Back to Journals » International Journal of Nanomedicine » Volume 13
DNA- and RNA-binding ability of oligoDapT, a nucleobase-decorated peptide, for biomedical applications
Authors Musumeci D , Roviello V, Roviello GN
Received 8 November 2017
Accepted for publication 19 January 2018
Published 1 May 2018 Volume 2018:13 Pages 2613—2629
DOI https://doi.org/10.2147/IJN.S156381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Thomas Webster
Domenica Musumeci,1,2 Valentina Roviello,3 Giovanni N Roviello1
1CNR-Institute of Biostructure and Bioimaging, Naples, Italy; 2Department of Chemical Sciences, University of Naples Federico II, Naples, Italy; 3Analytical Chemistry for the Environment and Centro Servizi Metereologici Avanzati, University of Naples Federico II, Naples, Italy
Background: Nucleobase-bearing peptides and their interaction with DNA and RNA are an important topic in the development of therapeutic approaches. On one hand, they are highly effective for modulating the nucleic-acid-based biological processes. On the other hand, they permit to overcome some of the main factors limiting the therapeutic efficacy of natural oligonucleotides, such as their rapid degradation by nucleases.
Methods and results: This article describes the synthesis and characterization of a novel thymine-bearing nucleoamino acid based on the l-diaminopropionic acid (l-Dap) and its solid phase oligomerization to α-peptides (oligoDapT), characterized using mass spectrometry, spectroscopic techniques, and scanning electron microscopy (SEM) analysis. The interaction of the obtained nucleopeptide with DNA and RNA model systems as both single strands (dA12, rA12, and poly(rA)) and duplex structures (dA12/dT12 and poly(rA)/poly(rU)) was investigated by means of circular dichroism (CD) and ultraviolet (UV) experiments. From the analysis of our data, a clear ability of the nucleopeptide to bind nucleic acids emerged, with oligoDapT being able to form stable complexes with both unpaired and double-stranded DNA and RNA. In particular, dramatic changes in the dA12/dT12 and poly(rA)/poly(rU) structures were observed as a consequence of the nucleopeptide binding. CD titrations revealed that multiple peptide units bound all the examined nucleic acid targets, with TLdap/A or TLdap/A:T(U) ratios >4 in case of oligoDapT/DNA and ~2 in oligoDapT/RNA complexes.
Conclusion: Our findings seem to indicate that Dap-based nucleopeptides are interesting nucleic acid binding-tools to be further explored with the aim to efficiently modulate DNA- and RNA-based biological processes.
Keywords: nucleopeptides, nucleic acid interaction, poly(rA) binding, circular dichroism
Introduction
A large number of chimeric compounds that bear nucleobases or other types of heteroaromatic rings connected to amino acid-containing moieties were discovered in various natural sources or are the result of synthetic efforts.1–5 Among these, of great interest are nucleoamino acids or nucleobase amino acids (NBAs). The natural (S)-willardiine and its analogs,6,7 for example, act as potent agonists of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptors or kainate receptors, whereas a large number of these synthetic monomers constitute the building blocks for the assembly of synthetic nucleobase-containing peptides (nucleopeptides), investigated for their potential applications in biomedicine and biomaterial fields.8–13
The range of applications of nucleopeptides recalls that of peptide nucleic acids (PNAs), the known DNA analogs, which are achiral, non-charged oligomers with a pseudo-peptide backbone, largely used in numerous diagnostic, antigene, and antisense strategies.14–20 The presence of both nucleobases and peptide backbone in nucleopeptides offers the possibility of binding nucleic acids or protein targets, modulating important cell regulatory processes, such as the viral reverse transcriptase inhibition,21–23 similar to other oligonucleotide (ODN) systems.24
NBA-containing peptides were used to select nucleic acid-related proteins derived from Saccharomyces cerevisiae and to detect the peptide/protein interactions on T7 phages displaying proteins.25 Moreover, the ability of nucleopeptide to deliver ODNs into cells was recently demonstrated treating HeLa cells with NBA-containing peptides incubated in the presence of a fluorescent Cy5-labeled hairpin DNA. Fluorescence microscopy studies evidenced how nucleopeptides deliver hairpin DNA to the cytosols of live cells, while cell viability assays showed their biocompatibility even at high dosages after 72 h of treatment.26
Despite all their favorable applications, one of the existing problems in the development of synthetic ODN analogs is the nonspecific interaction with DNA and RNA. A previous study reported that inserting short (R)-diethylene glycol (miniPEG) moieties in the PNA backbone can improve the specificity of the interaction.27 Alternatively, the insertion of chiral amino acids, possibly positively charged, into the PNA backbone provided both an improved water solubility and chirality, which has proven crucial in conferring high-sequence selectivity to artificial ODNs.8,27
Overall, the important role that NBA-containing structures play in biomedicine, as well as the possibility to realize new DNA- and RNA-binding nucleopeptides, prompted us to realize and investigate a new nucleobase-decorated artificial peptide, whose repeating unit is composed of a dipeptide moiety, based on the L-diaminopropionic acid (L-Dap), connected via a short linker to the thymine nucleobase (Figure 1).
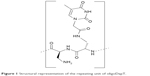  | Figure 1 Structural representation of the repeating unit of oligoDapT. |
The oligomerization of the building block depicted in Figure 1 leads to homothymine α-peptides (oligoDapT; Figure S1) in which the base-carrying L-Dap residues are alternated in the sequence with unmodified L-Dap, affording a cationic oligomer as a result of the protonation of the side chain amino groups at physiological pH. Thus, in addition to favorable H-bonding, potential electrostatic interactions are originated between oligoDapT and nucleic acids (Figure S2).
We here describe a convenient synthesis of oligoDapT, as well as the evaluation of its biomolecular recognition properties toward both single- and double-stranded DNA and RNA targets assessed by circular dichroism (CD) and ultraviolet (UV) spectroscopies.
Materials and methods
Materials
O-(7-aza-1H-Benzotriazol-1-yl)-N, N, N′, N′-tetramethyluronium hexafluorophosphate (HATU) and benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) were purchased from Novabiochem (Merck S.p.a., Milan, Italy), 9-fluorenylmethoxycarbonyl (Fmoc)-L-Dap (tert-butoxycarbonyl [Boc])-OH was purchased from Bachem (Weil am Rhein, Germany). Anhydrous N, N-dimethylformamide (DMF) was obtained from Labscan (Gliwice, Poland); piperidine from Biosolve (Valkenswaard, The Netherlands); high-performance liquid chromatography (HPLC) solvents and acetic anhydride (Ac2O) from Riedel-de Haën (Seelze, Germany); poly(rA), thymin-1-yl acetic acid (TCH2COOH), trifluoroacetic acid (TFA), 2,4,6-trimethylpyridine (TMP), and Rink-amide resin from Fluka (München, Germany); dichloromethane (DCM) and TFA (for HPLC) from Romil (Cambridge, UK); dA12 from Biomers.net GmbH (Ulm, Germany); and deuterated dimethyl sulfoxide (DMSO) from Sigma-Aldrich Co. (Milan, Italy).
1H NMR (nuclear magnetic resonance) and 13C NMR spectra were recorded at 25°C on a Varian 600 MHz (150 MHz) unit. Chemical shifts (δ) are given in parts per million (ppm), and all coupling constants (J) are given in hertz. 1H NMR chemical shifts are referenced to residual CHD2SOCD3 (δ = 2.49, quin). 13C NMR chemical shifts are referenced to the solvent (CD3SOCD3: δ = 39.5). Samples were centrifuged at 4,000 rpm on a Z 200 A centrifuge (Hermle from Del Chimica, Napoli, Italy).
Samples of both monomer and nucleopeptides underwent liquid chromatography–mass spectrometry (LC-MS) analysis (Figures S3 and S4, respectively) on an MSQ mass spectrometer (ThermoElectron, Milan, Italy) equipped with an electrospray ionization (ESI) source operating at 3 kV needle voltage and 320°C and with a complete Surveyor HPLC System, comprising an MS pump, an autosampler, and a photodiode array (PDA) detector, by using a Phenomenex (Castel Maggiore, Italy) Jupiter C18 300 Å (5 μm, 4.6 × 150 mm) column. We performed the gradient elution at 25°C (monitoring at 260 nm) building up a gradient starting with buffer A (0.05% TFA in water) and applying buffer B (0.05% TFA in acetonitrile) with a flow rate of 0.8 mL·min−1. We performed the semipreparative purifications on a Hewlett-Packard/Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA, USA), equipped with a PDA detector, and used a Phenomenex Jupiter C18 300 Å (10 μm, 4.6 × 250 mm) column. Gradient elution was performed at 25°C (monitoring at 260 nm) by using a gradient that started with buffer A’ (0.1% TFA in water) and applying buffer B’ (0.1% TFA in acetonitrile) with a flow rate of 4 mL·min−1. Samples were lyophilized from water in an FD4 freeze dryer (Heto Lab Equipment, Birkerød, Denmark) for 16 h.
Synthetic procedures
Synthesis of monomer 3
Commercial Fmoc/Boc-protected 2,4-diaminopropionic acid 1 (Fmoc-L-Dap(Boc)-OH: 50 mg, 0.12 mmol; Figure 2) was treated with a 1:1 TFA/DCM solution (2 mL) at 45°C, and the mixture was stirred for 1.5 h. Afterward, the solvent was removed in vacuo, and the crude mixture was treated with cold diethyl ether. After centrifugation, a white precipitate was recovered by filtration and repeated washings with diethyl ether, and was dried in vacuo. This product was dissolved in anhydrous DMF (1 mL) treated with N,N-diisopropylethylamine (DIEA; 0.9 equiv., 19 μL, 0.11 mmol) and TMP (0.6 equiv., 9 μL, 0.07 mmol), and then reacted with TCH2COOH (2: 2.2 equiv., 49 mg, 0.26 mmol), previously preactivated with HATU (2.0 equiv., 91 mg, 0.24 mmol) and DIEA (2.0 equiv., 42 μL, 0.24 mmol)/TMP (2.0 equiv., 32 μL, 0.24 mmol) in DMF (1 mL) for 2 min.
After 1.5 h, the solvent was removed in vacuo. The crude material was treated with water, sonicated, and filtered in order to remove the aqueous solution from the white precipitate. This solid was purified by semipeparative HPLC8 and the desired product (3) was obtained as a pure compound in good yield (47 mg, 0.25 mmol, 81% yield): LC-ESI-MS (Figure S3) m/z 493.92 (found), 493.50 (expected for [C25H24N4O7+H]+ =[M+H]+); m/z 515.85 (found), 515.48 (expected for [C25H24N4O7+Na]+); m/z 986.06 (found), 985.99 (expected for [2(C25H24N4O7)+H]+); δH (600 MHz, DMSO-d6; Figure S5) 11.28 (1H, s, NH thymine), 8.28 (1H, bs, NH amide), 7.89 (2H, d, J = 7.2, aromatic protons, CH Fmoc), 7.73 (2H, d, J = 7.8, aromatic protons, CH Fmoc), 7.60–7.33 (6H, m, aromatic protons, CH Fmoc, Fmoc-NH, CH thymine), 4.33–4.11 (6H, m, CH2 linker, FmocCH–CH2 and CHα), and 3.58–3.31 (2H, m, CH2NH), 1.74 (3H, s, CH3 thymine); δC (150 MHz, DMSO-d6; Figure S6) 171.97 (COOH), 167.43 (CH2CONH), 164.47 (thymine C-4), 156.08 (OCONH), 151.01 (thymine C-2), 143.80 (aromatic carbons, Fmoc), 142.28 (thymine C-6), 140.75 (aromatic carbons, Fmoc), 127.68 (aromatic carbons, Fmoc), 127.14 (aromatic carbons, Fmoc), 125.29 (aromatic carbons, Fmoc), 120.15 (aromatic carbons, Fmoc), 108.02 (thymine C-5), 65.79 (Fmoc CH2), 53.77 (CHα), 49.14 (CH2 acetyl linker), 46.62 (Fmoc CH), 39.92 (CH2NH), and 11.93 (thymine CH3).
Nucleopeptide synthesis
Solid-phase synthesis was performed in analogy with other literature reports28 on a Rink-amide-NH2 resin (0.55 mmol/g, 18 mg). After resin Fmoc cleavage by treatment with 25% piperidine in DMF over 30 min and repeated DMF washings of the resin, a mixture of Fmoc-L-Dap(Boc)-OH 1 or monomer 3 (300 μL of a 0.1 M solution in DMF, 30 μmol, 3 equiv.), PyBOP (50 μL of a 0.6 M solution in DMF, 30 μmol, 3 equiv.), and DIEA (10 μL, 60 μmol, 6 equiv.) was introduced into the reactor containing the free-NH2 resin and stirred for 20 min. Then, the coupling mixture was removed from the resin, and the solid support was washed repeatedly with DMF. Unreacted amino functions were capped by treating the resin with Ac2O (20%)/DIEA (5%) in DMF over 15 min. Finally, Fmoc was removed by treatment with 25% piperidine in DMF over 15 min. This procedure was repeated for the sequential insertion of L-Dap and L-DapT on the solid support. The overall yield, as estimated by UV Fmoc test, was 65% in case of the hexathymine derivative 5, whereas pentathymine nucleopeptide 4 was obtained in 89% yield. The deprotection and detachment of the nucleopeptides from the resin were realized by treatment with TFA/triisopropylsilane/H2O (95%/2.5%/2.5%) over 2 h. After nucleopeptide precipitation with cold diethyl ether, the samples were treated with milliQ H2O and purified by reversed-phase (RP)-HPLC using a gradient (10% [for 5 min] to 30%) of B’ in A’ over 30 min, collecting the peak at tR = 16.8 min.
After HPLC purification, the nucleopeptide samples were dissolved in a known amount of milliQ water and quantified by UV (T = 85°C, absorbance measured at λ = 260 nm). The used molar extinction coefficients of 43,000 cm−1·M−1 (4) and 51,600 cm−1·M−1 (5) were calculated starting from that corresponding to the thymine-containing PNA monomer (ie, 8,600 cm−1·M−1). ESI-MS characterization of oligoDapT 4 (Figure S4A): m/z: 1,795.59 (found), 1,795.76 (expected for [C68H99N33O26+2H]++); 899.72 (found), 898.85 (expected for [C68H99N33O26+3H]+++); and 600.20 (found), 599.26 (expected for [C68H99N33O26+4H]++++) and 5 (Figure S4B): m/z: 1,025.23 (found), 1,024.50 (expected for [C78H111N37O30+2H]++); and 683.24 (found), 683.34 (expected for [C78H111N37O30+3H]+++).
Scanning electron microscopy (SEM) analysis
Samples for microscopic observations were obtained using compounds in solution. After slow solvent evaporation of a droplet taken from a 167 μM solution in H2O, the sample of oligoDapT was coated with a conductive layer of Au–Pd and examined in Nova NanoSem 450 SEM from FEI Technologies Inc. (Hillsboro, OR, USA) at 2 and 3 kV in a high-vacuum mode. DNA (dA12) and RNA (poly(rA))/nucleopeptide complex solutions were obtained from the CD titrations with oligoDapT.
CD and UV studies
CD and UV studies were carried out in analogy with other literature studies.29–31 In particular, we obtained the CD spectra in a 200–320 nm wavelength range at 5°C on a J-810 spectropolarimeter (Jasco Europe S.R.L., Cremella, Italy) equipped with a Peltier PTC-423S/15, using a Hellma (Milan, Italy) tandem quartz cell (2 × 0.4375 cm). UV spectra were recorded on a UV–Vis Jasco model V-550 spectrophotometer equipped with a Peltier ETC-505T temperature controller.
Results and discussion
Synthesis of the thymine-containing peptides
The artificial peptides were obtained following the synthetic procedure described in Figure 2.
First, we synthesized monomer 3 by reacting commercial Fmoc/Boc-protected L-Dap monomer 1 with TCH2COOH (2) and characterized it by ESI-MS (Figure S3) and NMR spectroscopy (Figures S5 and S6). Subsequently, we used in a sequential manner 3 and 1 in order to synthesize in the solid phase, using Fmoc-chemistry protocol, two oligomers, ie, pentathymine 4 and hexathymine 5 (Figure 2). Fmoc UV measurements during the synthesis showed general high coupling efficiencies and, thus, overall good yields of the final nucleopeptides. After detachment from the resin and deprotection under acidic conditions, the constructs 4 and 5 were purified by RP-HPLC and characterized by ESI-MS (Figures S4). The obtained oligomers presented peptide chains in which half of the residues carried the DNA nucleobases, while the others were base free, thus ensuring positive charges to the overall structures under physiological conditions. This led to cationic nucleopeptides, highly soluble in aqueous solution, that had both base–base and ionic (−NH3+/−PO3−) binding possibilities toward natural nucleic acids.
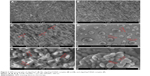
SEM analysis
SEM imaging of dried oligoDapT led to the micrographs shown in Figure 3.
Surface morphology of oligoDapT appears with a low roughness and some thread structures (Figure 3A). Several dense and long thread like structures, fiber-like, clearly appear (Figure 3B). At high magnifications, we found a fiber thickness ranging between 10 and 20 nm (Figure 3C) similar to that obtained by SEM, for self-assembling peptides rich in ionic and hydrophobic residues,32 or by TEM for aromatic peptides.33 In analogy with other nucleopeptide systems,26 here the interaction between aromatic moieties (nucleobases) has a role in the formation of the observed fibril-like structures.
Moreover, we analyzed by SEM the peptide/nucleic acid complexes to elucidate both morphology and size of the resulting structures. SEM of oligoDapT/DNA (Figure 3D and E) and oligoDapT/RNA (Figure 3F) revealed the absence of thread like structures and the formation of spherical morphologies (width 160–200 nm), in analogy with other literature reports on nucleic acid–peptide complexes or hybrids,34,35 arising from the nucleic acid/nucleopeptide binding, which induces the clustering of several nanofibers into larger aggregates.26
Nucleic acid-binding studies
Afterward, we studied the interaction of oligoDapT with nucleic acids, in both single- and double-stranded forms, by means of CD and UV spectroscopies. We recorded in a two-chamber quartz cuvette the sum of CD and UV spectra relative to the separated solutions of the nucleopeptide (4 μM concentration in thymine) and complementary single-stranded nucleic acids (dA12 DNA, rA12 RNA, and poly(rA) RNA, all at 4 μM concentration in adenine) in 10 mM phosphate buffer (pH 7.5) at 5°C and compared them with those obtained after the abovementioned solutions were mixed. Extensive changes in DNA and RNA CD spectra upon mixing the solutions of the two-cell reservoirs reflected strong oligoDapT/DNA and oligoDapT/RNA interactions (Figures 4A, 5A, and 6A).
We have found only minor UV changes (Figure 4B) for oligoDapT/dA12 mixed in a 1:1 ratio, suggesting a minor role of the base stacking in the formation of the TLdap/dA complex. On the other hand, the analogous oligoDapT/rA12 system largely involved nucleobase stacking as evidenced by the significant UV hypochromic effect (Figure 5B). An even more pronounced hypochromic effect was evidenced when poly(rA) was used instead of rA12 (Figure 6B). This RNA vs DNA selectivity, previously evidenced also for other L-diamino acid-based nucleopeptides by CD and surface plasma resonance (SPR) experiments,36 can be associated with additional H-bonds with RNA structures due to ribose 2′-OH moieties (absent in DNA) that reinforce the overall ODN/nucleopeptide structuring.
By titrating the oligoDapT/DNA solution with further amounts of the nucleopeptide, we observed that the CD profiles underwent further modifications in correspondence of TLdap/dA ratios higher than 2:1, which resulted in dramatic changes in both shape and intensity of the CD bands (Figure 4C). More in detail, a negative band at 274 nm and a positive one at 260 nm appeared, with the 260 nm band showing also a hypsochromic shift of its maximum upon titration (from 260 nm at TLdap/dA = 2 to 256 nm at TLdap/dA = 5). A substantial stabilization of the CD signal was observed at a 5:1 (TLdap/dA) ratio, indicating a multimeric oligoDapT/DNA complex.
Analogously, we performed a titration experiment also with rA12 and poly(rA); the latter is an RNA suggested as an important anticancer target.37,38 The stoichiometry of the obtained oligoDapT/RNA complexes was investigated by adding oligoDapT aliquots to the 1:1 (TLdap/rA) complex solutions until stabilization of the CD signal: for both rA12 and poly(rA), the formation of complexes with a 2/1 = TLdap/rA stoichiometry was evidenced (Figures 5A and 6A). However, upon titration with the nucleopeptide, more dramatic conformational changes in the poly(rA) with respect to rA12, reflected in strong CD profile changes, were observed.
To get more insight into the nature of the nucleopeptide/nucleic acid recognition, and in particular to establish the relative contribution of the electrostatic forces and sequence-specific recognition to the interaction, a binding experiment of oligoDapT with a noncomplementary poly(rU) strand was performed, and the obtained spectra were compared with those shown in Figure 6A for the poly(rA). CD experiments showed that the nucleic acid structure of the homouracil RNA underwent only negligible changes upon oligoDapT addition (Figure 6C), in contrast to the dramatic RNA structure perturbation observed for the oligoDapT/poly(rA) binding (Figure 6A). These findings indicate that the interaction is mainly driven by complementary base pairing, with the NH 3+/phosphate ionic recognition being only of secondary importance, ie, useful to reinforce the overall interaction but not the major driving force.
The importance of new drugs able to bind double-stranded DNA, as well as to interfere with double-stranded RNA-driven biological processes39–41 or act as stabilizers and carriers of double-stranded RNA drugs,42–46 prompted us to study the interaction of oligoDapT with dA12/dT12 and poly(rA)/poly(rU), chosen as double-stranded DNA and RNA model systems, respectively. First, the target complexes were prepared by mixing equimolar amounts of DNA and RNA single strands, heating the resulting solutions at 90°C for 10 minutes and then slowly cooling overnight. Subsequently, by using a two-chamber quartz cuvette, we recorded and compared the CD spectra relative to the sum of the separated solutions of oligoDapT and double-stranded nucleic acids with those recorded after mixing the solutions and also after further oligoDapT additions. CD changes in the DNA/DNA mixture spectrum were evident when a 1/1 TLdap/[dA:dT] ratio was achieved, with a stabilization of the signal only after further oligoDapT addition, corresponding to a 4/1 TLdap/[dA:dT] ratio. Moreover, a bathochromic shift (4 nm) for the positive band at ~280 nm was observed in the CD titration (Figure 7).
More dramatic changes were observed studying the interaction of oligoDapT with the RNA:RNA duplex, with a significant decrease in the overall CD signal intensity and a hypsochromic shift of the positive band at 266 nm, relative to the poly(rA):poly(rU) complex, to 259 nm in the spectrum of the ternary complex oligoDapT/RNA:RNA when a 2/1 TLdap/[rA:rU] ratio was reached (Figure 8). CD titration experiments suggested the formation of a complex with a 2/1 stoichiometry relative to the TLdap and rA:rU bases, since a certain CD signal stabilization was obtained for this mixture.
Taken together, the abovementioned findings demonstrated the ability of oligoDapT to bind all the nucleic acid systems selected for this study. Furthermore, CD titration experiments evidenced some differences in the binding stoichiometry of our nucleopeptide toward DNA vs RNA. In fact, the TLdap/A or TLdap/[A:T(U)] ratios were >4 in the case of oligoDapT/DNA and ~2 in the case of oligoDapT/RNA complexes.
Thermal stabilities of oligoDapT/nucleic acid complexes
Another aspect we have investigated is the thermal stability of the complexes formed by oligoDapT with DNA and RNA. To this aim, UV thermal denaturation experiments were performed (Figures S7–S11), whose results are summarized in Table 1.
  | Table 1 Melting temperatures for homothymine/homoadenine natural (DNA/DNA or RNA/RNA) and artificial (nucleopeptide/DNA and nucleopeptide/RNA) complexes |
In the case of DNA, our nucleopeptides 4 and 5, containing, respectively, 5 and 6 TLdap units, formed with the dA12 strand more stable complexes than that obtained with the natural and longer dT12 (Figures S7 and S8). Regarding RNA, we observed a Tm of 47.9°C for the poly(rU)/poly(rA) complex, whereas an almost 20°C more stable complex was formed, under the same experimental conditions, when poly(rA) bound oligoDapT (Table 1 and Figures S10 and S11). This stabilization is greater also than that relative to other literature nucleotide/RNA complexes.47 Moreover, oligoDapT forms more stable complexes with RNA than with DNA of the same nucleotide sequence, as observed comparing Tms of oligoDapT 4/dA12 and oligoDapT 4/rA12 (ΔTm ~10°C; Figure S9).
All the results obtained for the nucleopeptide/nucleic acid complexes can be explained considering the contribution of several factors to the recognition ability: 1) the H-bonding of complementary nucleobases; 2) the electrostatic interactions between the positive amino groups along the nucleopeptide chain and the negative phosphates of the nucleic acid strand; 3) the possibility for nucleopeptides to form multimeric structures also in complex with nucleic acids;26 and 4) the additional H-bonding possibilities involving the 2′-OH groups of the ribose ring (in RNA).
Conclusion
Here, we reported the synthesis of a novel thymine-bearing nucleoamino acid based on L-Dap and its convenient oligomerization in the solid phase to α-peptides (oligoDapT), characterized by ESI-MS and SEM analyses. Nucleic acid-binding studies, based on CD and UV experiments carried out on oligoDapT nucleopeptides with both single- and double-stranded DNA and RNA, evidenced sequence-specific interactions in all cases. Remarkable changes in the structures of the dA12/dT12 and poly(rA)/poly(rU) complexes were observed by CD spectroscopy upon oligoDapT addition. CD titrations revealed that multiple oligoDapT units bound all the examined nucleic acids, with TLdap/A or TLdap/[A:T(U)] ratios twofold higher for oligoDapT/DNA than for oligoDapT/RNA complexes. Additionally, UV melting experiments showed higher thermal stabilities of oligoDapT/nucleic acid complexes with respect to the natural DNA/DNA and RNA/RNA complexes and a higher stability of oligoDapT/RNA vs oligoDapT/DNA complexes, probably associated with the additional H-bonding given by 2′-OH in ribose moieties of RNA.
In future, we plan to synthesize oligoDap oligomers of different lengths containing all four nucleobases in order to evaluate their binding properties toward complementary nucleic acids and their ability to form suitable nucleopeptide-based biomaterials.48
Overall, the here-reported oligoDapT showed interesting properties exploitable in the context of molecular devices for potential biomedical applications; indeed, this is a nucleic acid binder able to target complementary DNA and RNA of interest in antigene, antisense, and double-stranded RNA targeting antiviral strategies, as well as to be used for the delivery of therapeutic ODNs, a recently evidenced property of nucleopeptides.26
Acknowledgment
We thank Prof Antonio Roviello and Mr Leopoldo Zona for their precious suggestions.
Disclosure
The authors report no conflicts of interest in this work.
References
Roviello GN, Gaetano SD, Capasso D, Franco S, Crescenzo C, Bucci EM. Pedone C RNA-binding and viral reverse transcriptase inhibitory activity of a novel cationic diamino acid-based peptide. J Med Chem. 2011;54:2095–2101. | ||
Weckenmann NM, Nachtsheim BJ. Enantioselective synthesis of D-α-(Uracil-5-yl) glycine derivatives and their racemization-free incorporation into a model peptide. Eur J Org Chem. 2015;30:6624–6630. | ||
Roviello GN, Ricci A, Bucci EM, Pedone C. Synthesis, biological evaluation and supramolecular assembly of novel analogues of peptidyl nucleosides. Mol Biosyst. 2011;7(5):1773–1778. | ||
Scognamiglio PL, Di Natale C, Perretta G, Marasco D. From peptides to small molecules: an intriguing but intricated way to new drugs. Curr Med Chem. 2013;20(31):3803–3817. | ||
Shi NQ, Gao W, Xiang B, Qi XR. Enhancing cellular uptake of activable cell-penetrating peptide-doxorubicin conjugate by enzymatic cleavage. Int J Nanomedicine. 2012;7:1613–1621. | ||
Dolman NP, More JC, Alt A, et al. Structure-activity relationship studies on N3-substituted willardiine derivatives acting as AMPA or kainate receptor antagonists. J Med Chem. 2006;49(8):2579–2592. | ||
Mizutani M, Jitsukawa K, Masuda H, Einaga H. Synthesis and X-ray structure of a unique multinuclear complex constructed by a nucleobase-amino acid DL-willardiine. Chem Comm. 1996;11(11):1389–1390. | ||
Roviello GN, Musumeci D. Synthetic approaches to nucleopeptides containing all four nucleobases, and nucleic acid-binding studies on a mixed-sequence nucleo-oligolysine. RSC Adv. 2016;6(68):63578–63585. | ||
Roviello GN, Roviello G, Musumeci D, Bucci EM, Pedone C. Dakin-West reaction on 1-thyminyl acetic acid for the synthesis of 1,3-bis(1-thyminyl)-2-propanone, a heteroaromatic compound with nucleopeptide-binding properties. Amino Acids. 2012;43(4):1615–1623. | ||
Roviello GN, Roviello V, Autiero I, Saviano M. Solid phase synthesis of TyrT, a thymine–tyrosine conjugate with poly(A) RNA-binding ability. RSC Adv. 2016;6:27607–27613. | ||
Roviello GN, Musumeci D, Pedone C, Bucci EM. Synthesis, characterization and hybridization studies of an alternate nucleo-epsilon/gamma-peptide: complexes formation with natural nucleic acids. Amino Acids. 2010;38(1):103–111. | ||
Roviello GN, Musumeci D, Moccia M, et al. dabPNA: design, synthesis, and DNA binding studies. Nucleos Nucleot Nucl Acids. 2007;26:1307–1310. | ||
Roviello GN, Musumeci D, Bucci EM, Pedone C. Synthesis and characterization of a novel ester-based nucleoamino acid for the assembly of aromatic nucleopeptides for biomedical applications. Int J Pharm. 2011;415(1–2):206–210. | ||
Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254(5037):1497–1500. | ||
Milano G, Musumeci D, Gaglione M, Messere A. An alternative strategy to synthesize PNA and DNA magnetic conjugates forming nanoparticle assembly based on PNA/DNA duplexes. Mol Biosyst. 2010;6(3):553–561. | ||
Roviello GN, Musumeci D, De Cristofaro A, et al. Alternate dab-aegPNAs: synthesis, nucleic acid binding studies and biological activity. Mol Biosyst. 2009;6(1):199–205. | ||
Amato F, Tomaiuolo R, Borbone N, et al. Design, synthesis and biochemical investigation, by in vitro luciferase reporter system, of peptide nucleic acids as new inhibitors of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. Med Chem Commun. 2014;5:68–71. | ||
Amato F, Tomaiuolo R, Nici F, et al. Exploitation of a very small peptide nucleic acid as a new inhibitor of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. Biomed Res Int. 2014;2014:610718. | ||
Pinto B, Rusciano G, D’Errico S, et al. Synthesis and label free characterization of a bimolecular PNA homo quadruplex. Biochim Biophys Acta. 2017;1861(5 Pt B):1222–1228. | ||
Roviello GN, Musumeci D, Bucci EM, et al. Evidences for complex formation between L-dabPNA and aegPNA. Bioorg Med Chem Lett. 2008;18(17):4757–4760. | ||
Roviello GN, Di Gaetano S, Capasso D, Cesarani A, Bucci EM, Pedone C. Synthesis, spectroscopic studies and biological activity of a novel nucleopeptide with Moloney murine leukemia virus reverse transcriptase inhibitory activity. Amino Acids. 2010;38(5):1489–1496. | ||
Roviello GN, Vicidomini C, Gaetano SD, Capasso D, Musumeci D, Roviello V. Solid phase synthesis and RNA-binding activity of an arginine-containing nucleopeptide. RSC Adv. 2016;6(17):14140–14148. | ||
Roviello GN, Crescenzo C, Capasso D, et al. Synthesis of a novel Fmoc-protected nucleoaminoacid for the solid phase assembly of 4-piperidyl glycine/L-arginine-containing nucleopeptides and preliminary RNA: interaction studies. Amino Acids. 2010;39(3):795–800. | ||
Musumeci D, Riccardi C, Montesarchio D. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules. 2015;20(9):17511–17532. | ||
Watanabe S, Tomizaki KY, Takahashi T, Usui K, Kajikawa K, Mihara H. Interactions between peptides containing nucleobase amino acids and T7 phages displaying S. cerevisiae proteins. Pept Sci. 2007;88(2):131–140. | ||
Du X, Zhou J, Li X, Xu B. Self-assembly of nucleopeptides to interact with DNAs. Interface Focus. 2017;7(6):20160116. | ||
Sahu B, Sacui I, Rapireddy S, et al. Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing γ-peptide nucleic acids with superior hybridization properties and water solubility. J Org Chem. 2011;76:5614–5627. | ||
Di Natale C, Scognamiglio PL, Cascella R, et al. Nucleophosmin contains amyloidogenic regions that are able to form toxic aggregates under physiological conditions. FASEB J. 2015;29(9):3689–3701. | ||
Adinolfi M, De Napoli L, Di Fabio G, Iadonisi A, Montesarchio D. Modulating the activity of oligonucleotides by carbohydrate conjugation: solid phase synthesis of sucrose-oligonucleotide hybrids. Org Biomol Chem. 2004;2(13):1879–1886. | ||
Musumeci D, Rozza L, Merlino A, et al. Interaction of anticancer Ru(III) complexes with single stranded and duplex DNA model systems. Dalton Trans. 2015;44(31):13914–13925. | ||
Roviello GN, Vicidomini C, Costanzo V, Roviello V. Nucleic acid binding and other biomedical properties of artificial oligolysines. Int J Nanomedicine. 2016;11:5897–5904. | ||
Zhang S, Holmes TC, Di Persio CM, Hynes RO, Su X, Rich A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16(18):1385–1393. | ||
Yuan D, Shi J, Du X, Zhou N, Xu B. Supramolecular glycosylation accelerates proteolytic degradation of peptide nanofibrils. J Am Chem Soc. 2015;137(32):10092–10095. | ||
Saleh T, Bolhassani A, Shojaosadati SA, Hosseinkhani S. Evaluation of cell penetrating peptide delivery system on HPV16E7 expression in three types of cell line. Iran J Biotechnol. 2015;13(1):55–62. | ||
Gour N, Kedracki D, Safir I, Ngo KX, Vebert-Nardin C. Self-assembling DNA-peptide hybrids: morphological consequences of oligonucleotide grafting to a pathogenic amyloid fibrils forming dipeptide. Chem Commun. 2012;48(44):5440–5442. | ||
Roviello GN, Musumeci D, D’Alessandro C, Pedone C. Binding ability of a thymine-functionalized oligolysine towards nucleic acids. Bioorg Med Chem. 2014;22(3):997–1002. | ||
Song G, Ren J. Recognition and regulation of unique nucleic acid structures by small molecules. Chem Commun. 2010;46(39):7283–7294. | ||
Roviello GN, Musumeci D, Roviello V, Pirtskhalava M, Egoyan A, Mirtskhulava M. Natural and artificial binders of polyriboadenylic acid and their effect on RNA structure. Beilstein J Nanotechnol. 2015;6:1338–1347. | ||
Saha B, Suresh Kumar G. Spectroscopic and calorimetric investigations on the binding of phenazinium dyes safranine-O and phenosafranine to double stranded RNA polynucleotides. J Photochem Photobiol B. 2016;161:129–140. | ||
Kabir A, Suresh Kumar G. Targeting double-stranded RNA with spermine, 1-naphthylacetyl spermine and spermidine: a comparative biophysical investigation. J Phys Chem B. 2014;118(38):11050–11064. | ||
Das A, Suresh Kumar G. Probing the binding of two sugar bearing anticancer agents aristololactam-β-(D)-glucoside and daunomycin to double stranded RNA polynucleotides: a combined spectroscopic and calorimetric study. Mol Biosyst. 2012;8:1958–1969. | ||
Hara RI, Maeda Y, Sakamoto T, Wada T. Double-stranded RNA-binding artificial cationic oligosaccharides stabilizing siRNAs with a low N/P ratio. Org Biomol Chem. 2017;15(7):1710–1717. | ||
Chen B, Yoo K, Xu W, Pan R, Han XX, Chen P. Characterization and evaluation of a peptide-based siRNA delivery system in vitro. Drug Deliv Transl Res. 2017;7(4):507–515. | ||
Lin YH, Huang JH, Liu Y, Belles X, Lee HJ. Oral delivery of dsRNA lipoplexes to German cockroach protects dsRNA from degradation and induces RNAi response. Pest Manag Sci. 2017;73(5):960–966. | ||
Sanitt P, Apiratikul N, Niyomtham N, et al. Cholesterol-based cationic liposome increases dsRNA protection of yellow head virus infection in Penaeus vannamei. J Biotechnol. 2016;228:95–102. | ||
Attasart P, Boonma S, Sunintaboon P, Tanwilai D, Pothikasikorn J, Noonpakdee WT. Inhibition of Plasmodium falciparum proliferation in vitro by double-stranded RNA nanoparticle against malaria topoisomerase II. Exp Parasitol. 2016;164:84–90. | ||
Roviello GN, Musumeci D, Castiglione M, Bucci EM, Pedone C, Benedetti E. Solid phase synthesis and RNA-binding studies of a serum-resistant nucleo-epsilon-peptide. J Pept Sci. 2009;15(3):155–160. | ||
Yuan D, Du X, Shi J, Zhou N, Zhou J, Xu B. Mixing biomimetic heterodimers of nucleopeptides to generate biocompatible and biostable supramolecular hydrogels. Angew Chem Int Ed Engl. 2015;54(19):5705–5708. |
Supplementary materials
This section provides
- a structural representation of a hexathymine oligoDapT;
- a structural representation of the interaction between oligoDapT and nucleic acids;
- liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) characterization of the thymine-based monomer 3 and nucleopeptides 4 and 5;
- 1H and 13C NMR spectra of the nucleoamino acid 3; and
- ultraviolet (UV) melting curves relative to oligoDapT/DNA, oligoDapT/RNA, and controls (DNA/DNA and RNA/RNA).
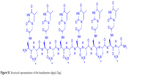  | Figure S1 Structural representation of the hexathymine oligo(L-Dap). |
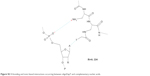  | Figure S2 H-bonding and ionic-based interactions occurring between oligoDapT and complementary nucleic acids. |
  | Figure S4 ESI-MS (positive mode) analyses of the thymine-based nucleopeptides 4 (A) and 5 (B). |
  | Figure S5 1H NMR spectrum of the nucleoamino acid 3 recorded at 25°C in deuterated DMF on a Varian 600 MHz unit. |
  | Figure S6 13C NMR spectrum of the nucleoamino acid 3 recorded at 25°C in deuterated DMF on a Varian 600 MHz (150 MHz) unit. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.