Back to Journals » OncoTargets and Therapy » Volume 8
Diagnostic value of SHOX2 DNA methylation in lung cancer: a meta-analysis
Authors Zhao Q , Guo T, Wang H, Zhang X, Zhang H, Wang Z, Yuan Z, Duan G
Received 12 August 2015
Accepted for publication 15 October 2015
Published 19 November 2015 Volume 2015:8 Pages 3433—3439
DOI https://doi.org/10.2147/OTT.S94300
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Qing-Tao Zhao,* Tao Guo,* Hui-En Wang,* Xiao-Peng Zhang, Hua Zhang, Zhi-Kang Wang, Zheng Yuan, Guo-Chen Duan
Department of Thoracic Surgery, Hebei General Hospital, Shijiazhuang, Hebei, People’s Republic of China
*These authors contributed equally to this work
Abstract: The diagnostic value of SHOX2 DNA methylation in patients with lung cancer remains controversial. Thus, we performed a systematic review and meta-analysis to assess diagnostic accuracy of SHOX2 DNA methylation in the lymph node, bronchial aspirates, pleural effusion, plasma, and tumor tissue for lung cancer. We conducted a comprehensive literature search in PubMed, Ovid, the Cochrane library, and Web of Science databases in May 2015. The diagnostic sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and summary receiver operating characteristic (SROC) curve were pooled using STATA 12.0 software. A total of 2,296 subjects included 1,129 lung cancer patients in eight studies were recruited in this meta-analysis. The summary estimates for SHOX2 DNA methylation in the diagnosis of lung cancer in these studies were pooled SEN =0.70 (95% confidence interval [CI]: 0.46–0.87), SPE =0.96 (95% CI: 0.91–0.99), PLR 20.01 (95% CI: 6.96–57.52), NLR 0.31 (95% CI: 0.15–0.64), and DOR 65.11 (95% CI: 13.10–323.61), and the area under the curve (AUC) was 0.96 (95% CI: 0.94–0.97). SHOX2 DNA methylation has greater diagnostic value in detecting lung cancer. In addition, considering the potential publication bias and high heterogeneity, further research studies with more well-designed and large sample sizes are needed in the future.
Keywords: lung cancer, SHOX2, meta-analysis, diagnostic test
Introduction
Lung cancer is the leading cause of cancer deaths worldwide. Lung cancer is the second most common cancer in both females and males representing approximately 15% of all cancer diagnoses.1 Despite research on the diagnosis of lung cancer and the use of increasingly advanced technology in its treatment, the prognosis of lung cancer is still poor. Diagnosis for lung cancer has become a major focus.
Early diagnosis and treatment can improve the 5-year survival rate and reduce mortality.2 However, most patients with lung cancer are found to be at stages III, IV when they are diagnosed for the first time. Low-dose spiral computed tomography can be a reliable screening tool for early detection of lung cancer.3,4 Even when signs, symptoms, and computed tomography findings are such that the clinical diagnosis of malignant lung neoplasms appears obvious, it often takes considerable effort and invasive ways (percutaneous needle biopsy or bronchoscopy biopsy) to obtain tissue material confirming the presence of lung cancer.5 However, studies demonstrated that the diagnosis cannot be confirmed in approximately 50% suspected lung cancer patients analyzed for the first time using invasive procedures.5–7 Therefore, a more effective way is needed to assist confirmatory diagnosis in order to avoid repeated percutaneous needle or bronchoscopy.
Short stature homeobox 2 (SHOX2) is a homologue to the short stature homeobox gene SHOX in humans. The SHOX2 gene, also called OG12, OG12X, or SHOT gene, is another gene of the SHOX gene family and is located at 3q25-q26.1.8 The SHOX2 gene is mainly expressed in the limb bud, gill arches, nose, heart, nervous system, and reproductive nodules of human embryos.9 Recently, several recent studies reported that hypermethylation of the SHOX2 DNA locus could be a candidate biomarker for lung cancer.10–14 SHOX2 DNA methylation has been described as a useful and powerful biomarker for detection of patients with lung cancer, based on the analysis of the lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration, bronchial aspirates, pleural effusion, plasma, and tumor tissue.10–14 However, the results are controversial. To fully understand the diagnostic performance of SHOX2 DNA methylation for lung cancer, we conducted a meta-analysis on the diagnostic sensitivity and specificity of SHOX2 DNA methylation for diagnosis of lung cancer.
Materials and methods
Search strategy
We conducted a comprehensive literature search in PubMed, Ovid, the Cochrane library, and Web of Science databases. The main search terms included: (SHOX2 or short stature homeobox 2 or SHOX2 DNA methylation) and (lung cancer or lung carcinoma or NSCLC or SCLC). All articles published till May 2015 were considered.
Inclusion criteria
All candidate studies were evaluated and extracted by two independent investigators (Qing-Tao Zhao and Tao Guo). The articles, which could not be excluded based on title and abstract, were retrieved for full-text review. If disagreement occurred, two investigators discussed and arrived at consensus with a third investigator (Hui-En Wang).
Studies were included in this meta-analysis if they met the following criteria: 1) the diagnostic potential of SHOX2 for lung cancer being studies; 2) study design being observational studies (cohort or case-control studies); 3) the patients being diagnosed with lung cancer by pathology; 4) sensitivity (SEN) and specificity (SPE) of SHOX2 being reported to provide sufficient information to construct 2×2 contingency tables.
Exclusion criteria
Studies were excluded from the meta-analysis for the following reasons: 1) abstracts, letters, reviews, expert opinions, case reports, or nonclinical studies; 2) studies had duplicate or overlapping data; 3) studies were not written in English; 4) no human studies.
Data extraction and quality assessment
All the studies were reviewed by two investigators (Qing-Tao Zhao and Tao Guo) independently based on titles and abstracts, author, year of publication, country of origin, sample size, assay methods, and diagnostic performance (SEN, SPE, true positive [TP], false-positive [FP], false-negative [FN], and true negative [TN]). We resolved disagreements by reaching a consensus through discussion.
The Quality Assessment of Diagnostic Accuracy Studies (QUADAS) was used to assess each study for the quality of the information reported.15 QUADAS is a quality assessment tool specifically developed for systematic reviews of diagnostic accuracy studies to assess bias in the study, including 14 questions (each of which is scored as yes, no, or unclear).
Statistical analysis
We used STATA software, version 12.0 (Stata Corporation, College Station, TX, USA) to perform the meta-analysis. The number of TP, TN, FP, and FN were retrieved from each article. The SEN, SPE, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) estimates with 95% confidence interval (CI) from each study were analyzed using a random-effect model and the bivariate summary receiver operating characteristic (SROC) curve was generated. The area under the curve (AUC) represents an analytical summary of the test performance and illustrates the trade-off between SEN and SPE. The between-study heterogeneity was evaluated by Q-test and I2 statistics. P<0.10 for Q-test or I2>50% indicates substantial heterogeneity. The leave-one-out sensitivity analysis was conducted to determine whether our assumptions or decisions have a major effect on the results of the review by omitting each study (one at a time). Furthermore, subgroup analyses were performed for sample size and types of biological sample. As publication bias is of concern for the meta-analysis of diagnostic studies, we tested for the potential presence of this bias using Deeks’ funnel plots.16 P<0.05 was considered statistically significant.
Results
Search results
A total of 37 titles and abstracts were preliminarily reviewed, of which eight studies were available for the meta-analysis, including 1,129 lung cancer patients who received SHOX2 DNA methylation tests.10–14,17–19 Figure 1 shows a flow diagram of the selection process. The characteristics of each study are shown in Table 1.
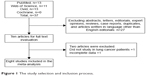  | Figure 1 The study selection and inclusion process. |
Assessment of methodological quality
Quality assessment based on QUADAS guidelines was conducted on all eight studies included for systematic review. The QUADAS scores of studies were from 10 to 13, which satisfy the majority of the standards.
Diagnostic accuracy analysis
The forest plot of SEN and SPE for SHOX2 DNA methylation assays in the diagnosis of lung cancer of the eight studies is shown in Figure 2. The SEN ranged from 0.12 to 0.96 (pooled: 0.70; 95% CI: 0.46–0.87) whereas SPE ranged from 0.84 to 1.00 (pooled, 0.96; 95% CI: 0.91–0.99) (Figure 2A and B). The PLR was 20.01 (95% CI: 6.96–57.52), NLR was 0.31 (95% CI: 0.15–0.64), and DOR was 65.11 (95% CI: 13.10–323.61).
The graph of the SROC curve is shown in Figure 3. It can demonstrate the trade-off between SEN and SPE values in multiple studies.20 SROC results showed that AUC of SHOX2 DNA methylation was 0.96 (95% CI: 0.94–0.97), indicating that SHOX2 DNA methylation may be able to differentiate lung cancer patients from non-lung-cancer patients with a high overall accuracy.21 The LRT_I2 statistic value was 91.99 (95% CI: 84.57–99.42), indicating that an evident heterogeneity was present in these eight studies. The LRTQ (χ2) statistic was 24.98 (P=0.000), indicating that the heterogeneity was likely the result of nonthreshold effects.
Subgroup analysis
Subgroup 1: the corresponding values of the subgroup with sample size ≥200 were 0.61 (95% CI: 0.58–0.65) for sensitivity and 0.89 (95% CI: 0.87–0.91) for specificity. It showed sensitivity of 0.70 (95% CI: 0.64–0.75) and specificity of 0.99 (95% CI: 0.97–1.00) in the sample size <200 subgroup. Subgroup 2: the corresponding values for bronchial aspirates were sensitivity of 0.69 (95% CI: 0.65–0.74) and specificity of 0.96 (95% CI: 0.93–0.97). It showed sensitivity of 0.43 (95% CI: 0.38–0.49) and specificity of 0.86 (95% CI: 0.83–0.89) in the pleural effusions subgroup. The corresponding values for the otherwise subgroup were sensitivity of 0.75 (95% CI: 0.70–0.80) and specificity of 0.94 (95% CI: 0.90–0.96). Subgroup 3: the corresponding values of the subgroup with Epi proLung BL were 0.66 (95% CI: 0.62–0.69) for sensitivity and 0.89 (95% CI: 0.87–0.91) for specificity. It showed sensitivity of 0.58 (95% CI: 0.53–0.63) and specificity of 0.96 (95% CI: 0.93–0.98) in the Roche and/or Qiagen subgroup (Table 2).
Sensitivity analysis
A single study involved in this meta-analysis was evaluated each time to reflect the influence of the individual data set to SEN and SPE. The results pattern was not impacted by single study (Table 2). The P-value for Q-test and the I2-value also showed that none of the single studies affected the heterogeneity of this meta-analysis.
Publication bias
We investigated the publication bias analysis for the meta-analysis. The funnel plots for publication bias showed no asymmetry (Figure 4). The result of Deeks’ test was 0.05, indicating that publication bias could exist in the meta-analysis.
  | Figure 4 The Deeks’ test of the diagnostic meta-analysis. |
Discussion
DNA methylation is an important manner of regulation at the epigenetic level for gene expression, cell growth, and cell differentiation. DNA methylation occurs mainly at the C-phosphate-G (CpG) site, in which cytosine is linearly connected to an adjacent guanine.22 Methylation of the cytosine within the CpG islands plays a key role in fundamental biological processes and human diseases and aberrant DNA methylation is a hallmark of human cancers.22–24 It has been found that the SHOX2 gene includes two large CpG islands, one at the 5′ end, covering a region of approximately 1 kb, the other at the 3′ end, covering approximately 0.5 kb.25 Studies have revealed that the methylation level of the SHOX2 gene CpG islands was significantly increased in tissues and cells of lung cancer compared with that from normal tissues and cells.10–14,17–19 Growing evidence suggests that SHOX2 DNA methylation is a promising auxiliary diagnostic biomarker for lung cancer, but with considerable controversial results.17,19,25 Therefore, in this study, we performed a systematic review and meta-analysis to assess diagnostic accuracy of SHOX2 DNA methylation in the lymph node, bronchial aspirates, pleural effusion, plasma, and tumor tissue of patients with lung cancer.
The results of the meta-analysis indicated that the pooled SEN and SPE of SHOX2 DNA methylation were 0.70 (95% CI: 0.46–0.87) and 0.96 (95% CI: 0.91–0.99), respectively. This means that 70% of the lung cancer patients had high SHOX2 levels, and 96% of non-lung-cancer patients have low SHOX2 DNA methylation levels. It has low sensitivity (70%) for diagnosing lung cancer. The reason for this discrepancy could be the inconsistent amount of methylated DNA obtained from samples. If a high amount of methylated DNA can be obtained from histological samples, the detection sensitivity is high. In contrast, for samples with much less or a trace amount of methylated DNA, such as pleural effusion samples, the sensitivity is expected to be lower. It is still better than the traditional serum-based biomarkers. For example, the sensitivities of CEA, CYFRA 21-1, SCC, and NSE were 47.5%, 47.5%, 49%, and 39.7% for diagnosing lung cancer, respectively.26 Thus, SHOX2 DNA methylation has higher sensitivity and specificity compared to conventional lung cancer biomarkers. It has higher sensitivity and specificity in effectively diagnosing lung cancer.
Glas et al27 found that the DOR combined the strengths of SEN and SPE as prevalence in dependent indicators and was useful from the statistical point of view in the assessment of the overall test accuracy in meta-analysis. The value of DOR ranges from 0 to ∞, with higher values indicating better discriminatory test performance.28 The DOR value of 6.66 indicates that the SHOX2 DNA methylation could be a useful biomarker for lung cancer patients’ diagnosis. AUC is calculated to evaluate accuracy of the selected indicator, and SROC is usually used to summarize overall test performance.29,30 To demonstrate excellent accuracy, the valve of AUC should be more than 0.97. An AUC of 0.93–0.96 is considered to be very good and 0.75–0.92 is good.21,31 In this study, we show that testing for methylated SHOX2 DNA shows good accuracy for the diagnosis of lung cancer, with an area under the ROC curve of 0.78. Overall, although the SEN is compromised, SHOX2 DNA methylation has a good SPE in the diagnosis of lung cancer. The PLR and NLR are more meaningful indicators of diagnostic accuracy.32,33 A good diagnostic test may have high PLR (PLR >5) and low NLR (NLR <0.2).34,35 However, the NLR value of 0.40 (95% CI: 0.37–0.44) did not meet the thresholds. The PLR value of 7.04 (95% CI: 5.72–8.65) demonstrated that lung cancer patients had approximately 7.04 times higher chance of testing positive than non-lung cancer, and this was relatively high for clinical purposes. SHOX2 DNA methylation was an effective biomarker for lung cancer diagnosis.
Exploring the sources of heterogeneity is one major purpose of meta-analysis.36,37 In this study, we found significant heterogeneity observed in our analysis, and the result of Spearman approach showed that heterogeneity could not be explained by a threshold effect. We surmised that the heterogeneity was attributed to the sample, etiology, and different stages of lung cancer patients. We speculated that the limited number of eligible studies was the main factor that made subgroup analysis not possible. However, these hypotheses need to be investigated in the future study.
This meta-analysis had some limitations. First, we only included eight studies that have a smaller number of cases. Therefore, the results of the trials in a pooled analysis were not robust. More studies are needed for future analyses. Second, we did not calculate some covariates because sufficient raw data were not available from the selected articles. These probable covariates included tumor type, ethnicity, and TNM staging. Third, this meta-analysis was based on published studies; the exclusion of unpublished data is generally associated with an overestimation of the true effect, thus resulting in a publication bias. Fourth, only articles published in the English language were included in this meta-analysis, and all studies included in our meta-analysis were from Western countries. This would have introduced some bias to the analysis.
In conclusion, this meta-analysis demonstrated that SHOX2 DNA methylation is a valuable diagnostic biomarker for lung cancer diagnosis. More well-designed research studies with strictly defined controls are needed to elucidate further the accuracy of SHOX2 DNA methylation for diagnosis indicator of lung cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. | ||
Howlader N, Mariotto AB, Woloshin S, Schwartz LM. Providing clinicians and patients with actual prognosis: cancer in the context of competing causes of death. J Natl Cancer Inst Monogr. 2014;2014(49):255–264. | ||
Wood DE, Kazerooni E, Baum SL, et al. Lung cancer screening, version 1.2015: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2015;13(1):23–34. | ||
Field JK, Baldwin D, Brain K, et al. CT screening for lung cancer in the UK: position statement by UKLS investigators following the NLST report. Thorax. 2011;66(8):736–737. | ||
Roth K, Hardie JA, Andreassen AH, Leh F, Eagan TM. Predictors of diagnostic yield in bronchoscopy: a retrospective cohort study comparing different combinations of sampling techniques. BMC Pulm Med. 2008;8:2. | ||
Schreiber G, McCrory DC. Performance characteristics of different modalities for diagnosis of suspected lung cancer: summary of published evidence. Chest. 2003;123(Suppl 1):S115–S128. | ||
Shital P, Rujuta A, Sanjay M. Transbronchial needle aspiration cytology (TBNA) in endobronchial lesions: a valuable technique during bronchoscopy in diagnosing lung cancer and it will decrease repeat bronchoscopy. J Cancer Res Clin Oncol. 2014;140(5):809–815. | ||
De Baere E, Speleman F, Van Roy N, De Paepe A, Messiaen L. Assignment of SHOX2 (alias OG12X and SHOT) to human chromosome bands 3q25-->q26.1 by in situ hybridization. Cytogenet Cell Genet. 1998;82(3–4):228–229. | ||
Clement-Jones M, Schiller S, Rao E, et al. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Genet. 2000;9(5):695–702. | ||
Darwiche K, Zarogoulidis P, Baehner K, et al. Assessment of SHOX2 methylation in EBUS-TBNA specimen improves accuracy in lung cancer staging. Ann Oncol. 2013;24(11):2866–2870. | ||
Schmidt B, Liebenberg V, Dietrich D, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. | ||
Ilse P, Biesterfeld S, Pomjanski N, Fink C, Schramm M. SHOX2 DNA methylation is a tumour marker in pleural effusions. Cancer Genomics Proteomics. 2013;10(5):217–223. | ||
Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6(10):1632–1638. | ||
Schneider KU, Dietrich D, Fleischhacker M, et al. Correlation of SHOX2 gene amplification and DNA methylation in lung cancer tumors. BMC Cancer. 2011;11:102. | ||
Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. | ||
Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. | ||
Dietrich D, Kneip C, Raji O, et al. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol. 2012;40(3):825–832. | ||
Dietrich D, Jung M, Puetzer S, et al. Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant and malignant pleural effusions. PLoS One. 2013;8(12):e84225. | ||
Ilse P, Biesterfeld S, Pomjanski N, Wrobel C, Schramm M. Analysis of SHOX2 methylation as an aid to cytology in lung cancer diagnosis. Cancer Genomics Proteomics. 2014;11(5):251–258. | ||
Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat Med. 1993;12(14):1293–1316. | ||
Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. | ||
Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. | ||
Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. | ||
Tan Q, Wang G, Huang J, et al. Epigenomic analysis of lung adenocarcinoma reveals novel DNA methylation patterns associated with smoking. Onco Targets Ther. 2013;6:1471–1479. | ||
Song L, Yu H, Li Y. Diagnosis of lung cancer by SHOX2 Gene Methylation Assay. Mol Diagn Ther. 2015;19(3):159–167. | ||
Chu XY, Hou XB, Song WA, Xue ZQ, Wang B, Zhang LB. Diagnostic values of SCC, CEA, Cyfra21-1 and NSE for lung cancer in patients with suspicious pulmonary masses: a single center analysis. Cancer Biol Ther. 2011;11(12):995–1000. | ||
Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. | ||
Pai M, McCulloch M, Enanoria W, Colford JJ. Systematic reviews of diagnostic test evaluations: what’s behind the scenes? ACP J Club. 2004;141(1):A11–A13. | ||
Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg. 2005;79(1):16–20. | ||
Walter SD. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med. 2002;21(9):1237–1256. | ||
Chen Z, Liu HB, Yu CH, Wang Y, Wang L, Song Y. Diagnostic value of mutation-specific antibodies for immunohistochemical detection of epidermal growth factor receptor mutations in non-small cell lung cancer: a meta-analysis. PLoS One. 2014;9(9):e105940. | ||
McGee S. Simplifying likelihood ratios. J Gen Intern Med. 2002;17(8):646–649. | ||
Gallagher EJ. Clinical utility of likelihood ratios. Ann Emerg Med. 1998;31(3):391–397. | ||
Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM. Systematic reviews of diagnostic test accuracy. Ann Intern Med. 2008;149(12):889–897. | ||
Deeks JJ. Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323(7305):157–162. | ||
Coory MD. Comment on: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010;39(3):932. | ||
Naaktgeboren CA, van Enst WA, Ochodo EA, et al. Systematic overview finds variation in approaches to investigating and reporting on sources of heterogeneity in systematic reviews of diagnostic studies. J Clin Epidemiol. 2014;67(11):1200–1209. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




