Back to Journals » OncoTargets and Therapy » Volume 12
Diagnostic value of circular RNAs as effective biomarkers for cancer: a systematic review and meta-analysis
Authors Tan H , Gan L , Fan X, Liu L, Liu S
Received 8 December 2018
Accepted for publication 27 February 2019
Published 10 April 2019 Volume 2019:12 Pages 2623—2633
DOI https://doi.org/10.2147/OTT.S197537
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Hong Tan,1,* Li Gan,2,* Xiaoming Fan,3 Limin Liu,4 Shan Liu3
1Department of General Surgery, Chengdu Integrated TCM & Western Medicine Hospital (Chengdu First People’s Hospital), Chengdu, 610041, China; 2School of Medicine, University of Electronic Science and Technology of China, Key Laboratory for Human Disease Gene Study, Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital, Chengdu, 610054, China; 3Department of Laboratory Medicine, Affiliated Hospital of University of Electronic Science and Technology, Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, Chengdu, 610072, China; 4Jiangsu Institute of Hematology, the First Affiliated Hospital of Soochou University, Institute of Blood and Marrow Transplantation, Suzhou, 215006, China
*These authors contributed equally to this work
Background: Increasing evidence has identified circular RNAs (circRNAs) as ideal molecular biomarkers for cancer diagnosis, therapy, and prognosis. However, the overall diagnostic efficiency of circRNAs remains unclear. Thus, this meta-analysis aimed to comprehensively evaluate the diagnostic accuracy of circRNA expression profiles for cancer.
Methods: A literature search of online databases was conducted to identify all eligible studies. The quality of the studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 tool. All statistical analyses were executed using STATA 14.0, Meta-DiSc 1.4, and Review Manager 5.2 software.
Results: A total of 32 studies, involving 2,400 cases and 2,295 controls, were included in the diagnostic meta-analysis. The pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and area under the curve were 0.79 (95% CI: 0.73–0.84), 0.73 (95% CI: 0.67–0.79), 2.9 (95% CI: 2.5–3.5), 0.29 (95% CI: 0.24–0.36), 10 (95% CI: 8–13), and 0.83 (95% CI: 0.79–0.86), respectively. The overall analysis suggested that circRNAs are useful diagnostic biomarkers for cancer. Subgroup analysis indicated that plasma samples had a better diagnostic performance than cancer tissue samples for cancer detection. Studies involving ≥100 cases or gastric cancer showed higher sensitivities than those including <100 cases or other cancers.
Conclusion: This meta-analysis revealed that circRNAs were significantly correlated with cancer diagnosis. In addition, circRNAs had good diagnostic accuracy and might serve as effective diagnostic biomarkers for cancer.
Keywords: circular RNAs, cancer, diagnosis, biomarkers, meta-analysis
Introduction
Circular RNAs (circRNAs), a novel class of endogenous noncoding RNAs (ncRNAs), are generated from back-splicing events and are characterized by a covalently closed continuous loop without 5′ caps and 3′ poly (A) tails.1,2 Owing to the closed continuous loop structure, circRNAs can escape exonuclease-mediated degradation; therefore, they are more stable in blood or plasma than are microRNAs (miRNAs) and long noncoding RNAs (lncRNAs).3 CircRNAs acting as ‘‘miRNA sponges” are involved in the initiation and progression of several types of cancer by binding to miRNAs.4 Moreover, recent studies have revealed that some circRNAs play significant roles in various kinds of cancer, including gastric cancer (GC), hepatocellular carcinoma (HCC), breast cancer (BC), colorectal cancer (CRC), and lung adenocarcinoma (LAC), among others.5 These findings indicate that circRNAs have the potential to serve as novel noninvasive diagnostic biomarkers for various cancers.
However, due to small sample sizes and study design limitations, research evidence for the diagnostic accuracy of circRNAs in cancer is inaccurate and inadequate. To address these shortcomings, we conducted a comprehensive systematic analysis of data from all relevant publications to investigate the relationship between circRNAs and cancer diagnosis.
Methods
Literature search strategy
All potential literature in this meta-analysis was independently retrieved and screened by two researchers (GL and LS). A comprehensive and systematic search was conducted of the PubMed, Web of Science, Cochrane Library, Embase, CNKI, and IEEE online databases to identify eligible studies indexed through November 25, 2018. The search terms were as follows: “circular RNAs OR circRNAs” AND “cancer OR carcinoma OR tumor OR neoplasm” AND “sensitivity OR specificity OR ROC curve OR AUC OR diagnosis”. In addition, the reference lists of the included articles were manually reviewed to identify additional relevant studies. As this study was based on previously published studies, no ethical approval or patient consent was required.
Selection criteria of reported research
The selection process in this meta-analysis was executed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.6 All eligible studies fulfilled the following inclusion criteria. 1) Studies had a definite diagnosis of human cancer made using circRNAs. 2) All cancer cases were confirmed by pathological examination. 3) CircRNAs expression was detected in serum, plasma, or cancer tissue with quantitative reverse transcription–polymerase chain reaction (qRT–PCR) or other methods. 4) Studies provided sufficient diagnostic parameters to calculate true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). The exclusion criteria included the following: 1) studies that did not satisfy the abovementioned inclusion criteria; 2) studies that were duplicate articles, reviews, animal studies, editorials, case reports, comments, and meta-analyses; 3) studies lacking sufficient data to construct a diagnostic 2 × 2 table.
Data extraction and quality assessment
Two researchers (GL and TH) carefully reviewed the full texts of all eligible studies and independently extracted the relevant data, including the first author’s name, publication year, country, cancer type, circRNA profiles, specimen, detection method, sample size, cutoff values, area under the curve (AUC), sensitivity and specificity, etc. Furthermore, 2 × 2 tables were created using TP, FP, TN, and FN. Any disagreement among the authors was resolved through discussions with a third author (LS) until a consensus was reached.
Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) was used to appraise the risk of bias and applicability of the included studies using Review Manager 5.2 software.7 The QUADAS-2 tool consists of four key domains: patient selection, index test, reference standard, and flow and timing. The risk of bias and concerns regarding applicability were evaluated as “low”, “high”, or “unclear”.
Statistical analysis
Statistical analysis of the diagnostic tests was executed using Stata 14.0, Meta-DiSc 1.4, and Review Manager 5.2. Q tests and I2 statistics were used to estimate the heterogeneity caused by a non-threshold effect among the included articles. Either P<0.10 or I2>50% suggested the existence of substantial heterogeneity; in this study, a random-effects model was applied to quantify the pooled sensitivity, specificity, diagnostic odds ratio (DOR), positive likelihood ratio (PLR), negative likelihood ratio (NLR), and AUC, with corresponding 95% confidence intervals (CIs). Otherwise, a fixed-effects model was used. Spearman correlation analysis was conducted to verify the threshold effects. Moreover, subgroup analysis and meta-regression were applied to trace the sources of heterogeneity. Sensitivity analysis was performed to assess the stability of our analysis. Publication bias was evaluated with Deeks’ funnel plots. All tests were two-sided and P<0.05 was considered statistically significant.
Results
Literature search and selection of studies
A total of 290 articles were systematically retrieved from the online databases. A total of 231 records remained after duplicates were removed. First, we roughly screened the titles and abstracts and eliminated 161 publications that were irrelevant to the topic. The remaining 70 articles were further examined by careful review of the full text; as a result, 46 articles were excluded. Finally, a total of 32 eligible studies from 24 articles8-31 involving 3,016 participants were included in the meta-analysis. The flow diagram is shown in Figure 1.
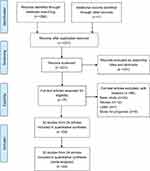 | Figure 1 Flow diagram of the study selection process. |
Study characteristics
The main characteristics of the studies are listed in Table 1. This diagnostic meta-analysis analyzed 32 eligible studies from 24 articles involving 2,400 cases and 2,295 controls. All cancer cases were confirmed pathologically and the controls consisted of adjacent nontumorous tissue, noncancer tissue, or plasma from unaffected subjects. Specifically, tissue, plasma or saliva specimens were collected before treatment including radiotherapy, chemotherapy, and targeted therapy. The expression of circRNAs was detected by qRT–PCR. All studies referred to five different cancer types: gastric (GC, n=16), liver (HCC, n=7), breast (BC, n=3), colorectal (CRC, n=3), lung (LAC, n=2), and oral squamous cell carcinoma (OSCC, n=1).
 | Table 1 Main characteristics of the studies included in the meta-analysis |
Quality assessment
A quality assessment of the eligible studies was performed using QUADAS-2 (Figure 2). The figure depicts the relatively moderate quality of the 32 included studies. Most studies had either low or unclear risks of bias due to a lack of information on patient selection, exclusion criteria, or pre-specified thresholds.
 | Figure 2 Quality assessment of the included studies according to QUADAS-2. |
Diagnostic accuracy
Heterogeneity among studies was evaluated by examining the threshold and non-threshold effects. In our study, the Spearman correlation coefficient and P-value were 0.658 and 0.218, respectively, suggesting that there was no threshold effect. Heterogeneity owing to non-threshold effects was then assessed with Q-tests and I2 statistics. There was significant heterogeneity in the pooled sensitivity (I2=89.57%, P<0.001) and specificity (I2=90.88%, P<0.001); thus, a random-effects model was applied to analyze the diagnostic parameters. The pooled sensitivity and specificity were 0.79 (95% CI: 0.73–0.84) and 0.73 (95% CI: 0.67–0.79), respectively (Figure 3). In addition, the pooled PLR, NLR, and DOR were 2.9 (95% CI: 2.5–3.5), 0.29 (95% CI: 0.24–0.36), and 10 (95% CI: 8–13), respectively. The summary receiver operator characteristic (SROC) curve is shown in Figure 4; the AUC was 0.83. These results indicated that circRNAs have potential diagnostic value for several cancers.
 | Figure 3 Forest plots of sensitivity and specificity of circRNAs for cancer diagnosis. (A) Pooled sensitivity for circRNAs. (B) Pooled specificity for circRNAs. |
 | Figure 4 Summary receiver operator characteristic curve for cancer diagnosis. |
Subgroup analysis and meta-regression
To explore the potential sources of heterogeneity, subgroup analyses were first performed based on specimen (tissue vs other), case size (≥100 vs <100), and cancer type (GC vs other) (Table 2). Studies using other samples (plasma or saliva) had better diagnostic accuracy than those using tissue samples for cancer diagnosis, with increased sensitivity (0.75 vs 0.84), decreased NLR (0.33 vs 0.23), increased DOR (9 vs 12), and increased AUC (0.81 vs 0.84). Additionally, studies involving ≥100 cases or GC showed higher sensitivities but lower specificities than those involving <100 cases or other cancers. However, meta-regression analyses indicated that no methodological covariates affected the diagnostic accuracy of circRNAs (all joint P>0.05).
 | Table 2 Results of subgroup analysis |
We also analyzed subgroups according to cancer type, and the main results were summarized in Figure 5. Significant heterogeneity was observed in the GC group (I2=57.8%, P=0.002). No significant heterogeneity was observed in the HCC (I2=23.0%, P=0.254), CRC (I2=13.9%, P=0.313), LAC (I2=0.0%, P=0.379), and BC (I2=0.0%, P=0.517) subgroups. The results suggest that cancer type may act as potential sources of heterogeneity.
 | Figure 5 Subgroup analyses according to cancer type. |
Sensitivity analysis
To further explain the heterogeneity of individual studies, we performed a sensitivity analysis by removing individual studies. As shown in Figure 6, no outlier study was identified and the results were relatively stable and reliable.
 | Figure 6 Sensitivity analysis of the overall pooled study. |
Publication bias
Deeks’ funnel plot asymmetry tests were applied to estimate publication bias in the meta-analysis (shown in Figure 7). The results confirmed the lack of significant publication bias across the overall combined diagnostic studies (P=0.68, >0.05).
 | Figure 7 Deeks’ funnel plot to assess publication bias.Abbreviation: ESS, effective sample size. |
Discussion
Owing to their closed continuous loop structure, circRNAs are more stable in the extracellular space compared to linear RNAs. This feature makes circRNAs advantageous for use as molecular markers of cancer. The current study comprehensively assessed the diagnostic efficacy of circRNAs for several cancers. The pooled effect showed the relatively high level of diagnostic accuracy of circRNAs, suggesting their potential as effective biomarkers for cancer diagnosis.
CircRNAs have recently been identified as a family of naturally occurring endogenous noncoding RNAs that may regulate gene expression in mammals.32 With the widespread application of bioinformatics and next-generation sequencing technologies, a large number of circRNAs have been sequenced and entered into a database of 32,914 human exonic circRNAs.33 In addition, some circRNAs are related to cancer diagnosis. Previous studies have shown that circRNAs play a crucial role in transcriptional or posttranscriptional regulation of gene expression.34 Moreover, circRNAs, acting as efficient microRNA (miRNA) sponges, can specifically bind to miRNAs and compete with endogenous RNA, strongly suppressing microRNA activity and potentially contributing to tumor progression.3 Furthermore, circRNAs are associated with RNA binding proteins, which play crucial roles in cancer development through dysregulation of transcription or expression.35 Most importantly, circRNAs are more abundant and stable than the corresponding linear RNAs. Recently, a fusion circRNA (F-circEA) was discovered to monitor fusion genes involved in tumorigenesis, which could be a potential novel “liquid biopsy” biomarker in non-small cell lung cancer.36 Li et al first reported on exosomes enriched with stable circRNAs and proposed their potential of circRNAs as a new class of cancer biomarkers.37 These findings suggest that circRNAs may be a promising diagnostic biomarker for cancer.
Our study, comprising 3,016 participants (2,400 cases and 2,295 controls) is the most comprehensive meta-analysis to assess the diagnostic value of circRNAs for various cancers. Two meta-analyses have been published on the diagnostic value of circRNAs. Li et al38 and Wang et al39 reported circRNA AUCs of 0.793 and 0.79, respectively. Compared to their results, our study observed a higher diagnostic efficiency (AUC=0.83). In our meta-analysis, we first conducted a quality assessment of the 32 enrolled studies, which revealed relatively moderate quality. The overall pooled sensitivity, specificity, and AUC of circRNAs for cancer diagnosis were 0.79 (95% CI: 0.73–0.84), 0.73 (95% CI: 0.67–0.79), and 0.83 (95% CI: 0.79–0.86), respectively; higher values than those for traditional plasma-based biomarkers such as CEA and CA19-9.40 In addition, the pooled DOR of circRNAs was 10, suggesting a powerful discriminating capacity of circRNAs for cancer diagnosis. Together, these findings suggest that circRNAs might be effective biomarkers for cancer diagnosis.
The pooled results indicated that there was significant heterogeneity that could impact the accuracy among the overall studies. The Spearman correlation coefficient was 0.658 (P=0.218), suggesting that the threshold effect was not the source of heterogeneity. We further performed meta-regression and subgroup analysis. Studies with plasma samples had better diagnostic accuracy, while ≥100 cases or GC showed higher sensitivities than those with <100 cases or other cancers. The results implied that specimen, case size, and cancer type might influence the diagnostic accuracy; however, the differences were not statistically significant. Sensitivity analysis was also conducted to analyze the heterogeneity, but no outlier studies were found. The heterogeneity could derive from confounding factors and differences in methodology.
The present meta-analysis has several limitations. First, there was significant heterogeneity among the included studies. Although we performed subgroup analysis and meta-regression to explore the sources of heterogeneity, the results did not fully explain the potential heterogeneity. Second, only studies conducted in Asia were included; therefore, the results for other ethnicities might be missed, which may lead to population selection bias. Therefore, additional higher-quality, multicenter, and well-designed studies are required to confirm our findings.
Conclusion
The results of our meta-analysis suggest the potential of circRNAs as biomarkers for the diagnosis of several cancers, with good diagnostic efficiency (AUC=0.83). However, the application of circRNAs for cancer diagnosis requires further validation.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (No. 81603018) and the Clinical and Translational Science Foundation of Sichuan Provincial People’s Hospital (No. 2017LY03).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2013;32(7):923–925. doi:10.1038/emboj.2013.53
2. Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–1287. doi:10.1101/gr.202895.115
3. Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. doi:10.1186/s12943-017-0598-7
4. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
5. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. doi:10.1186/s13045-016-0370-2
6. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi:10.1136/bmj.b2651
7. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi:10.7326/0003-4819-155-8-201110180-00009
8. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi:10.1038/bjc.2016.451
9. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi:10.1111/febs.14132
10. Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi:10.1016/j.cca.2017.10.011
11. Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32(4):e22333. doi:10.1002/jcla.2018.32.issue-4
12. Fu L, Wu S, Yao T, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018;32(2). doi:10.1002/jcla.22239.
13. Li WH, Song YC, Zhang H, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. doi:10.1155/2017/4587698
14. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi:10.3233/CBM-150552
15. Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025.
16. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi:10.1002/1878-0261.12045
17. Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi:10.2147/OTT.S147378
18. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96. doi:10.1007/s00109-017-1600-y
19. Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95(22):e3811. doi:10.1097/MD.0000000000004864
20. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi:10.1016/j.cca.2015.02.018
21. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi:10.1016/j.cca.2017.01.025
22. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi:10.3748/wjg.v23.i34.6330
23. Lu R, Shao Y, Ye G, Xiao B, Guo J. Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(6):1010428317704175. doi:10.1177/1010428317704175
24. Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. doi:10.3233/CBM-170379
25. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3):e22281. doi:10.1002/jcla.2018.32.issue-3
26. Lu J, Zhang PY, Xie JW. et al. Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal;2018:e22726. doi:10.1002/jcla.22726
27. Zhang C, Zhang C, Lin J, Wang H. Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(1):290–300. doi:10.1159/000495230
28. Shao Y, Chen L, Lu R, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4):1010428317699125. doi:10.1177/1010428317699125
29. Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379. doi:10.1002/jcla.2018.32.issue-5
30. Zhao SY, Wang J, Ouyang SB, Huang ZK, Liao L. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell Physiol Biochem. 2018;47(6):2511–2521. doi:10.1159/000491624
31. Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8(35):58405–58416. doi:10.18632/oncotarget.16881
32. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6(6):1167–1176.
33. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi:10.1038/srep34985
34. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
35. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014
36. Tan S, Gou Q, Pu W, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Research. 2018;28(6):693–695. doi:10.1038/s41422-018-0033-7
37. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research. 2015;25(8):981–984. doi:10.1038/cr.2015.82
38. Li Y, Zeng X, He J, et al. Circular RNA as a biomarker for cancer: A systematic meta-analysis. Oncol Lett. 2018;16(3):4078–4084. doi:10.3892/ol.2018.9125
39. Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18(1):303. doi:10.1186/s12885-018-4242-8
40. Wu J, Li G, Wang Z, et al. Circulating MicroRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers. 2015;2015:435656. doi:10.1155/2015/105358
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
