 2021;7(1):113. doi:10.1038/s41420-021-00491-3
49. Panico P, Ostrosky-Wegman P, Salazar AM. The potential role of COVID-19 in the induction of DNA damage. Mutat Res Rev Mutat Res. 2022;789:108411. doi:10.1016/j.mrrev.2022.108411
50. Laphanuwat P, Jirawatnotai S. Immunomodulatory Roles of Cell Cycle Regulators. Front Cell Dev Biol. 2019;7:23. doi:10.3389/fcell.2019.00023
51. Troseid M, Dahl TB, Holter JC, et al. Persistent T-cell exhaustion in relation to prolonged pulmonary pathology and death after severe COVID-19: results from two Norwegian cohort studies. J Intern Med. 2022. doi:10.1111/joim.13549
52. Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi:10.1038/leu.2010.6
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.