Back to Journals » Infection and Drug Resistance » Volume 11
Distribution of sasX, pvl, and qacA/B genes in epidemic methicillin-resistant Staphylococcus aureus strains isolated from East China
Authors Kong H, Fang L, Jiang R, Tong J
Received 6 October 2017
Accepted for publication 28 November 2017
Published 9 January 2018 Volume 2018:11 Pages 55—59
DOI https://doi.org/10.2147/IDR.S153399
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Haishen Kong,1,2 Lingmei Fang,3 Rujin Jiang,4 Jixiang Tong2
1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 2Key Laboratory of Clinical In Vitro Diagnostic Techniques of Zhejiang Province, Department of Laboratory Medicine, First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 3Clinical Laboratory, Chunan First People’s Hospital, Zhejiang Province People’s Hospital Chunan Branch, Hangzhou, China; 4Clinical Laboratory, Yuhang Hospital of Traditional Chinese Medicine, Hangzhou, China
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen. Various virulence and antiseptic-resistant factors increase the pathogenicity of MRSA strains and allow for increased infection rates.
Purpose: The purpose of this study was to investigate the prevalence and distribution of virulence-associated and antiseptic-resistant genes from epidemic MRSA strains isolated from East China.
Methods: A newly designed multiplex PCR assay was used to assess whether the virulence-associated genes sasX and pvl and the chlorhexidine tolerance gene qacA/B were present in 189 clinical isolates of MRSA. Multilocus sequence typing (MLST) and Staphylococcal protein A (spa) typing of these isolates were also performed. The frequency of these genes in isolates with epidemic sequence types (STs) was investigated.
Results: Twenty STs and 36 spa types with five epidemic clones (ST5-t311, ST59-t437, ST5-t002, ST239-t030, and ST239-t037) were identified. The prevalence of sasX, pvl, and qacA/B in all isolates was 5.8%, 10.1%, and 20.1%, respectively. The prevalences of these genes in isolates with ST5, ST59, ST239, and other ST genetic backgrounds were all significantly different (P<0.001). Isolates that had the highest frequency of sasX, pvl, or qacA/B were ST239 (33.3%), ST59 (28.9%), and ST5 (34.1%), respectively. The gene distribution pattern from all of the isolates showed that sasX–pvl–qacA/B+, sasX–pvl+qacA/B–, and sasX+pvl–qacA/B– were closely associated with epidemic clones ST5-t311, ST59-t437, and ST239-t037, respectively.
Conclusion: There are significant differences in the prevalence of virulence-associated and antiseptic-resistant genes in epidemic MRSA strains. Using this information, more effective control and prevention strategies for nosocomial MRSA infections can be developed.
Keywords: MRSA, MLST, virulence genes, sasX, pvl, qacA/B, multiplex PCR
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as one of the most important nosocomial pathogens, causing considerable morbidity and mortality in hospitals.1 Information regarding the molecular characteristics and distribution of virulence determinants and antiseptic-resistant genes of MRSA is essential for controlling and treating the diseases caused by this medically important pathogen. The novel S. aureus cell wall-anchored protein gene, sasX, is a critical factor for promoting nasal colonization, immune evasion, and virulence.2 The recent spread of sasX from sequence type (ST) 239 to invasive clones belonging to other STs suggests that this ST shift may be a driving force behind the Asian MRSA epidemic.2 In addition, an association between the presence of Panton–Valentine leukocidin (PVL), an S. aureus toxin that causes neutrophil lysis, and the onset of invasive disease in communities has emerged worldwide. However, in a study by Shallcross et al, the PVL encoding gene, pvl, was consistently associated with skin and soft tissue infections and rarely associated with invasive disease.3 Additional data are needed to reconcile the differences between these studies.
Chlorhexidine gluconate use in the prevention and decolonization strategies is being increasingly employed to control MRSA transmission, but increased tolerance to these treatments has been reported.4 Decontamination with chlorhexidine gluconate is only effective for patients colonized with strains that lack the antiseptic-resistant genes qacA/B.5 qacA and qacB genes are closely related, and their sequences only differ by seven nucleotides. Therefore, in this study, the presence of these genes was reported as a single gene.
The presence of both virulence factors and antiseptic resistance genes within a bacterial strain has been previously reported in Gram-negative and Gram-positive bacteria.6,7 Quickly identifying these attributes in bacterial strains can have a major impact on treatment and decontamination strategies. In this study, we described a multiplex PCR assay capable of detecting three genes involved in either MRSA virulence or antiseptic resistance, sasX, pvl, and qacA/B simultaneously. The prevalence and distribution of these genes among epidemic clones from 189 MRSA strains isolated from Eastern China was also determined.
Materials and methods
MRSA isolates
One hundred and eighty-nine clinical MRSA isolates were collected from two tertiary care hospitals and two secondary hospitals located in Eastern China between January 2016 and May 2017. Clinical sources of the isolates included the respiratory tract (n=76), skin and soft tissue (n=31), blood cultures (n=40), sterile body fluid (n=24), and urinary tract (n=18). Bacterial identification and oxacillin resistance were performed using the Vitek 2 Compact Automated Microbiology System (BioMérieux, Durham, NC, USA) in a clinical microbiology laboratory accredited by the College of American Pathologists and certified by the Clinical Laboratory Improvement Act.
DNA extraction
All isolates were cultured on blood agar and incubated overnight at 37°C. DNA was isolated using a QIAamp DNA Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. The isolated DNA was used as the template for all PCRs.
Gene tying
Multilocus sequence typing (MLST) was performed on all isolates by sequencing the internal fragments of seven housekeeping genes (arcC, aroE, glpF, gmK, pta, tpi, and yqiL). The sequence profile and ST of each allele were determined according to the MLST database (http://saureus.mlst.net).8 The allelic profiles were assigned by comparing the sequences at each locus with those of the known alleles in the S. aureus MLST database and then were defined as the STs. Staphylococcal protein A (spa) typing involved PCR amplification, and subsequent sequencing of the highly variable X region in spa using the previously described primers spa-1113f and spa-1514r.9 These spa sequences were analyzed and assigned a unique spa type using the spa database (http://spa.ridom.de/).10
Multiplex PCR assay for sasX, pvl, and qacA/B detection
The specific primers used for the identification of sasX, pvl, and qacA/B genes were listed in Table 1. Three genes were detected by multiplex PCR assay simultaneously in a single tube. This amplification was performed using 12.5 µL Qiagen Multiplex PCR Master kit (Qiagen); 1.5, 1.0, or 0.5 µL of each upstream or downstream primer for sasX, pvl, and qacA/B, respectively; 2 µL of template DNA; and enough distilled water for a final volume of 25 µL. No-template control, sterile water instead of template DNA was incorporated in each run under the following conditions: 5 minutes of denaturation at 94°C, followed by 35 cycles for 30 seconds at 94°C, 30 seconds at 56°C, and 30 seconds at 72°C, with a final 10 minutes elongation step at 72°C. The PCR products were analyzed on a 1.5% agarose gel and further confirmed by DNA sequencing. Sequence comparisons with publicly available sequence data were performed using BLAST analyses (http://www.ncbi.nlm.nih.gov/BLAST/).11
  | Table 1 Primers used in this study |
Validation of multiplex PCR
Conventional single PCR assays were also performed for sasX, pvl, and qacA/B, as previously described.2,9,12 The results from the single and multiplex PCR methods were compared, and 80 of the 189 clinical MRSA isolates were validated by both the conventional and multiplex PCR assays.
Statistical analysis
Statistical analyses were performed using the SPSS version 16.0 statistical software package (SPSS Inc., Chicago, IL, USA). The descriptive data were presented as percentages for the categorical data. Pearson’s chi-square test or Fisher’s exact test were used to determine whether differences in the frequency of sasX, pvl, and qacA/B exist among isolates with different ST types. A P-value of ≤0.05 was considered statistically significant.
Results
Genotyping of MRSA isolates
Among the 189 MRSA isolates studied, 20 different STs and 36 spa types were identified. The three major STs were ST5 (88, 46.6%), ST59 (45, 23.8%), and ST239 (18, 9.5%). The remaining STs had frequencies ranging from 0.5% to 3.7% and were ST398, ST630, ST22, ST1, ST965, ST7, ST88, ST944, ST764, ST6, ST338, ST1611, ST2580, ST3355, ST3360, ST3361, and a new ST type. In addition, the most common spa types were t311 (71, 37.6%), t437 (28, 14.8%), t002 (14, 7.4%), t030 (10, 5.3%), t163 (8, 4.2%), and t037 (7, 3.7%). The remaining 30 spa types accounted for 27.0% of the total isolates, with none of the types accounting for more than 3.2%. The most common genotype was ST5-t311 (66, 34.9%), followed by ST59-t437 (22, 11.6%), ST5-t002 (14, 7.4%), ST239-t030 (10, 5.3%), and ST239-t037 (6, 3.2%).
Validation of results of the multiplex PCR assay with clinical isolates
Comparisons between the data from the single and the multiplex PCR assays showed that 80 clinical isolates were completely concordant (100%). Of these 80 isolates, two harbored sasX, seven harbored pvl, 19 harbored qacA/B, three harbored both sasX and qacA/B, and 49 were negative for all three genes.
Prevalence and distribution of virulence determinants and antiseptic-resistant genes
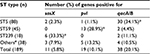
Of the 189 clinical isolates of MRSA, the sasX, pvl, and qacA/B genes were found in 11 (5.8%), 19(10.1%), and 38 (20.1%) of the isolates, respectively. The sasX and qacA/B genes were detected concomitantly in five (2.6%) of the isolates. Similarly, sasX and pvl were both found in one (0.5%) of the isolates. No isolates that harbored all three genes simultaneously were identified. Isolates were sorted into four groups, based on gene typing: ST5, ST59, ST239, and other ST types (which excluded the ST5, ST59, and ST239 groups), and further grouping analysis was performed. The results showed that the incidence rates of sasX, pvl, and qacA/B were all significantly different in isolates with ST5, ST59, ST239, and other ST genetic backgrounds (P<0.001). The sasX gene occurred with a maximum frequency in ST239 (33.3%), pvl had the highest frequency in ST59 (28.9%), and qacA/B occurred with the highest frequency in ST5 (34.1%). The sasX gene was not detected among 45 ST59 isolates, pvl was not detected among 18 ST239 isolates, and was found in only one (1.1%) of the 88 ST5 isolates. The prevalence of the three genes from the MRSA isolates harboring different ST types is summarized in Table 2.
Five gene distribution patterns, based on the presence of sasX, pvl, and qacA/B in the isolates, were determined (Table 3). These patterns included sasX–pvl–qacA/B+ (33, 17.5%), sasX–pvl+qacA/B– (18, 9.5%), sasX+pvl–qacA/B– (5, 2.6%), sasX+pvl–qacA/B+ (5, 2.6%), and sasX+pvl+qacA/B– (1, 0.5%). Furthermore, high incidence of sasX-pvl-qacA/B+, sasX-pvl+qacA/B–, and sasX+pv-qacA/B– was found in ST5-t311, ST59-t437 and ST239-t037 strains, respectively (75.8%, 72.2%, and 80.0%, respectively). The sasX, pvl, and qacA/B genes were not found in any of the remaining 127 isolates.
Discussion
MRSA is considered the most significant multidrug-resistant organism that causes infections, and the global spread of MRSA has become a major problem worldwide.13,14 Analysis of the distribution of virulence determinants and antiseptic-resistant genes of MRSA clones is valuable for understanding the evolution and dissemination of MRSA in different geographic regions. In this study, a multiplex PCR assay that targets the sasX and pvl virulence genes and the chlorhexidine-resistant gene, qacA/B, was evaluated using clinical isolates. Results of the present study showed that the multiplex PCR assay was accurate, simple, and reliable. To our knowledge, this is the first study demonstrating the simultaneous amplification and subsequent analyses of the presence of sasX, pvl, and qacA/B in MRSA isolates.
In previous reports, from other groups, the prevalence of the sasX gene in MRSA isolates revealed significant regional differences, with incidence rates ranging from 0% to 39%.2,15–18 These studies also showed that the ST239 strains carried the sasX gene with the highest frequencies (up to 99.1%).17 In addition, Li et al observed the recent spread of sasX from ST239 to invasive clones belonging to other STs.2 Our results indicate that the sasX gene frequency (5.8%) of MRSA in East China is lower than that previously found at three different teaching hospitals in China (21%–39%, from 807 strains) and in Chinese children (10.7%, from 243 MRSA strains).2,15 Our study also demonstrated that ST239 is dominant in the sasX-positive isolates and that the spread of sasX to other STs such as ST5, ST22, ST965, and ST3361 occurs. This study is the first to observe the presence of sasX in ST22, ST965, and ST3361 isolates. The sasX gene has not been detected among the prevalent clone of community isolate ST59, indicating that sasX-positive clones spread predominantly within the hospital setting. Moreover, our study showed that the ST59 strains carried the pvl gene with the highest frequencies (28.9%). Meanwhile, pvl has not been detected among ST239 isolates, indicating that pvl-positive clones spread predominantly within the community setting. This finding was similar to the results from previous studies.3,19
Chlorhexidine gluconate, a water-soluble cationic bisbiguanide, has been widely used as an antiseptic agent for infection control. The correlation between the presence of qacA/B and chlorhexidine resistance has not been definitely determined.20 However, the qacA/B carrier rate was found to be higher in S. aureus isolates with higher chlorhexidine minimal bactericidal concentrations.20 In this study, we found that the highest incidence of qacA/B was in ST5 clone (34.1%), which was the predominant clone in the MRSA isolates from this region.21 This finding was different from Batra et al’s study which showed that carriage of qacA/B in an outbreak of ST239 MRSA strains was higher than that of other ST types.5 Survival of these strains in a hospital environment must be considered since the high prevalence of the qacA/B gene might be due to either selective pressure imposed by the use of quaternary ammonium compounds (QACs) or cross-resistance and co-resistance between QACs and antibiotics.22 The qacA/B genes are typically located on a transposon of a transmissible multidrug-resistant plasmid such as pSK1.5.23 Therefore, surveillance of antiseptic-resistant MRSA could provide important information on the control of nosocomial infections. This is the first study to simultaneously report the prevalence of sasX, pvl, and qacA/B in Chinese MRSA strains. We found significant differences in the prevalence of these genes in specific ST types. The ST239 isolate type had the highest prevalence of sasX, isolates resembling ST59 had the highest rates of pvl, and ST5-type isolates had a higher incidence of qacA/B than other ST types. Even so, understanding the distributional differences of virulence determinants and antiseptic-resistant genes in MRSA isolates is still limited and warrants continued intensive study.
Acknowledgments
This study was supported by grants from the Department of Science and Technology of Zhejiang Province (2016C33133), the Medical and Health Research Project of Hangzhou (20140633B56), and the Health Science and Technology Project of Hangzhou (2014B38).
Author contributions
HK designed the experiments and wrote the manuscript. LF, RJ, and JT performed research and analyzed data. All the authors have read, critically revised, and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. | ||
Li M, Du X, Villaruz AE, et al. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med. 2012;18:816–819. | ||
Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13:43–54. | ||
Simor AE. Staphylococcal decolonisation: an effective strategy for prevention of infection? Lancet Infect Dis. 2011;11:952–962. | ||
Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth J. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2010;50:210–217. | ||
Sheng WH, Wang JT, Lauderdale TL, Weng CM, Chen D, Chang SC. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: emphasis on chlorhexidine susceptibility. Diagn Microbiol Infect Dis. 2009;63:309–313. | ||
Grande Burgos MJ, Fernández Márquez ML, Pérez Pulido R, Gálvez A, Lucas López R. Virulence factors and antimicrobial resistance in Escherichia coli strains isolated from hen egg shells. Int J Food Microbiol. 2016;238:89–95. | ||
MLST Database [homepage on the Internet]. Mark Enright. Available from: http://saureus.mlst.net/. Accessed July 10, 2017. | ||
Larsen AR, Stegger M, Sorum M. spa typing directly from a mecA, spa and pvl multiplex PCR assay-a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin Microbiol Infect. 2008;14:611–614. | ||
spa Database [homepage on the Internet]. Ridom GmbH. Available from: http://www.spa.ridom.de/. Accessed July 20, 2017. | ||
BLAST Database [homepage on the Internet]. National Institutes of Health. Available from: http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed July 15, 2017. | ||
Vali L, Davies SE, Lai LLG, Dave J, Amyes SGB. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother. 2008;6:524–532. | ||
Song JH, Hsueh PR, Chung DR. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66:1061–1069. | ||
Klein EY, Sun L, Smith DL, Laxminarayan R. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. Am J Epidemiol. 2013;177:666–674. | ||
Li SP, Sun J, Zhang JZ, et al. Comparative analysis of the virulence characteristics of epidemic methicillin–resistant Staphylococcus aureus (MRSA) strains isolated from Chinese children: ST59MRSA highly expresses core gene–encoded toxin. APMIS. 2014;122:101–114. | ||
Tekeli A, Ocal DN, Ozmen BB, Karahan ZC, Dolapci I. Molecular characterization of methicillin-resistant Staphylococcus aureus bloodstream isolates in a Turkish university hospital between 2002 and 2012. Microb Drug Resist. 2016;22:564–569. | ||
Chen CJ, Huang YC, Su LH, et al. Molecular epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus bloodstream isolates in Taiwan, 2010. PLoS One. 2014;9:e101184. | ||
Nair R, Hanson BM, Kondratowicz K, et al. Antimicrobial resistance and molecular epidemiology of Staphylococcus aureus from Ulaanbaatar, Mongolia. PeerJ. 2013;1:e176. | ||
Otter JA, French GL. Community-associated methicillin-resistant Staphylococcus aureus: the case for a genotypic definition. J Hosp Infect. 2012;81:143–148. | ||
Ho CM, Li CY, Ho MW, Lin CY, Liu SH, Lu JJ. High rate of qacA- and qacB-positive methicillin-resistant Staphylococcus aureus isolates from chlorhexidine-impregnated catheter-related bloodstream infections. Antimicrob Agents Chemother. 2012;56:5693–5697. | ||
Kong H, Yu F, Zhang W, Li X, Wang H. Molecular epidemiology and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus strains in a tertiary hospital in China. Front Microbiol. 2017;8:838. | ||
Wang C, Cai P, Zhan Q, Mi Z, Huang Z, Chen G. Distribution of antiseptic-resistance genes qacA/B in clinical isolates of methicillin-resistant Staphylococcus aureus in China. J Hosp Infect. 2008;69:393–409. | ||
Noguchi N, Suwa J, Narui K, et al. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med Microbiol. 2005;54:557–565. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.