Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 10
Diet and lifestyle factors associated with miRNA expression in colorectal tissue
Authors Slattery ML, Herrick JS, Mullany LE, Stevens JR, Wolff RK
Received 21 July 2016
Accepted for publication 27 August 2016
Published 20 December 2016 Volume 2017:10 Pages 1—16
DOI https://doi.org/10.2147/PGPM.S117796
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Martha L Slattery,1 Jennifer S Herrick,1 Lila E Mullany,1 John R Stevens,2 Roger K Wolff1
1Department of Internal Medicine, The University of Utah, Salt Lake City, 2Department of Mathematics and Statistics, Utah State University, Logan, UT, USA
Abstract: MicroRNAs (miRNAs) are small non-protein-coding RNA molecules that regulate gene expression. Diet and lifestyle factors have been hypothesized to be involved in the regulation of miRNA expression. In this study it was hypothesized that diet and lifestyle factors are associated with miRNA expression. Data from 1,447 cases of colorectal cancer to evaluate 34 diet and lifestyle variables using miRNA expression in normal colorectal mucosa as well as for differential expression between paired carcinoma and normal tissue were used. miRNA data were obtained using an Agilent platform. Multiple comparisons were adjusted for using the false discovery rate q-value. There were 250 miRNAs differentially expressed between carcinoma and normal colonic tissue by level of carbohydrate intake and 198 miRNAs differentially expressed by the level of sucrose intake. Of these miRNAs, 166 miRNAs were differentially expressed for both carbohydrate intake and sucrose intake. Ninety-nine miRNAs were differentially expressed by the level of whole grain intake in normal colonic mucosa. Level of oxidative balance score was associated with 137 differentially expressed miRNAs between carcinoma and paired normal rectal mucosa. Additionally, 135 miRNAs were differentially expressed in colon tissue based on recent NSAID use. Other dietary factors, body mass index, waist and hip circumference, and long-term physical activity levels did not alter miRNA expression after adjustment for multiple comparisons. These results suggest that diet and lifestyle factors regulate miRNA level. They provide additional support for the influence of carbohydrate, sucrose, whole grains, NSAIDs, and oxidative balance score on colorectal cancer risk.
Keywords: colorectal cancer, carbohydrate, miRNA, NSAIDs, oxidative balance, sucrose
Introduction
MicroRNAs (miRNAs) are small non-protein-coding RNA molecules that regulate gene expression either by posttranscriptionally suppressing messenger RNA (mRNA) translation or by causing mRNA degradation.1–6 We know that miRNAs play a critical role in regulation of proliferation, differentiation, apoptosis, and stress response and are involved in the majority of physiological processes.7,8 While we are beginning to understand the role of miRNAs in various physiological functions, our understanding of what regulates miRNA expression is minimal. However, some studies have shown that some diet and other lifestyle factors such as specific dietary components, oxidative stress, and aspirin and nonsteroidal anti-inflammatory drugs (NSAIDs) alter miRNA expression.9–13
One of the factors most consistently inversely associated with colorectal cancer (CRC) risk is use of aspirin/NSAIDs. Likewise, dietary factors such as antioxidants have been associated with CRC risk.14–17 We and others have shown that the oxidative balance score (OBS) is associated with CRC,18–20 with CRC risk lowest among those with a score that is higher in antioxidants and lower in prooxidant factors. The role that these diet and lifestyle factors have on miRNA expression is limited, especially as it applies to colorectal tissue.
In this study, we examine the linear association between diet and lifestyle factors and miRNA expression in colon and rectal tissues. Our hypothesis is that lifestyle factors commonly associated with CRC risk alter miRNA expression profiles. We test associations for colon and rectal cancers separately, since risk factors often differ by tumor location. We evaluate associations of diet and lifestyle factors with miRNA expression for normal colon and rectal mucosa as well as for the difference of miRNA expression between paired carcinoma and normal colorectal tissue to help determine if associations influence the disease process. If miRNA expression profiles are altered by diet and lifestyle factors, it would strengthen the biological support for these associations and provide avenues for cancer prevention.
Methods
Study participants
Study participants came from two population-based case–control studies that included all incident colon and rectal cancers between 30 and 79 years of age who resided along the Wasatch Front in Utah or were members of the Kaiser Permanente Medical Care Program (KPMCP) in Northern California. Participants were White, Hispanic, or Black for the colon cancer study; the rectal cancer study also included Asians and American Indians not living on reservations.21,22 Cases had to have tumor registry verification of a first primary adenocarcinoma of the colon or rectum and were diagnosed between October 1991 and September 1994 for the colon cancer study and between June 1997 and May 2001 for the rectal cancer study. Detail study methods have been described earlier.23 The study was approved by the Institutional Review Board of the University of Utah and at KPMCP and all participants provided written informed consent.
miRNA processing
RNA was extracted from formalin-fixed paraffin-embedded tissue. Both normal mucosa adjacent to the carcinoma tissue and matched carcinoma tissue were used. Normal mucosa tissue served as a control. The Agilent Human miRNA Microarray V19.0 (Agilent Technologies, Santa Clara, CA, USA) was used given the number of miRNAs, its high level of reliability (repeatability coefficient was 0.98 in our data), and the amount of RNA needed to run the platform. The microarray contains probes for 2,006 unique human miRNAs. About 100 ng total RNA was labeled with Cy3 and hybridized to the Agilent Microarray and were scanned on an Agilent SureScan microarray scanner model G2600D. Data were extracted from the scanned image using Agilent Feature Extract software v.11.5.1.1 (Agilent Technologies). Data were required to pass stringent quality control (QC) parameters established by Agilent that included tests for excessive background fluorescence, excessive variation among probe sequence replicates on the array, and measures of the total gene signal on the array to assess low signal. If samples failed to meet quality standards for any of these parameters, the sample was relabeled, hybridized to arrays, and scanned. If a sample failed QC assessment a second time, the sample was deemed to be of poor quality and was excluded from down-stream analysis.
Diet and lifestyle data
Data were collected by trained and certified interviewers using laptops. All interviews were audiotaped as previously described and reviewed for QC purposes.24 The referent period for the study was 2 years prior to diagnosis. As part of the study questionnaire, information was collected on regular use and current use of aspirin and NSAIDs and on physical activity during the referent period and for 10 and 20 years prior to diagnosis. Physical activity was obtained for the physical intensity of activity performed as well as the frequency and duration of activity. Body size information, including height (measured at the time of interview), weight (recalled for referent period), and waist and hip circumference measurements, were also collected. Dietary information was obtained for the referent period using an extensive diet history questionnaire adapted from the validated CARDIA diet history.25 Foods were converted to nutrients using the Nutrition Coding Center Nutrient Data System Version 19 (Nutrition Coordinating Center, University of Minnesota, Minneapolis, MN, USA) as well as being grouped into categories of similar foods. Foods units were standard servings per day, which was half cup of fruit, vegetable, or dairy product; meat servings were 2–3 oz of meat; grain products were half cup of rice-type grains or one slice of bread. Prudent and western dietary patterns were developed based on the principal component analysis.26 Our prudent dietary pattern was heavily loaded toward diets high in fruits, vegetables, whole grains, fish, and chicken, whereas the western dietary pattern was highly loaded toward red meat, processed meats, and refined grains and high-sugar-high-fat foods. Additional questions were asked about meat consumption, doneness, and preparation methods that were combined and used to create a mutagen index score.27
Statistical methods
Of the 2,006 unique human miRNAs assessed, 1,278 were expressed in colorectal carcinoma tissue. To minimize differences in miRNA expression that could be attributed to the array, amount of RNA, location on array, or other factors that could erroneously influence expression, total gene signal was normalized by multiplying each sample by a scaling factor28 (http://genespring-support.com/files/gs_12_6/GeneSpring-manual.pdf), which was the median of the 75th percentiles of all the samples divided by the individual 75th percentile of each sample. We limited our analysis to miRNAs that were expressed in at least 20% of the samples in the tissue(s) of interest. Data were assessed for colon and rectal cancer separately, and the number of miRNAs analyzed varied from 766 to 817 depending on tumor site and tissue type (ie, carcinoma or normal mucosa). Our sample consisted of 1,447 subjects with both miRNA expression data for carcinoma and paired normal mucosa as well as diet and lifestyle variables.
We assessed long-term vigorous physical activity, body mass index (kilogram per square meter) during referent year, waist circumference, and hip circumference. Assessment of aspirin/NSAID use included recent use (ie, using NSAIDs during the referent period or not) and ever regular use. We assessed 28 dietary variables including energy intake, western dietary pattern, prudent dietary pattern, mutagen index, total fat, total trans-fatty acid, total carbohydrates, sucrose, animal protein, vegetable protein, vitamins B12, C, D, and E, calcium, folic acid, dietary fiber, carotenoids, β-carotene, lutein + zeaxanthin, lycopene, and servings per day of dairy, fruit, vegetables, meat, processed meat, whole grains, and refined grains. All variables were analyzed as continuous variables unless they were collected as categorical (ie, NSAIDs variables). To summarize risk associated with multiple exposures, we developed an OBS that consisted of 13 diet and lifestyle factors that were prooxidants (dietary iron and polyunsaturated fat and cigarette smoking) and antioxidants (vitamin C, vitamin E, selenium, β-carotene, lycopene, lutein + zeaxanthin, vitamin D, calcium, and folic acid and NSAID use).18 To create the OBS, these diet and lifestyle factors were assigned values of 2 for low levels of exposure for each prooxidants or high exposure to antioxidants (low risk), 1 for intermediate levels of exposure, and 0 for high levels of exposure to prooxidants and low exposure to antioxidants (high risk). The individual scores for the 13 variables were then combined to obtain the OBS. Higher summary score corresponded to greater oxidative balance.
We examined lifestyle variables to determine if there was an association between each lifestyle variable and miRNA expression by fitting a linear model to the log2-transformed expression levels and adjusting for age at diagnosis, study center, and sex. We examined miRNA expression in both normal mucosa and the difference between miRNA expressions in carcinoma and in the normal colonic mucosa. p-values were generated using the bootstrap method by creating a distribution of 10,000 F statistics derived by resampling the residuals from the null hypothesis model of no association between the lifestyle variables and the miRNAs using the boot package in R. Associations were considered important if the false discovery rate (FDR) q-value was <0.11.29 We standardized the β coefficient presented in order to compare the results across the miRNA and lifestyle factors by subtracting the mean of the mRNA and dividing the result by the standard deviation of the mRNA before calculating the slope. If the lifestyle variable in the regression model was continuous, then we applied the same technique to it as well.
Results
The study population consisted of 892 cases of colon cancer and 555 cases of rectal cancer (Table 1). The mean age for colon cancer cases was 64.7 years and for rectal cancer cases was 61.8 years. The majority of study participants were males and most reported never having used aspirin/NSAIDs (subsequently referred to as NSAIDs) on a regular basis.
There were few diet and lifestyle factors associated with miRNA expression in either normal colonic mucosa or with differential miRNA expression between carcinoma and normal mucosa when applying an FDR q-value of <0.1. Body mass index, waist and hip circumference, and physical activity showed no associations with miRNA expression with the FDR q-value at <0.1 as did most dietary factors analyzed. Only three dietary factors, carbohydrate intake, servings per day of whole grains, and sucrose intake, were associated with miRNA expression. For carbohydrate intake and sucrose intake, there were 250 and 198 miRNAs significantly differentially expressed between colon carcinoma and normal mucosa tissue by level of dietary intake. Of these miRNAs, 166 were significantly differentially expressed for both carbohydrate intake and sucrose intake (Table 2 shows the top 85 miRNAs based on q-value and Table S1 shows all miRNAs significantly differentially expressed for both carbohydrate and sucrose intake). Although the adjusted q-values indicated more significant associations with carbohydrate intake, the β coefficients for the two variables were very similar. There were 32 miRNAs uniquely associated with sucrose intake (Table 3). The top 53 miRNAs associated uniquely with carbohydrate intake (Table 3) all had a q-value of <0.05. Evaluation of miRNA expression in whole grains showed 99 miRNAs differentially expressed by level of intake in normal colonic mucosa with an FDR of <0.1 (Table 4).
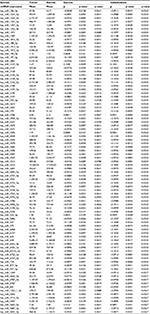  | Table 2 Colon cancer differential miRNA expression (top 85) associated with both sucrose and carbohydrate intake Abbreviation: miRNA, micro RNA. |
  | Table 3 Differential miRNA expression between colon carcinoma and normal colonic mucosa uniquely associated with either sucrose or carbohydrate intake Abbreviation: miRNA, micro RNA. |
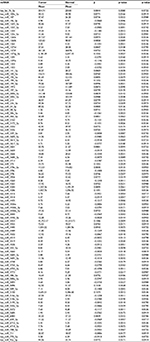  | Table 4 miRNA expression in normal colonic mucosa associated with whole grain intake Abbreviation: miRNA, micro RNA. |
Associations with miRNAs were similar for all indicators of NSAIDs use and for both normal colonic mucosa and differential miRNA expression between carcinoma and normal colonic mucosa. Table 5 shows the top 85 of the 135 miRNAs differentially expressed in normal colonic mucosa by recent NSAID use versus no recent NSAID use. OBS was only associated with miRNA expression for rectal tissue (Table 6 for top dysregulated miRNAs); 137 miRNAs were differentially expressed between rectal carcinoma and paired normal rectal mucosa based on OBS level.
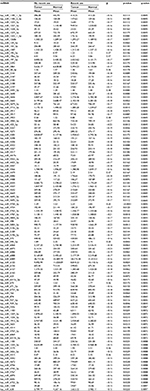  | Table 5 miRNA expression in normal colonic mucosa associated with recent NSAIDs use Abbreviations: miRNA, micro RNA; NSAIDs, nonsteroidal anti-inflammatory drugs. |
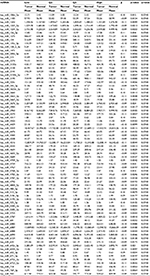  | Table 6 Rectal cancer differential miRNA expression between carcinoma and normal rectal mucosa associated with OBS Abbreviations: miRNA, micro RNA; OBS, oxidative balance score; Q, quartile. |
Discussion
We assessed 34 diet and lifestyle variables with miRNA expression levels in colorectal tissue and observed that only five of these factors altered miRNA expression level after adjusting for multiple comparisons (FDR q-value <0.1). These variables fell into two categories: 1) dietary carbohydrate, sucrose, and whole grains that could be operating through an insulin-related pathway and 2) NSAIDs and OBS that could be influencing miRNA expression level because of their role in inflammation and oxidative stress. Although others have suggested that diet and lifestyle factors could alter disease risk through their impact on miRNA expression,13,30 we have been able to test this hypothesis broadly within a large population-based study with detailed diet and lifestyle data along with miRNA expression data.
In evaluating these results, there are several considerations, such as miRNA expression in this study is from colorectal carcinoma and normal colorectal mucosa, and miRNA expression could be different in other tissue types. Perhaps the largest limitation of the study relates to the interpretation of results. While we can show that factors such as carbohydrate intake are associated with miRNA expression after adjustment for multiple comparisons, it is much more difficult to determine the specific biological mechanism associated with alteration in miRNA expression. For instance, the 250 miRNAs differentially expressed by level of carbohydrate intake are associated with 7,152 unique validated target genes. It is difficult to determine the relative importance of the multitude of pathways associated with these genes that relate specifically with carbohydrate intake. However, we can say that some lifestyle factors do influence miRNA expression levels, giving credence to reports of these factors in disease processes. We have also compared our current findings to our previous findings from these data for miRNAs that were differentially expressed between carcinoma and normal colorectal mucosa.23,31 We observed that 223 of the 250 miRNAs associated with carbohydrate intake, 175 of the 198 miRNAs associated with sucrose intake, 75 of the 99 miRNAs associated with whole grain intake, 121 of the 135 miRNAs associated with recent NSAID use, and 116 of the 137 miRNAs associated with OBS were also differentially expressed between carcinoma and normal colorectal mucosa, suggesting a role in tumor development.
Dietary factors have been cited as being important regulators of miRNAs.9,30 Studies have primarily been done in mice and have focused on targeted miRNAs. Reported associations have been found between dietary folate and let-7a, miR-21, miR-23, miR-130, miR-190, miR-17-92, and miR-122 in liver samples and between retinoic acid and let-7a, miR-15a/miR-16-1, and miR-23 in acute promyelocytic leukemia.9 Others have reported associations between miRNAs and polyphenols such as the antioxidant resveratrol with miR-663, miR-155, miR-21, miR-181b, and miR-30c2 in breast tissue cells.32 We did not replicate these findings. In a review by Garcia-Segura et al,30 carbohydrates were cited as being associated with miR-29c and miR-21. In our study, miR-21-3p was associated with carbohydrates. One controlled study using colorectal cells showed that starch consumption upregulated expression of the miR-17-92 cluster.33 We did not see associations within this miR cluster. Sucrose was associated with dysregulated miRNAs in a similar manner that carbohydrate intake was associated with miRNA expression. It has been proposed that sucrose metabolism downregulates expression of miR-15634 and that miR-398 and miR-408 are responsive to sucrose levels.35 Again, we did not see associations between these miRNAs and dietary sucrose level in our data. Although there was overlap of nine miRNAs that differentially expressed by level of carbohydrate intake and by level of whole grain intake, the direction of the associations was different.
NSAIDs have been examined with miRNAs in a few studies. Celecoxib has been associated with miR-222 levels in breast tissue in mice,36 and miR-271 has been associated with an NSAID and reactive oxygen species pathway.11 Other studies have focused on COX-2 expression and miRNAs and have shown that miR-101a and miR-199a are associated with higher COX-2 expression and that miR-10b and miR-21 had a high influence on Cox-2 expression. None of these miRNAs were associated with recent NSAID use in our study population, although miR-199a was associated with ever using NSAIDs.
Inflammation and oxidative stress are key elements in the CRC carcinogenic process. We developed an OBS to account for dietary and lifestyle factors that could act together to influence CRC risk.18 Oxidative stress also has been examined with miRNA expression in a limited number of studies.10 miR-200a has been associated with oxidative stress in breast cells; miR-155 has been linked to inflammatory and oxidative stress pathways; miR-21, miR-125b, miR-196, and miR-210 have been linked to inflammatory cytokines and signaling pathways.10 Others have cited miR-181a, miR-205, miR-1, miR-21, miR-24, miR-25, miR-185, miR-214, miR-133, miR-145, and miR-495 as being modulated by reactive oxygen species.37 Of these, only miR-200a-5p was associated with OBS in our data.
While we have found associations between a limited number of diet and lifestyle factors and miRNA expression levels, we have failed to replicate other findings that have been cited in the literature. These differences could stem from several sources, the primary reason being our study is the only study conducted in humans, while others have relied on mouse models and cell lines and were usually conducted in noncolorectal tissue. Additionally, while others have targeted a few miRNAs, we have incorporated a platform of over 2,000 miRNAs. This methodological difference has resulted in our level of adjustment being considerable, while other studies have no or minimal adjustment for multiple comparisons. Our data are based on recall of diet and lifestyle factors from cases, mainly for a referent period of 2 years to diagnosis. While other referent periods may be important, more distant referent periods would represent a time less temporal to the time of the miRNA expression. We do however believe that our data are excellent, in that results obtained from this study in terms of risk are similar to several other large cohort studies. However, if there is bias toward the null in our recalled data, it could influence our ability to detect associations. Other factors such as potentially different effects by age of participant are possible. Although we adjusted for age to control for confounding, we did not conduct separate age-stratified analysis. Our sample size, although large, would be too small for detailed subgroup analysis. Likewise, we have used an Agilent platform and have previously compared platform results to those obtained from quantitative polymerase chain reaction. Our results were in 100% agreement in direction of association and fold changes calculated from data on the Agilent platform and that obtained from quantitative polymerase chain reaction.31
Conclusion
In summary, we have shown that carbohydrate intake, sucrose intake, NSAID use, and OBS are associated with miRNA expression level. Additionally, most of these miRNAs were differentially expressed between colorectal carcinoma and normal mucosa, suggesting a role in CRC. We believe that these findings lend support to the hypothesis that miRNAs are regulated by diet and lifestyle factors. It is possible that other diet and lifestyle factors could be important in control settings, which we were unable to detect at the population level. We urge other researchers to replicate these findings utilizing laboratory-based studies to better understand the functional significance of these findings. These findings, if replicated, could provide further support for these diet and lifestyle factors in cancer prevention.
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. The authors acknowledge Sandra Edwards for data oversight, Dr Bette Caan and the staff at the KPMCP for their contribution to data collection, Dr Wade Samowitz for miRNA slide review, Erica Wolff and Michael Hoffman for miRNA analysis, Brett Milash and the Bioinformatics Shared Resource of the Huntsman Cancer Institute and University of Utah for miRNA and mRNA bioinformatics data processing, and Daniel Pellatt for statistical assistance. This study was supported by NCI grants CA163683 and CA48998 from the National Cancer Institute at the National Institutes of Health.
Author contributions
MLS designed research; RW and MLS conducted research; JSH, LEM, and JRS analyzed data and performed statistical analysis; MLS wrote paper and had primary responsibility for the final content; all authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | ||
Murray BS, Choe SE, Woods M, Ryan TE, Liu W. An in silico analysis of microRNAs: mining the miRNAome. Mol Biosyst. 2010;6(10):1853–1862. | ||
Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288(3–4):77–87. | ||
Gartel AL, Kandel ES. miRNAs: little known mediators of oncogenesis. Semin Cancer Biol. 2008;18(2):103–110. | ||
Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37(Web Server issue):W356–W362. | ||
Drusco A, Nuovo GJ, Zanesi N, et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014;9(6):e96670. | ||
Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer – a brief overview. Adv Biol Regul. 2015;57:1–9. | ||
Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–5856. | ||
Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66(8):477–482. | ||
Zhang C, Shu L, Kong AT. MicroRNAs: new players in cancer prevention targeting Nrf2, oxidative stress and inflammatory pathways. Curr Pharmacol Rep. 2015;1(1):21–30. | ||
Pathi SS, Jutooru I, Chadalapaka G, et al. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol Cancer Res. 2011;9(2):195–202. | ||
Parasramka MA, Dashwood WM, Wang R, et al. MicroRNA profiling of carcinogen-induced rat colon tumors and the influence of dietary spinach. Mol Nutr Food Res. 2012;56(8):1259–1269. | ||
Parasramka MA, Ho E, Williams DE, Dashwood RH. MicroRNAs, diet, and cancer: new mechanistic insights on the epigenetic actions of phytochemicals. Mol Carcinog. 2012;51(3):213–230. | ||
Slattery ML, Benson J, Curtin K, Ma KN, Schaeffer D, Potter JD. Carotenoids and colon cancer. Am J Clin Nutr. 2000;71(2):575–582. | ||
Slattery ML, Edwards SL, Anderson K, Caan B. Vitamin E and colon cancer: is there an association? Nutr Cancer. 1998;30(3):201–206. | ||
Stone WL, Papas AM. Tocopherols and the etiology of colon cancer. J Natl Cancer Inst. 1997;89(14):1006–1014. | ||
Satia-Abouta J, Galanko JA, Martin CF, Potter JD, Ammerman A, Sandler RS. Associations of micronutrients with colon cancer risk in African Americans and whites: results from the North Carolina Colon Cancer Study. Cancer Epidemiol Biomarkers Prev. 2003;12(8):747–754. | ||
Slattery ML, Lundgreen A, Welbourn B, Wolff RK, Corcoran C. Oxidative balance and colon and rectal cancer: interaction of lifestyle factors and genes. Mutat Res. 2012;734(1–2):30–40. | ||
Goodman M, Bostick RM, Gross M, Thyagarajan B, Dash C, Flanders WD. Combined measure of pro- and anti-oxidant exposures in relation to prostate cancer and colorectal adenoma risk: an update. Ann Epidemiol. 2010;20(12):955–957. | ||
Goodman M, Bostick RM, Dash C, Terry P, Flanders WD, Mandel J. A summary measure of pro- and anti-oxidant exposures and risk of incident, sporadic, colorectal adenomas. Cancer Causes Control. 2008;19(10):1051–1064. | ||
Slattery ML, Potter J, Caan B, et al. Energy balance and colon cancer – beyond physical activity. Cancer Res. 1997;57(1):75–80. | ||
Slattery ML, Caan BJ, Benson J, Murtaugh M. Energy balance and rectal cancer: an evaluation of energy intake, energy expenditure, and body mass index. Nutr Cancer. 2003;46(2):166–1671. | ||
Slattery ML, Herrick JS, Pellatt DF, et al. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016;37(3):245–261. | ||
Edwards S, Slattery ML, Mori M, et al. Objective system for interviewer performance evaluation for use in epidemiologic studies. Am J Epidemiol. 1994;140(11):1020–1028. | ||
Liu K, Slattery M, Jacobs D Jr, et al. A study of the reliability and comparative validity of the cardia dietary history. Ethn Dis. 1994;4(1):15–27. | ||
Slattery ML, Boucher KM, Caan BJ, Potter JD, Ma KN. Eating patterns and risk of colon cancer. Am J Epidemiol. 1998;148(1):4–16. | ||
Kampman E, Slattery ML, Bigler J, et al. Meat consumption, genetic susceptibility, and colon cancer risk: a United States multicenter case-control study. Cancer Epidemiol Biomarkers Prev. 1999;8(1):15–24. | ||
Agilent Technologies Inc. Agilent GeneSpring User Manual. Santa Clara, California: Agilent Technologies Inc; 2013. | ||
Storey JD. A direct approach to false discovery rates. J Royal Stat Soc. 2002;64(3):479–498. | ||
Garcia-Segura L, Perez-Andrade M, Miranda-Rios J. The emerging role of MicroRNAs in the regulation of gene expression by nutrients. J Nutrigenet Nutrigenomics. 2013;6(1):16–31. | ||
Pellatt DF, Stevens JR, Wolff RK, et al. Expression profiles of miRNA subsets distinguish human colorectal carcinoma and normal colonic mucosa. Clin Transl Gastroenterol. 2016;7:e152. | ||
Lancon A, Michaille JJ, Latruffe N. Effects of dietary phytophenols on the expression of microRNAs involved in mammalian cell homeostasis. J Sci Food Agric. 2013;93(13):3155–3164. | ||
Humphreys KJ, Conlon MA, Young GP, et al. Dietary manipulation of oncogenic microRNA expression in human rectal mucosa: a randomized trial. Cancer Prev Res. 2014;7(8):786–795. | ||
Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67. | ||
Ren L, Tang G. Identification of sucrose-responsive microRNAs reveals sucrose-regulated copper accumulations in an SPL7-dependent and independent manner in Arabidopsis thaliana. Plant Sci. 2012;187:59–68. | ||
Wong TY, Li F, Lin SM, Chan FL, Chen S, Leung LK. Celecoxib increases miR-222 while deterring aromatase-expressing breast tumor growth in mice. BMC Cancer. 2014;14:426. | ||
Magenta A, Dellambra E, Ciarapica R, Capogrossi MC. Oxidative stress, microRNAs and cytosolic calcium homeostasis. Cell Calcium. 2016;60(3):207–217. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

