Back to Journals » Infection and Drug Resistance » Volume 13
Distribution and Frequency of rpoB Mutations Detected by Xpert MTB/RIF Assay Among Beijing and Non-Beijing Rifampicin Resistant Mycobacterium tuberculosis Isolates in Bangladesh
Authors Uddin MKM, Rahman A , Ather MF, Ahmed T, Rahman SMM , Ahmed S , Banu S
Received 1 December 2019
Accepted for publication 7 February 2020
Published 10 March 2020 Volume 2020:13 Pages 789—797
DOI https://doi.org/10.2147/IDR.S240408
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mohammad Khaja Mafij Uddin, 1,* Arfatur Rahman, 1, 2,* Md Fahim Ather, 1 Tanvir Ahmed, 1 Syed Mohammad Mazidur Rahman, 1 Shahriar Ahmed, 1 Sayera Banu 1
1Infectious Diseases Division, icddr,b, Dhaka, Bangladesh; 2Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University (Parkville Campus), Parkville VIC 3052, Australia
*These authors contributed equally to this work
Correspondence: Sayera Banu
Infectious Diseases Division, icddr,b, 68, Shaheed Tajuddin Ahmed Sarani, Dhaka 1212, Bangladesh
Email [email protected]
Background: Rifampicin resistance (RR) is a key indicator of multidrug-resistant tuberculosis (MDR-TB) and 95% of the RR is associated with the mutation in the 81-bp rifampicin resistance determining region (RRDR) of the rpoB gene of Mycobacterium tuberculosis complex (MTBC). The Xpert MTB/RIF (Xpert) assay uses five overlapping molecular beacon probes (A-E) complementary to RRDR region that detect MTBC and mutations associated with RR. The objective of the study was to investigate the distribution and frequency of mutations detected by Xpert assay among Beijing and non-Beijing RR-TB isolates.
Methods: A total of 205 randomly selected RR-TB specimens detected by Xpert assay were included in this study. A portion of specimens was further subjected to culture, MTBDRplus test and the positive culture isolates were genotyped by spoligotyping.
Results: We found that the most frequent mutation occurred at probe E (S531L) binding region in both Beijing and non-Beijing isolates (61.9% and 66.9%, respectively). The Beijing family had higher mutation rates than non-Beijing (19.0% vs 12.4%) at probe B (D516V) while the non-Beijing family had higher mutations at probe D (H526D or H526Y) than the Beijing (13.2% vs 10.7%) family. Mutations at probes Aand C were less common in both Beijing and non-Beijing isolates. There was no significant difference (P=0.36) in the occurrence of mutations at different probes between Beijing and non-Beijing isolates.
Conclusions: The study results revealed that the most frequent mutation occurs in the region of probe E and the least common mutations at probe A and C among both Beijing and non-Beijing RR-TB cases. This first insight into the probe mutation variation and frequencies among the RR-TB cases in Bangladesh forms the baseline information for further investigation.
Keywords: Mycobacterium tuberculosis, rifampicin resistant, GeneXpert, probe mutations, spoligotyping
Introduction
Tuberculosis (TB) remains one of the major causes of death worldwide. The World Health Organization (WHO) reported 10.0 million new cases and an estimated 1.6 million deaths globally in 2017. The development of multidrug-resistant TB (MDR-TB) is becoming one of the major threats to global TB control program.1 Bangladesh ranks seventh among 30 high TB burden countries with 225/100,000 TB incident cases and 45/100,000 mortality rate.2 According to the Global TB report, MDR-TB/Rifampicin resistant TB (RR-TB) occurred at a frequency of 1.6% and 29% among new and previously treated cases, respectively, in Bangladesh.1 First nationwide drug resistance surveillance in 2011 showed that the overall MDR-TB rate was 7% and this rate was 1.4% and 28.5% among the new and previously treated cases, respectively.3 Another study showed the frequency of MDR-TB among new and re-treatment cases were 2.3% and 13.8%, respectively.4 Prevalence of HIV in Bangladesh is less than 0.1% among the adults aged 15–49 years. A study conducted in Bangladesh showed that the prevalence of TB/HIV co-infection was 0.1% among the enrolled patients.5 MDR-TB is defined as the type of mycobacterial infection that is resistant to at least Rifampicin (RIF) and Isoniazid (INH). RIF is the most potent bactericidal agent that impedes the initiation of transcription by binding to the β-subunit of DNA dependent RNA polymerase (rpoB).6,7 Resistance to RIF is the key indicator of MDR-TB as more than 90% of the RR isolates are also resistant to other available drugs.8,9 Previous studies have shown that 95% of the RR is associated with the mutation in the 81-bp hot spot region (codons 507–533) of the rpoB gene which is also known as Rifampicin Resistance Determining Region (RRDR).6,10,11 Many earlier studies have identified RR by the sequence analysis of the rpoB gene. The vast majority of the mutations (90%) occur in codons 516, 526 and 531 within the RRDR.7,12
Current molecular diagnostic technologies recommended by WHO including Xpert MTB/RIF (Xpert) assay and the Line Probe Assay (LPA) can simultaneously detect Mycobacterium tuberculosis complex (MTBC) and drug resistance in both pulmonary and extra-pulmonary specimens. The Xpert assay (Cepheid, Sunnyvale, CA, USA) is a cartridge-based, automated hemi-nested real-time PCR system that uses five overlapping molecular beacon probes (A-E) targeting the RRDR of rpoB gene to detect MTBC along with the mutations conferring RR. The probes hybridizes to the sequence of the RRDR of rpoB gene: A (codons 507–511), B (codons 512–518), C (codons518–523), D (codons 523–529) and E (codons 529–533).13 MTBC is detected when at least two of the five probes show a cycle threshold (Ct) ≤ 38. Any strain showing no hybridization of one or more probes or when the difference between first and last Ct is more than 3.5 is considered as RR case. Genotype MTBDRplus (LPA; Hain Life Sciences, Germany), is based on DNA strip technology in combination with multiplex PCR system, for the rapid detection of MTBC and mutations associated with RIF and INH. In Genotype MTBDRplus assay, the probe mutations causing RR are located in the RRDR of rpoB gene which comprises eight overlapping wild type (WT) probes and four mutated probes (MUT). In this assay, dropout of WT probes and hybridization of MUT probes is the basis of RIF and/or INH resistance.
The distribution and frequency of mutations of the rpoB gene vary in the different geographical regions. Globally, many studies revealed that the most common mutation associated with RIF is S531L. Studies conducted in Russia, Kazakhstan, and Germany showed the prevalence of S531L mutation higher among the Beijing isolates than the non-Beijing.14–16 On the other hand, a study from Thailand reported mutation at codon 531 is the prevalent one, both in Beijing and non-Beijing isolates.17 Previous studies in Bangladesh have shown that Beijing genotypes along with other lineages (EAI, T1, CAS, LAM, and MANU) are circulating all over the country and also MDR-TB isolates are associated with Beijing genotypes.18,19 Data regarding frequency and distribution of probe mutations among the Beijing and non-Beijing isolates are scarce. The present study aimed to investigate the distribution and frequency of mutations conferring RR among Beijing and non-Beijing isolates detected by Xpert assay.
Materials and Methods
Study Setting and Data Collection
This study was a part of “Surveillance of multi-drug resistant (MDR) and extensively drug-resistant (XDR) tuberculosis in Bangladesh” where sputum specimens were collected from the 14 different health-care facilities across the country. All participants, and legal guardians of participants under the age of 18 years provided written consent, and that this study was conducted in accordance with the Declaration of Helsinki. The trained field staff was responsible for the collection of data including age, sex, alcohol consumption, smoking, diabetes and TB history. The sputum specimens were subjected to acid-fast bacilli (AFB) smear microscopy, culture, Xpert and MTBDRplus tests. As our aim of the study was to investigate the pattern of the rpoB mutations using probe dropout in Xpert assay among the Beijing and non-Beijing MDR-TB isolates, we included only the RR isolates detected by Xpert assay. A total of 205 RR isolates were randomly selected during the period of October 2011 to December 2015 for analysis. This study was approved by the Research Review Committee and the Ethical Review Committee of icddr,b.
Sample Processing and Culture
All specimens were processed with widely used N-Acetyl-L-Cysteine (NALC)-Sodium Hydroxide (NaOH) method where specimens were digested and decontaminated with 4% NaOH and 2.9% Sodium-Citrate along with 1% NALC.20 After centrifugation, pellets were then inoculated on Lowenstein-Jensen (L-J) media, incubated at 37°C for up to 8 weeks and examined weekly for the visible colony.
Xpert MTB/RIF Assay
The Xpert assay was performed according to the manufacturer’s instructions.21 Briefly, sample reagent was added to untreated sputum specimens in a 2:1 ratio, inverted the samples twice during 15-mins incubation period at room temperature, 2 mL of liquefied specimens were transferred to Xpert Cartridge (Version 3.0), loaded the cartridge into the Xpert machine and the results were viewed in the Xpert software.
GenoType MTBDRplus Assay
The GenoType MTBDRplus assay (Version 2.0) was performed on processed specimens according to the manufacturer’s instructions.22 The three common steps of this assay are: 1) DNA extraction, 2) multiplex PCR amplification and 3) hybridization of the PCR product with specific probes coated on a membrane strip. The MTBDRplus strips consist of 6 control probes and additional 21 probes for the detection of mutations specific for rpoB, inhA and katG genes.
Spacer Oligonucleotide Typing (Spoligotyping)
Spoligotyping was performed according to the standard protocol23 using a commercially available kit (Isogen Biosciences, Netherlands). Briefly, bacterial DNA was amplified by specific primers and the amplified products were hybridized with nitrocellulose membrane that is covalently linked with 43 synthetic oligonucleotides corresponding to 43 spacers. Enhanced chemiluminescence system (Amersham, UK) was used to identify the hybridized products and SITVITWEB- database was used for determination of the spoligotypes and phylogenetic clades.24
Statistical Analysis
Data were entered and analyzed using the Statistical Package for Social Sciences (SPSS) version 20.0 (IBM Corp, USA). Chi-square tests or Fisher’s exact tests were performed to identify any statistically significant probe mutations among the Beijing and non-Beijing lineages. P-value < 0.05 was considered to be statistically significant.
Results
Association of Demographic Characteristics with the Probe Dropouts
Probe mutations detected by Xpert assay were compared with the available demographic and clinical data for 205 RR-TB patients (Table 1). Frequency of probe E dropout was higher for all categories (age, sex, TB history), followed by B or D. Among 205 cases, 136 (66.3%) were male and 69 (33.7%) were female. The frequencies of different probe mutation for male and female were: probe A 3.7% (5/136) & 1.45% (1/69), probe B 17.6% (24/136) & 10.1% (7/69), probe D 14.7% (20/136) & 7.24% (5/69) and probe E 57.3% (78/136) & 79.7% (55/69) respectively. The results revealed that the mutations at probe A, B, and D region were almost two times frequent among the isolates from male than the female (3.7% vs 1.45%, 17.6% vs 10.1% and 14.7% vs 7.24% in male and female, respectively), whereas the isolates from female had significantly (p=0.001) higher mutation at probe E than male (79.7% vs 57.3%). There were five cases of mutation at probe C site among the isolates from male but no such cases found in female. According to treatment history, most of the patients (202/205, 98.5%) were previously treated as TB patients. Among these cases, the mutations were found across the RRDR region covered by all probes: 64.4% (130/202) had probe E mutation followed by 15.3% (31/202), 12.4% (25/202), 2.9% (6/202) and 2.5% (5/202) for probe B, D, A and C, respectively. The combination of multiple probes dropout (5/202, 2.5%) was found only in the previously treated cases. All the new cases (3/205, 1.5%) had mutations only at probe E region. Among the isolates from different age groups, the frequency of mutation at probe E site was significantly higher (p= <0.001) for younger groups (15–44 years); 72.9% in 15–24 years and 72.7% in 25–34 50% in 35–44 years. Isolates from older age groups (>55 years) harbored higher frequencies of mutations at probe D (6/21, 28.6%) than their younger counterparts (Table 1).
 |
Table 1 Association of Demographic Profile and Mutations at Different Probe Sites Among the RR-TB Cases |
Distribution of Probe Dropout in Xpert Assay Among Beijing and Non-Beijing Isolates
Spoligotyping was performed on 205 isolates, where 84 were identified as Beijing and 121 were non-Beijing families. Different probe mutations found in Xpert assay for Beijing were compared with the mutations among non-Beijing isolates (Table 2). The substitution of serine with leucine (S531L) at probe E binding site was the most frequent mutation found among both Beijing (52/84; 61.9%) and non-Beijing isolates (81/121; 66.9%). The second most common mutation was at probe B binding site for Beijing (16/84; 19.0%) and probe D for non-Beijing isolates (16/121; 13.2%). Mutations at the region of probe A (3.6% vs 2.5%) and probe C (1.2% vs 3.3%) were less common in both Beijing and non-Beijing isolates, respectively. Simultaneous mutations in the region of two probes were also found in five cases: A & E in one case, B & E in one case and A & B in one case of Beijing family, while A & D in two cases only in the non-Beijing isolates. We did not find any significant difference (p=0.36) in the occurrence of mutations at different probe binding regions between Beijing and non-Beijing isolates (Table 2).
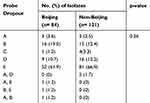 |
Table 2 Pattern of Probe Dropouts in Xpert Assay Among Beijing and Non-Beijing RR-TB Isolates |
Spoligo Patterns and Associated Probe Dropout
The spoligotyping analysis of 121 non-Beijing isolates further distributed as T1 (35), CAS (22), LAM and EAI (18 each); and H3 and S (1 each) and Orphan/unidentified (26). Among the CAS lineages, 63.7% had mutations in probe E site, followed by probe B (27.2%) and probe D (9.1%). Mutation at probe E binding site was lower (50%) than the average (65%) among EAI lineages and 27.8% had probe D, 16.6% probe C and 5.6% of probe A mutation. More than 94% of LAM strains had probe E binding site mutation which is higher than any other lineages but no cases with probe A, B and C mutation were found within this lineage. Probe B dropout (27.2%) was higher in CAS than any other lineage, followed by T1 (17.2%) and orphan (11.5%). Among the T1 lineages, 62.9% had probe E mutation followed by probe B and D, respectively. Besides, two cases of double mutations (probe A and D) were found only within T1 spoligotypes (Table 3).
 |
Table 3 Detection of Probe Dropout Among Different Lineages in RR-TB Cases |
Distribution of Wild Type Probe Dropout or Mutant Probe Binding in MTBDRplus Assay Among Beijing and Non-Beijing Isolates
All of the Beijing and non-Beijing isolates were also tested with GenoType MTBDRplus assay (Table 4). The substitution of serine with leucine at codon 531 of the rpoB gene was the most frequent mutation conferring RR found among both Beijing (63%) and non-Beijing isolates (69.4%). Mutation at codon 526, substitution of histidine with aspartic acid (H526D) was higher in non-Beijing (12.4%) than Beijing isolates (2.4%) while mutation at codon 526, the substitution of histidine with tyrosine (H526Y) was higher in Beijing (11.9%) than the non-Beijing isolates (3.3%). The rate of occurrence of mutation at codon 516, the substitution of aspartic acid with valine (D516V) was 14.3% and 9.0% among Beijing and non-Beijing isolates, respectively (Table 4). Specific probe mutations for 13 (6.3%) RR cases could not be identified. Mutant probes hybridization for these isolates were missing along with the absence of wild type bands which indicated that RR occurred due to mutations of RRDR region other than the mutant probes used in this assay. In MTBDRplus assay, mutation rate at katG was higher in non-Beijing than the Beijing isolates (85.9% vs 72.6%) whereas mutation rate at inhA was similar for both Beijing and non-Beijing isolates (5.9% vs 6.6%). Interestingly, simultaneous mutation at inhA and katG was higher in Beijing than non-Beijing isolates (11.9% vs 2.5%). A total of 14 isolates, those were resistant in conventional drug susceptibility testing but no mutation was found at these two regions (data not shown).
 |
Table 4 Frequency of Specific Mutations Based on MTBDRplus Probe Binding Among the Beijing and Non-Beijing Isolates (n=205) |
Comparison of Probe Mutations in Xpert Assay with MTBDRplus
As Xpert and MTBDRplus determine the mutation in the same RRDR region using WT probe dropout or corresponding mutant probe binding (MTBDRplus), the results are comparable. So, the RR results found in Xpert assay were compared with MTBDRplus assay. There were strong agreement among two assays for mutation detection at the region of probe C, D and E (100%, 96% and 97.7%, respectively), having only four isolates collectively to show discrepancies. Maximum numbers of inconsistencies were found in the region of probe B and A (22.6% and 16.7% disagreement, respectively). The comparison revealed 94% overall concordance between these two molecular techniques. Distinctly, both Beijing and non-Beijing isolates showed 92.6% and 94.1% agreements, respectively (Table 5).
 |
Table 5 Comparison of Specific Probe Mutations Based on Xpert and MTBDRplus Assay Among the Beijing and Non-Beijing Isolates (n=200) |
Geographical Distribution of Isolates with Different Probe Mutations
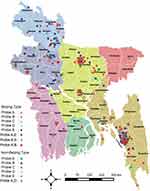
The geographical distribution of probe mutations among RR-TB cases is shown in Figure 1. RR-TB cases with probe E mutation were distributed throughout the country but most frequently observed in Chittagong and Dhaka divisions. Isolates with probe B and D mutations were mostly found in Chittagong and Rajshahi divisions. The combination of two probe mutations (AE, BE, AD) were found mostly in the Chittagong division.
Discussion
MDR-TB is becoming a global threat worldwide. To stop the spread of MDR-TB, many rapid and reliable genetic methods have been developed to identify MDR-TB. In most of the previous studies, the mechanism of RR or the point mutation was identified by rpoB gene sequencing.11,25 In the present study, we aimed to explore the distribution and frequency of probe mutation causing RR based on Xpert assay among Beijing and non-Beijing MTB isolates. According to spoligotyping, TB isolates can be classified into Beijing and non-Beijing lineages where the later one can be further classified into other lineages. It is demonstrated that the virulence of Beijing isolate is higher than the non-Beijing isolate due to its enhanced pathogenicity, increased transmissibility and a higher level of drug resistance. Additionally, isolates of modern Beijing lineage are likely to display a higher level of virulence compared to the strains of ancient non-Beijing sub-lineage by attaining an increased level of mutations in the virulence-associated genes.26 Moreover, the accelerated rate of general mutations also leads to the acquisition of MDR among both the Beijing and non-Beijing lineages.27,28
In the present study, the mutation patterns of all the spoligotypes were further investigated upon targeting the 81 bp RRDR region of the rpoB gene. The most frequent mutation (S531L) occurred at probe E site for both Beijing and non-Beijing isolates (62% and 67%, respectively). Moreover, mutations at codon 516 and 526 are also common (varies from 0% to 30%) which are located in the region of probe B and D, respectively.
In this study, the Beijing lineage had higher frequencies of mutation at probe B region (D516V) than the non-Beijing isolates (19.04% and 12.4%, respectively). On the other hand, we found that the non-Beijing isolates had moderately higher mutation rates at probe D (H526D/Y) than the Beijing isolates (13.2% and 10.72%, respectively), hence, the mutation at probe B was considered as secondary cause of RR among Beijing, while non-Beijing isolates had secondary mutation at probe D. There was no significant difference (p=0.36) in incidence of probe mutation determined by Xpert assay among Beijing and non-Beijing lineages. Sequence analysis of 66 RR-TB cases from East Asian countries including China, Japan, Korea, and Taiwan showed similar findings where the point mutation at codon S531L (corresponds to probe E) was higher both in Beijing and non-Beijing isolates (51% and 50% respectively).29 They also found a higher mutation rate at D516V (corresponds to probe B) among Beijing isolates than non-Beijing and mutation at H526D (corresponds to probe D) was higher among non-Beijing compared to Beijing isolates. Another study conducted in Thailand revealed that there was no significant difference in the occurrence of mutations at codon 526 (p=0.148) or 516 (p=0.257) between the Beijing and non-Beijing isolates.17
In the current study, very few mutations were found in the region of probe A and probe C in both Beijing and non-Beijing isolates. This result indicates that these regions are less susceptible to mutations conferring RR. Again, simultaneous mutations of two probes were found in five cases: three (A&E, B&E, A&B) in Beijing isolates and two (A&D) in non-Beijing isolates. This happened among retreatment cases, as it is proved that retreatment cases of TB harbor increased the proportion of mixed bacilli than primary cases of TB.30
In MTBDRplus assay, a total of eight probes (WT1-WT8) covered the 95 bp region of the rpoB gene which also encompasses the 81 bp RRDR region targeted by Xpert assay.31 Thus, the mutation patterns of Beijing and non-Beijing lineages found in Xpert assay was further evaluated by MTBDRplus assay, where sound punctual mutations were identified between them. Overall 94% agreement was found among these two methods which corresponds with other studies where the similar level of agreements were observed.8,32 The results along with the review of other literature demonstrate that the most common mutation responsible for RR is positioned in the probe E region. Along with Bangladesh, two neighboring countries, India and Myanmar, also demonstrate RR strains with probe E mutation. The in- and out-migration within these three countries favors the transmission of pathogens among the people.33 The limitations of the study were: i) rpoB gene sequencing was not performed to identify the specific mutations among the discrepant cases; ii) distribution of the mutated probe among the geographical location of the country varied due to the sample size and procedure of the selection of presumptive MDR-TB cases.
Conclusion
In conclusion, the study results and review of the literature revealed that most frequent mutation occurs in the region of probe E and less common mutations at probe A and C among Beijing and non-Beijing RR-TB cases. These data provide the first insight into the probe mutation variation and frequencies among the RR-TB cases in Bangladesh. Also, Xpert assay can be used as an alternative epidemiological tool to study RR resistant TB and which ultimately facilitates the early treatment initiation to halt further transmission of RR-TB.
Acknowledgments
This study was supported by the United States Agency for International Development (USAID), Washington DC, USA. icddr,b acknowledges with gratitude the commitment of USAID to its research efforts. icddr,b is also grateful to the Governments of Bangladesh, Canada, Sweden and the UK for providing core/unrestricted support. The authors are thankful to all surveillance physicians, nurses, health workers and participating health facilities. They are also grateful to all the patients who were participated in this study to make it successful. They would like to extend their gratitude to all laboratory staff for their excellent work on this study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Global Tuberculosis report, 2018.
2. Annual Report, National TB control Programme, Bangladesh. 2017.
3. Kamal S, Hossain A, Sultana S, et al. Anti-tuberculosis drug resistance in Bangladesh: reflections from the first nationwide survey. Int J Tuberculosis Lung Dis. 2015;19(2):151–156. doi:10.5588/ijtld.14.0200
4. Banu S, Rahman MT, Ahmed S, et al. Multidrug-resistant tuberculosis in Bangladesh: results from a sentinel surveillance system. Int J Tuberculosis Lung Dis. 2017;21(1):12–17. doi:10.5588/ijtld.16.0384
5. National Guidelines on TB/HIV Management Program Collaboration & Implementation Manual; 2nd edition; 2016. Directorate General of Health Services Ministry of Health and Family Welfare Dhaka, Bangladesh. Available from: http://www.ntp.gov.bd/ntp_dashboard/magazines_image/21-TB-HIVGuidelines%202nd%20edition.pdf. Accessed February 27, 2020.
6. Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104(6):901–912. doi:10.1016/S0092-8674(01)00286-0
7. Kocagoz T, Saribas Z, Alp A. Rapid determination of rifampin resistance in clinical isolates of Mycobacterium tuberculosis by real-time PCR. J Clin Microbiol. 2005;43(12):6015–6019. doi:10.1128/JCM.43.12.6015-6019.2005
8. Rahman A, Sahrin M, Afrin S, et al. Comparison of Xpert MTB/RIF assay and GenoType MTBDRplus DNA probes for detection of mutations associated with rifampicin resistance in Mycobacterium tuberculosis. PLoS One. 2016;11(4):e0152694. doi:10.1371/journal.pone.0152694
9. Mani C, Selvakumar N, Narayanan S, et al. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J Clin Microbiol. 2001;39(8):2987–2990. doi:10.1128/JCM.39.8.2987-2990.2001
10. Wang S, Zhao B, Song Y, et al. Molecular characterization of the rpoB gene mutations of Mycobacterium tuberculosis isolated from China. J Tuberculosis Res. 2013;1(01):1. doi:10.4236/jtr.2013.11001
11. Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tubercle Lung Dis. 1998;79(1):3–29. doi:10.1054/tuld.1998.0002
12. Pang Y, Lu, J, Wang, Y, Song, Y, Wang, S, Zhao, Y. Study of the rifampin mono-resistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;57(2):
13. Lawn SD, Nicol MP. Xpert® MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future Microbiol. 2011;6(9):1067–1082. doi:10.2217/fmb.11.84
14. Hillemann D, Kubica T, Agzamova R, et al. Rifampicin and isoniazid resistance mutations in Mycobacterium tuberculosis strains isolated from patients in Kazakhstan. Int J Tuberculosis Lung Dis. 2005;9(10):1161–1167.
15. Lipin M, Stepanshina VN, Shemyakin IG, et al. Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in Russia. Clin Microbiol Infect. 2007;13(6):620–626. doi:10.1111/j.1469-0691.2007.01711.x
16. Hillemann D, Kubica T, Rusch-Gerdes S, et al. Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob Agents Chemother. 2005;49(3):1229–1231. doi:10.1128/AAC.49.3.1229-1231.2005
17. Prammananan T, Cheunoy W, Taechamahapun D, et al. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect. 2008;14(5):446 453. doi:10.1111/j.1469-0691.2008.01951.x
18. Uddin MKM, Ahmed M, Islam MR, et al. Molecular characterization and drug susceptibility profile of Mycobacterium tuberculosis isolates from Northeast Bangladesh. Infect Genet Evol. 2018;65:136–143. doi:10.1016/j.meegid.2018.07.027
19. Banu S, Uddin MKM, Islam MR, et al. Molecular epidemiology of tuberculosis in rural Matlab, Bangladesh. Int J Tuberculosis Lung Dis. 2012;16(3):319–326. doi:10.5588/ijtld.11.0426
20. Uddin MKM, Chowdhury M, Ahmed S, et al. Comparison of direct versus concentrated smear microscopy in detection of pulmonary tuberculosis. BMC Res Notes. 2013;6(1):291. doi:10.1186/1756-0500-6-291
21. Organization, W.H. Automated Real-Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB. World Health Organization; 2013.
22. HL, S. GenoType MTBDRplus VER 2.0 Molecular Genetic Assay for Identification of the M. tuberculosis Complex and Its Resistance to Rifampicin and Isoniazid from Clinical Specimens and Cultivatedsamples; 2012.
23. Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–914. doi:10.1128/JCM.35.4.907-914.1997
24. Demay C, Liens B, Burguière T, et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12(4):755–766. doi:10.1016/j.meegid.2012.02.004
25. Almeida Da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. 2011;66(7):1417–1430. doi:10.1093/jac/dkr173
26. Kato-Maeda M, Shanley, C.A, Ackart, D, et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin Vaccine Immunol. 2012;19(8):
27. de Steenwinkel JE, Ten Kate MT, de Knegt GJ, et al. Drug susceptibility of Mycobacterium tuberculosis Beijing genotype and association with MDR TB. Emerg Infect Dis. 2012;18(4):660. doi:10.3201/eid1804.110912
28. Ford CB, Shah RR, Maeda MK, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45(7):784. doi:10.1038/ng.2656
29. Qian L, Abe C, Lin T-P, et al. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J Clin Microbiol. 2002;40(3):1091–1094. doi:10.1128/JCM.40.3.1091-1094.2002
30. Fang R, Li X, Li J, et al. Mixed infections of Mycobacterium tuberculosis in tuberculosis patients in Shanghai, China. Tuberculosis. 2008;88(5):469–473. doi:10.1016/j.tube.2008.02.002
31. Machowski EE, Kana BD. Genetic mimetics of Mycobacterium tuberculosis and methicillin resistant Staphylococcus aureus as verification standards for molecular diagnostics. J Clin Microbiol. 2017;55(12):
32. Racheal SD-M, Zephaniah D, Reggie M, et al. Diagnosis of multi-drug resistant tuberculosis mutations using Hain line probe assay and GeneXpert: a study done in Zimbabwe. Br J Med Med Res. 2015;5(8):1044. doi:10.9734/BJMMR
33. Rahim Z, Nakajima C, Raqib R, et al. Molecular mechanism of rifampicin and isoniazid resistance in Mycobacterium tuberculosis from Bangladesh. Tuberculosis. 2012;92(6):529–534. doi:10.1016/j.tube.2012.07.005
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.