Back to Journals » International Journal of Nanomedicine » Volume 11
Diagnosis of prostate cancer using anti-PSMA aptamer A10-3.2-oriented lipid nanobubbles
Authors Fan X , Guo Y, Wang L , Xiong X, Zhu L , Fang K
Received 17 May 2016
Accepted for publication 11 July 2016
Published 12 August 2016 Volume 2016:11 Pages 3939—3950
DOI https://doi.org/10.2147/IJN.S112951
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Xiaozhou Fan,1 Yanli Guo,1 Luofu Wang,2 Xingyu Xiong,1 Lianhua Zhu,1 Kejing Fang1
1Department of Ultrasound, Southwest Hospital, Third Military Medical University, Chongqing, People’s Republic of China; 2Department of Urology, Daping Hospital, Institute of Surgery Research, Third Military Medical University, Chongqing, People’s Republic of China
Abstract: In this study, the lipid targeted nanobubble carrying the A10-3.2 aptamer against prostate specific membrane antigen was fabricated, and its effect in the ultrasound imaging of prostate cancer was investigated. Materials including 2-dipalmitoyl-sn-glycero-3-phosphocholine, 1,2-dipalmitoyl-sn-glycero-3-phosphatidic acid, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine, 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol, carboxyl-modified 1,2-distearoyl-sn-glycero-3-phosphoethanolamine, and polyethyleneglycol-2000 were for mechanical oscillation, and nanobubbles were obtained through the centrifugal flotation method. After mice were injected with nanobubbles, abdominal color Doppler blood flow imaging significantly improved. Through left ventricular perfusion with normal saline to empty the circulating nanobubbles, nanobubbles still existed in tumor tissue sections, which demonstrated that nanobubbles could enter tissue spaces via the permeability and retention effect. Fluorinated A10-3.2 aptamers obtained by chemical synthesis had good specificity for PSMA-positive cells, and were linked with carboxyl-modified 1,2-distearoyl-sn-glycero-3-phosphoethanolamine lipid molecules from the outer shell of nanobubbles via amide reaction to construct targeted nanobubbles. Gel electrophoresis and immunofluorescence confirmed that targeted nanobubbles were fabricated successfully. Next, targeted nanobubbles could bind with PSMA-positive cells (C4-2 cells), while not with PSMA-negative cells (PC-3 cells), using in vitro binding experiments and flow cytometry at the cellular level. Finally, C4-2 and PC-3 xenografts in mice were used to observe changes in parameters of targeted and non-targeted nanobubbles in the contrast-enhanced ultrasound mode, and the distribution of Cy5.5-labeled targeted nanobubbles in fluorescent imaging of live small animals. Comparison of ultrasound indicators between targeted and non-targeted nanobubbles in C4-2 xenografts showed that they had similar peak times (P>0.05), while the peak intensity, half time of peak intensity, and area under the curve of ½ peak intensity were significantly different (P<0.05). In PC-3 xenografts, there were no differences in these four indicators. Fluorescent imaging indicated that targeted nanobubbles had an aggregation ability in C4-2 xenograft tumors. In conclusion, targeted nanobubbles carrying the anti-PSMA A10-3.2 aptamer have a targeted imaging effect in prostate cancer.
Keywords: molecular imaging, contrast-enhanced ultrasound, aptamer, prostate specific membrane antigen, targeted imaging
Introduction
With the in-depth understanding of ultrasound contrast agents (UCAs) and the development of material science, the outer shell of UCAs can be constructed of materials such as lipid components, denatured albumin (ALB), and polymers based on different needs.1 Of all, the lipid outer shell is the most classic and most appropriate component of diagnostic contrast agents, due to the common features of excellent elasticity, easy modification of lipid molecules, and non-toxicity to humans.2 Based on these traits, groups in lipid molecules are easily linked with ligands specific to molecules of interest, such as compounds with small molecular weight and antibodies. In this way, outer shell components of targeted lipid UCAs were fabricated, which is also a key part in ultrasound molecular imaging.3 However, current studies about ultrasound molecular imaging are mainly applied in targeted ultrasound diagnosis of thrombosis, inflammation, and tumors.4–7 The newly emerging lipid ultrasound nanobubble (NB) is a UCA with a particle size smaller than 1,000 nm, which can overcome the limitations of traditional ultrasound microbubbles exclusively in blood vessels.8 Nano-scale UCAs in tumor tissues can utilize the permeability and retention effects of tumors, such as larger endothelial gaps, lack of a basement membrane, and poor lymph drainage in tumors, to enter extravascular spaces to achieve imaging of tumor parenchyma.9,10
PSMA is a membrane protein specifically expressed by prostate cancer cells. Its expression has a positive correlation with lethality, for example, it is highly expressed in metastatic prostate cancer and androgen-independent prostate cancer.11 Many ligands against PSMA have been developed, such as monoclonal antibodies, engineered antibodies, and small-sized substances including nanobodies in our previous studies.12,13 The aptamer is a nucleotide or deoxynucleotide sequence that can bind to other substances. Of all, RNA aptamers composed of nucleotides have advantages of diverse structures and strong specificity. Moreover, fluorinated aptamers can resist nucleases in the human body and be easily linked with therapeutic gene fragments. Therefore, aptamers have been applied extensively in studies on molecular imaging and gene therapy.14–18 Simultaneously, the aptamer as an element of the targeted probe can avoid immunogenicity and its high purity can also guarantee its non-toxicity and safety compared with other ligands.19–21 Early aptamers including A10 and A9 were screened from the synthesized RNA aptamer library, and can both effectively bind to PSMA.22,23 However, they both consist of 79 nucleotides, which leads to a very high molecular weight and severely restricts their role as targeting molecules. Researchers have continued to shorten their sequences in their subsequent experiments and the A10-3 aptamer molecule with only 59 nucleotides was developed. A10-3 was further shortened to obtain the A10-3.2 aptamer with only 37 nucleotides. Its molecular weight was significantly reduced, but it still has very good binding affinity to PSMA.24,25
On the basis of previous studies on NBs carrying anti-PSMA monoclonal antibodies and nanobodies, this study aimed to couple the A10-3.2 aptamer as a ligand with lipid NBs and to study its imaging effect in prostate cancer. The purpose was not only to provide ultrasound molecular probes with better safety, small particle size, strong penetration and specificity for prostate cancer, but also to provide methods for relevant studies on targeted ultrasound NBs carrying aptamers.
Materials and methods
Preparation of NBs
The lipid components, including 2-dipalmitoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Alabaster, AL, USA), 1,2-dipalmitoyl-sn-glycero-3-phosphatidic acid (Corden Pharma, Liestal, Switzerland), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (Corden Pharma), 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (Corden Pharma), polyethyleneglycol-2000 (PEG-2000) and carboxyl-modified 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE-PEG2000-COOH; NANOCS, Boston, MA, USA), were dissolved at a certain ratio in a hydration solution containing glycerin:phosphate-buffered saline (PBS) at 1:9 (v/v) and incubated overnight on a horizontal shaker at 37°C. The solution was transferred to a penicillin bottle the next day, and the air in the head of the penicillin bottle was replaced with perfluoropropane (C3F8) (Research Institute of Physical and Chemical Engineering of Nuclear Industry, Tianjin, People’s Republic of China). The bottle was shaken using the horizontal reciprocating mechanical oscillation method with an operating frequency greater than 4,500 rpm for 80 seconds using an ST-B series amalgamator (AT&M Biomaterials Co., Beijing, People’s Republic of China). The obtained suspension was sequentially centrifuged at 300× g and 300 rpm for 3 minutes each to discard the lipid materials that were not encapsulated into NBs and microbubbles in the upper layer. During the preparation of NBs carrying DiI dye (Beyotime Institute of Biotechnology, Shanghai, People’s Republic of China), a certain amount of DiI staining material was added into the hydration solution. The other steps were the same as the preparation of NBs.
Construction of in vitro cell culture and tumor xenograft animal models
PC-3 prostate cancer cells from the American Type Culture Collection (ATCC, Manassas, VA, USA) and C4-2 prostate cancer cells from ViroMed Laboratories at Johns Hopkins were cultured in RPMI 1640 culture medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in a 5% CO2 and 37°C incubator. During subcutaneous implantation of animal xenograft tumors, 200 μL of tumor cells at the logarithmic growth period at the concentration of 2.5×107/mL and 200 μL of BD Matrigel (BD Biosciences, San Jose, CA, USA) were mixed and subcutaneously injected into 4–5-week-old BALB/c-nu nude mice (HFK Bioscience Co., Beijing, People’s Republic of China). Animals were continuously housed in a clean environment. Animal experiments involved in this study were all approved by the Animal Ethics Committee of the Third Military Medical University, and performed in accordance with the International Guiding Principles for Biomedical Research involving Animals-1985.
Imaging and penetration ability of NBs in nude mice
After mice were anesthetized and immobilized, the 50 mm broadband array probe L12-5 was used in the iu22 ultrasound diagnosis apparatus (Royal Dutch Philips Electronics Ltd., Amsterdam, the Netherlands). The abdominal blood flow condition of mice was observed under color Doppler blood flow imaging (CDFI). Next, 2×106 NBs from the above procedure were injected through the retro-orbital venous sinus. After observing contrast-enhanced ultrasound (CEUS) of abdominal organs, CDFI was performed again to observe abdominal blood flow in the same mice.
Two million NBs encapsulated with DiI dye were injected into nude mice bearing a PC-3 prostate cancer xenograft, and real-time CEUS acquisition was performed in the tumor regions. After the imaging effect was stabilized, normal saline was applied for perfusion through the left ventricle of nude mice to empty ultrasound NBs into blood vessels. Next, xenograft tumor tissues and heart tissues were isolated for frozen sections. Some sections were used for hematoxylin-eosin (HE) staining to observe the distribution of NBs under a light microscope, and others were stained with 4′,6-diamidino-2-phenylindole to stain cell nuclei, and then used to observe the distribution of DiI NBs under a Zeiss 780 laser scanning confocal microscope (LSCM) (Carl Zeiss Meditec AG, Jena, Germany).
Synthesis and the specificity of the A10-3.2 aptamer
The A10-3.2 aptamer against PSMA and the aptamer modified with infrared dye Cy5.5 at the 5′ terminus, 5′-Cy5.5-GGGAGGACGAUGCGGAUCAGCCAUGUUUACGUCACUCCU-spacer-NH2-3′ with 2′-fluoro pyrimidines (RiboBio Co. Ltd, Guangzhou, People’s Republic of China), were chemically synthesized.25 The co-localization of anti-PMSA monoclonal antibodies (Abcam, Cambridge, MA, USA) and Cy5.5-labeled aptamer molecules in C4-2 cells and PC-3 cells was detected under the LSCM. In addition, Cy5.5-labeled aptamer molecules were incubated with C4-2 cells or PC-3 cells at 4°C for 30 minutes. After several washes with PBS, single-cell fluorescence detection was performed using a FACSCalibur flow cytometer (BD Biosciences) to measure the binding capacity between A10-3.2 aptamer and two types of cells.
Preparation and validation of targeted NBs
On the basis of obtaining the above NBs containing the DSPE-PEG2000-COOH lipid molecule, 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC; Solarbio, Beijing, People’s Republic of China) and 0.1 M N-hydroxysuccinimide (NHS; Solarbio) were added at the ratio of 1:1 (v/v) and reacted for 15 minutes on a shaker. The synthesized A10-3.2 aptamer molecules at 10 mM were then added, and the reaction was carried out at room temperature for 30 minutes. After the coupling reaction was finished, the blocking solution was added, and targeted NBs were obtained using the centrifugal flotation method. The NBs without the addition of EDC and NHS for reaction were used as non-targeted NBs. Next, non-targeted NBs and targeted NBs were burst under the condition of 1 W/cm2 ultrasonic power. The A10-3.2 aptamer molecules and the resulting components from both non-targeted and targeted NBs were used as three samples and subjected to 3% agarose electrophoresis to observe their band locations. In addition, the Cy5.5-labeled aptamers were constructed onto the surface of NBs using the same method. The conditions of fluorescence carried by these two types of NBs were observed and compared under an LSCM to determine again whether targeted NBs were successfully constructed.
A Malvern Zetasizer Nano ZS90 detector (Malvern Instruments, Malvern, UK) was used to measure the particle size and the dispersion degree of non-targeted NBs and targeted NBs according to the principle of dynamic light scattering. The signal intensity under CEUS (mechanic index of 0.12) of non-targeted NBs and targeted NBs at four different concentrations (1.0×108/mL, 5.0×107/mL, 2.5×107/mL, and 5.0×106/mL) was compared in an in vitro agarose model. In addition, red blood cells from Sprague Dawley rats were collected, and then targeted NBs at three different concentrations (1.0×107/mL, 5.0×107/mL, and 1.0×108/mL) were added into red blood cells at the 2% volume fraction and co-incubated for 30 minutes. The hemolysis condition was observed under naked eyes and a light microscope using double-distilled water as the positive control and normal saline as the negative control.
Validation of the targeting capacity of NBs in vitro at the cell level
After coverslips (1×1 cm) were placed in a 24-well plate, C4-2 or PC-3 cells at the logarithmic growth phase were added. Then cells were fixed in 4% paraformaldehyde and blocked in phosphate buffer with 3% bovine serum albumin after overnight incubation. These two types of cells were incubated with targeted NBs or non-targeted NBs at 4°C for 1 hour. After washing with PBS, the binding between NBs and cells was observed under a light microscope. In addition, the Cy5.5-labeled targeted NBs prepared previously were incubated with either C4-2 cells or PC-3 cells on ice for 1 hour. After washing with PBS, the binding efficiencies between these two types of cells and targeted NBs were detected using a flow cytometer.
Ultrasound imaging and fluorescence imaging of live small animals in tumor-bearing mice
When the volume of the subcutaneous tumor in tumor-bearing nude mice reached approximately 1 cm3, five C4-2 tumor-bearing mice and five PC-3 tumor-bearing nude mice were randomly collected and were anesthetized by intraperitoneal injection of 1% pentobarbital sodium. After the superficial probe L12-5 under the B-type ultrasound in the iu22 ultrasound diagnosis apparatus was used to search for the maximum section of xenograft tumors, the probe was then fixed. Next, the imaging mode was switched to “contrast/tissue” hybrid imaging under CEUS mode of the superficial organ. The mechanic index was adjusted to 0.12, and the focus center was located at the center of the xenograft tumor. Real-time collection was initiated before injection. Injection of 200 μL of targeted NBs or non-targeted NBs at the concentration of 5×107/mL (the total amount of injected NBs was 1×107/mL) was performed in the same nude mouse through the retro-orbital venous sinus. The interval between completion of ultrasound imaging of one type of NB and the initial injection of another type of NB was longer than half an hour. During this period, the built-in manual flash technology in the apparatus was used to destruct residual NBs in the body to ensure that the imaging effects of these two types of NBs would not interfere with each other. The time-intensity curve of the sectional area of the xenograft tumor was acquired using QLab8.1 offline software (Philips, Amsterdam, the Netherlands). The four ultrasound indicators (including peak time, peak intensity, half time, and area under the curve of ½ peak intensity) were extracted to compare imaging conditions of targeted NBs and non-targeted NBs in PSMA-positive C4-2 and PMSA-negative PC-3 xenograft tumors.
Cy5.5-labeled targeted NBs (1×108) were injected into the above two different types of tumor-bearing nude mice. The changes in fluorescence intensity in PC-3 xenograft tumors and C4-2 xenograft tumors at different time points (0 seconds, 5 seconds, 10 seconds, 30 seconds, 60 seconds, and 300 seconds) were acquired using the surface reflection fluorescence imaging in the IVIS Spectrum in vivo imaging system (PerkinElmer Inc., Waltham, MA, USA).
Detection of PSMA expression in xenograft tumors
PC-3 and C4-2 xenograft tissues were collected, fixed in paraformaldehyde, embedded in paraffin, and sectioned using a microtome. The immunohistochemistry procedure for tissue sections was then performed. After deparaffinization, sections were blocked in 3% H2O2 for at least 10 minutes. Antigen retrieval was performed in citric acid at high temperature and high pressure. Sections were incubated with a rabbit anti-human PSMA monoclonal antibody (Abcam) (1:400 dilution) at 4°C overnight. Next, sections were incubated with a horseradish peroxidase-labeled goat anti-rabbit secondary antibody (Beyotime Institute of Biotechnology) at room temperature for 1 hour. In all the procedures, sections were washed with PBS after each step. Finally, the results were developed with the DAB reagent, and cell nuclei were counterstained using hematoxylin. Sections were mounted using neutral balsam and observed under a light microscope.
Statistical analysis
Histograms were generated using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis was performed using SPSS 22.0 (IBM Corporation, Armonk, NY, USA). The in vitro imaging effects of targeted NBs and non-targeted NBs were compared using the non-parametric Mann–Whitney U-test. The four ultrasound imaging indicators of the two NBs are presented as the mean ± standard deviation. All ultrasound parameters were compared and analyzed using the paired-sample Student’s t-test. P<0.05 indicated statistical significance.
Results
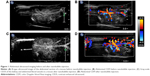
CDFI and permeability of NBs in mice
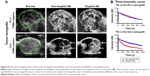
A total of 5×106 NBs were injected through the retro-orbital venous sinus, and abdominal CDFI in nude mice was acquired before and after NB injection (Figure 1). With the conventional B-type ultrasound, the liver and kidney structures of nude mice could be delineated; however, large blood vessels in the abdomen were not clear. The CDFI showed that ultrasound imaging clearly displayed the liver, kidney, and large blood vessels in the abdomen; however, small blood vessels with lower flow rates were not clear. Through the injection of NBs, CDFI clearly displayed the perfusion of blood in the kidney and the morphology of small blood vessels. In addition, blood vessel branches in the kidney, abdominal aorta, inferior vena cava, and iliac artery and vein branches were well delineated.
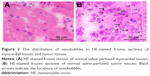
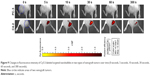
NBs encapsulated with DiI dye in blood vessels were then emptied through perfusion of normal saline in the left ventricle, and the distribution of NBs was observed in frozen sections of PC-3 xenograft tissues (Figure 2). In the HE-stained frozen sections, there were no NBs in myocardial tissues (Figure 2A), while tumor parenchymal cells were surrounded with many NBs (Figure 2B). LSCM showed that myocardial tissue spaces did not have DiI-labeled NBs (Figure 3A–C), while DiI-labeled NBs existed in tumor tissue spaces (Figure 3D–F). The above results all confirmed that NBs could enter tissue spaces through vascular endothelial gaps in tumors.
Validation of specificity of aptamers
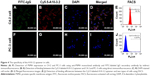
Two techniques, including immunocytochemistry and flow cytometry, both confirmed that chemically synthesized A10-3.2 aptamer could specifically bind to positive PSMA-expressing cells. In C4-2 cells, the anti-PSMA monoclonal antibody and Cy5.5-labeled A10-3.2 aptamer co-localized on the cell membrane of C4-2 cells (Figure 4A–D). Meanwhile, flow cytometry indicated that the number of labeled C4-2 cells was 80.37%±3.90% after co-incubation with aptamer molecules (Figure 4E). In contrast, PC-3 cells did not bind with anti-PSMA monoclonal antibody or A10-3.2 aptamer (Figure 4F–I), and the number of labeled PC-3 cells was only 0.21%±0.13% through flow cytometry (Figure 4J).
Identification and characteristics of targeted NBs
The key for preparation of targeted NBs is to determine whether NBs carry A10-3.2 aptamer molecules. Aptamer molecules and two lipid materials obtained from burst non-targeted NBs and targeted NBs were loaded for electrophoresis. The results showed that the location of targeted NB components was different from that of aptamer molecules, while there was no obvious band of non-targeted NBs. This result was consistent with our prediction, indicating that aptamer molecules had already coupled to lipid materials, and the band location of outer shell materials from targeted NBs had been changed (Figure 5A). In addition, observation under LSCM showed that the outer shells of targeted NBs contained the red fluorescence of Cy5.5 used for labeling the A10-3.2 aptamer molecules (Figure 5B and C), while the surface of non-targeted NBs without adding EDC and NHS did not carry the Cy5.5-modified A10-3.2 aptamer. These two methods both confirmed that the amide reaction could construct aptamer molecules onto the surface of NBs.
The particle sizes of NBs were measured with the Malvern instrument. The particle size of non-targeted NBs was 519.4±74.6 nm, and the polydispersity index was 0.105. The particle size of targeted NBs was 576.6±40.2 nm, and the polydispersity index was 0.180. They were evenly distributed under a light microscope and displayed the typical shell–core structure. Their imaging conditions at different concentrations under CEUS mode were observed in in vitro agarose models. The results showed that the imaging signals of two NBs at the same concentration were not significantly different (P=0.843, P>0.05) (Figure 5D and E).
The result of hemolysis experiments showed that targeted NBs did not have significant effects on the number or morphology of red blood cells, even though the concentration reached up to 1.0×108/mL. The appearance and number were similar to that of normal saline in the negative control, indicating that targeted NBs did not have an obvious hemolysis function (Figure 6A–C).
In vitro validation of binding between targeted NBs and cells
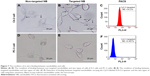
The binding of targeted and non-targeted NBs to the two types of cells was observed under a light microscope and assayed with flow cytometry. The microscopy showed that targeted NBs attached to the surroundings of PSMA-positive C4-2 cells, while non-targeted NBs predominantly did not attach to C4-2 cells (Figure 7A and B). At the same time, flow cytometry showed that after co-incubation with targeted NBs carrying Cy5.5 dye, the proportion of labeled C4-2 cells was 54.27%±4.20% (Figure 7C). In contrast, there was no obvious attachment of targeted NBs or non-targeted NBs to the surroundings of PSMA-negative PC-3 cells (Figure 7D and E), and the number of labeled PC-3 cells was only 0.50%±0.34% (Figure 7F). Although the black dots in the inner core of PC-3 cells seemed like NBs, they were just some refractive substances in cell nuclei, which could be clearly observed when the microscope was slightly adjusted. So far we have observed no lipid UCA internalization in cancer cells while some studies suggested UCAs could benefit the drug delivery or be internalized into macrophages.26,27
CEUS and the in vivo fluorescence imaging in the xenograft tumors
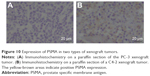
The time-intensity curves of targeted NBs and non-targeted NBs in the two types of prostate cancer xenografts were analyzed, and four parameters (the peak time, peak intensity, half time of peak intensity, and area under the curve of ½ peak intensity) were calculated. The results showed that the peak intensity, half time of peak intensity, and area under the curve of ½ peak intensity were significantly different between targeted NBs and non-targeted NBs (P<0.05), while the peak time did not have a significant difference (P>0.05) in the PSMA-positive C4-2 xenograft. However, there were no ultrasound parameters with a significant difference in the PSMA-negative PC-3 xenograft (P>0.05) (Figure 8 and Table 1). Fluorescence imaging of live small animals showed that Cy5.5-labeled targeted NBs aggregated in the C4-2 xenograft for a longer time, but there was no significant aggregation in the PC-3 xenograft. These results indicated that targeted NBs could specifically recognize tumors with PSMA expression (Figure 9). Finally, immunohistochemistry further confirmed that the C4-2 prostate cancer xenograft expressed PSMA, while the PC-3 tumor did not express PSMA (Figure 10).
Discussion
Prostate cancer is a malignant tumor, which severely threatens the health of elderly males. Traditional imaging technologies (ultrasound, computed tomography, and magnetic resonance imaging [MRI]) do not have very high sensitivity or specificity in the diagnosis of prostate cancer.28 Ultrasound imaging is the conventional method for the screening of prostate diseases, but it has higher missed diagnosis rates in smaller lesions. Although CEUS, in the recent decade, is used as an effective means to increase vascular imaging in tumors, it still lacks specificity in the diagnosis of prostatitis and prostate cancers. The emergence of molecular imaging brings hope for specific imaging of prostate cancer. Some ligands with specific targets can connect with a variety of imaging substances, such as nuclides, UCAs, quantum dots, and magnetic particles, to constitute targeted molecular probes. After intravenous injection, they can specifically bind to molecules expressed on human tissues and cells to achieve molecular imaging of lesioned tissues in vivo.29–31 PSMA has high levels of specific expression in both androgen-dependent prostate cancer and androgen-independent prostate cancer, and has become a commonly used target in the molecular imaging of prostate cancer. Currently, a variety of ligands against PSMA have been extensively applied in fluorescence imaging, MRI, and molecular nuclear medicine. For example, 111In-labeled anti-PSMA monoclonal antibodies have been used in clinical nuclear medicine imaging.32,33 In the ultrasound molecular imaging field, the reported molecular probes are mainly micron-scale UCAs targeting receptors on the vascular endothelia. The most successful of these is the BR55, which targets VEGFR and has been reported in tumor diagnosis and evaluations of anti-vascular therapy.34,35 In the ultrasound molecular imaging of tumor parenchyma, targeted ultrasound microbubbles at the micron scale cannot penetrate tumor blood vessels to enter tumor tissues; therefore, there are fewer relevant studies on the direct specific binding between ultrasound microbubbles and tumor cells to achieve targeted imaging.
To overcome the limitations of the above ultrasound molecular imaging technologies, our previous studies constructed ultrasound NBs carrying anti-PSMA monoclonal antibodies and anti-PSMA nanobodies.12,13 We confirmed that these NBs both had the ability to penetrate tumor blood vessels to enter tumor parenchyma. Compared to non-targeted NBs, these two types of NBs both had a certain value in the targeted diagnosis of prostate cancer xenografts in animals. However, our previous studies also had the following shortcomings. 1) Using the biotin-avidin connection method is not applicable to clinical research. 2) Nanobodies screened from non-immune nanobody phage display libraries have lower affinity (dissociation constant KD =519 nM). The reported PSMA-targeting aptamers have higher affinity and are easy to synthesize and modify. Moreover, few studies about ultrasound lipid NBs to carry them for use as molecular probes were reported in ultrasound molecular imaging.12,13,36 Therefore, this study linked the anti-PSMA aptamer A10-3.2 with ultrasound NBs through the amide reaction, and further established its targeted diagnostic performance in PSMA-positive prostate cancer.
In this study, light microscopy and LSCM results of the NBs with a particle size of 519.4±74.6 nm showed that NBs could enter tissue spaces through vascular gaps in tumors but could not enter myocardial tissue spaces. This result confirmed that the NBs had the ability to penetrate tumor blood vessels to reach tumor parenchyma. This finding was consistent with results in other studies on the penetration ability of nano-scale UCAs, which provided a basis for the entry of targeted NBs carrying aptamers into tumor parenchyma, and thus allowing them to specifically target tumor cells.37,38 In our further study, we will seek a solution to observe the important dynamic process in which NBs enter tumor parenchyma through vascular gaps under live imaging with a fluorescence microscope. On the basis of specific binding between the synthesized A10-3.2 aptamers and PSMA-expressing C4-2 prostate cancer cells in our study, targeted NBs carrying A10-3.2 aptamers were subsequently constructed via the amide reaction successfully. The particle size of these targeted NBs was 576.6±40.2 nm. In vitro experiments in the cell level and in vivo experiments in the xenograft both confirmed that they had the ability to specifically bind to C4-2 cells and specifically aggregate in tumor tissues. Moreover, the results about imaging features showed that targeted NBs had higher peak intensity, longer half time of peak value, and larger area under the curve of ½ peak intensity than non-targeted NBs in C4-2 xenografts. Of all four indicators, the area under the curve reflected the duration that UCAs remained in target xenograft areas and the total amount of UCAs. These differences confirmed that NBs carrying PSMA aptamers bound to specific antigens on the surface of tumor cells, while non-targeted NBs did not specifically bind to target areas. In addition, fluorescence imaging of live small animals further indicated the specific binding of targeted NBs in tumor target areas. It is worth noting that there was no observed fluorescence signal in PC-3 xenograft tumors in small animals, which might have been caused by a combination of reasons, eg, that the blood flow was not rich in PC-3 xenograft tumors, tumors did not express PSMA targets, and the amount of Cy5.5 dye carried by targeted NBs was low.
Therefore, the successfully constructed targeted NBs carrying the A10-3.2 aptamer in this study had the following advantages. 1) The particle size was small. The mean particle diameter of targeted NBs in this study was 576.6 nm, smaller than the 644.3 nm of the NBs carrying monoclonal antibodies in our previous studies. Although the size of previously reported NBs carrying nanobodies was only about 487.6 nm, the screening process for nanobodies (with their strong specificity) was more complicated and difficult. 2) The penetration was strong. They could penetrate blood vessels in tumors to enter tissue spaces in tumors. 3) They had better safety profiles. Lipid substances were used as the outer shell, and fluorocarbon gas was used as the inner core. In addition, targeted NBs were connected using an amide reaction. Regarding material safety, they could be used clinically. 4) They had strong specificity. In vitro and in vivo experiments both confirmed that NBs carrying the anti-PSMA aptamer specifically bound to tumor cells and specifically aggregated in tumor tissues. Currently, there are still no in vivo reports on this type of targeted NB.39
We will continue in-depth studies on the following subjects and improve the effect of this targeted NB in experimental animals with the goal of clinical translation. 1) The PSMA-positive and -negative cells used with the same in vitro concentrations of the two types of NBs were used for imaging in the mice. Although this setting could avoid wrong conclusions to some extent, using mutant aptamer molecules as the nonsense control and using non-targeted NBs that carry mutant aptamer molecules as the negative control would be more convincing. 2) In order to diagnose prostate cancer in molecular imaging more sufficiently, it is necessary to seek a new target molecule in prostate cancer types with no or few PSMA. Furthermore, the potential of “PSMA-guided” ultrasound diagnostics in prostate cancer will also be faced with other possibilities of PSMA-specific imaging, such as positron emission tomography, MRI, and multimodal imaging. 3) Using small animal ultrasound diagnostic apparatuses with built-in CEUS for detection in small animal experiments can increase the resolution of images to obtain better imaging signals.
Conclusion
In this study, we combined our previous findings with the advantages of the small molecular weight and excellent specificity of aptamers, and then used the safe amide reaction connection method to construct targeted NBs carrying the anti-PSMA aptamer A10-3.2. These were safe and had excellent penetration ability, strong specificity, and potential for clinical translation.39,40 This study can not only provide a basis for ultrasound molecular imaging of prostate cancer, but can also provide methods for constructing targeted ultrasound probes carrying aptamers. In addition, because the aptamer is a gene fragment in itself, it can be intercalated with anthracyclines or can be connected to other gene fragments, which can lay the foundation for future construction of targeted NBs carrying drugs (or genes) in targeted therapy under ultrasound irradiation.41–44
Acknowledgments
This work was supported by International Science & Technology Cooperation Program of China (No 2015DFA30920), National Natural Science Foundation of China (No 81571732), and Science & Technology (International Science & Technology Cooperation) Research Base Construction Program of Chongqing (No cstc2014gjhz110004).
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
Paefgen V, Doleschel D, Kiessling F. Evolution of contrast agents for ultrasound imaging and ultrasound-mediated drug delivery. Front Pharmacol. 2015;6:197. | ||
Pu C, Chang S, Sun J, et al. Ultrasound-mediated destruction of LHRHa-targeted and paclitaxel-loaded lipid microbubbles for the treatment of intraperitoneal ovarian cancer xenografts. Mol Pharm. 2014;11(1):49–58. | ||
Freise AC, Wu AM. In vivo imaging with antibodies and engineered fragments. Mol Immunol. 2015;67(2 Pt A):142–152. | ||
Hoyt K, Warram JM, Wang D, Ratnayaka S, Traylor A, Agarwal A. Molecular Ultrasound Imaging of Tissue Inflammation Using an Animal Model of Acute Kidney Injury. Mol Imaging Biol. 2015;17(6):786–792. | ||
Alonso A, Artemis D, Hennerici MG. Molecular imaging of carotid plaque vulnerability. Cerebrovasc Dis. 2015;39(1):5–12. | ||
Anderson CR, Hu X, Zhang H, et al. Ultrasound molecular imaging of tumor angiogenesis with an integrin targeted microbubble contrast agent. Invest Radiol. 2011;46(4):215–224. | ||
Dixon AJ, Kilroy JP, Dhanaliwala AH, et al. Microbubble-mediated intravascular ultrasound imaging and drug delivery. IEEE Trans Ultrason Ferroelectr Freq Control. 2015;62(9):1674–1685. | ||
Wang D, Yang K, Gao YH, Tan KB, Liu Z. Preparation and characterization of a nanoscale ultrasound contrast agent. Clin Imaging. 2010;34(4):288–292. | ||
Xing Z, Wang J, Ke H, et al. The fabrication of novel nanobubble ultrasound contrast agent for potential tumor imaging. Nanotechnology. 2010;21(14):145607. | ||
Cai WB, Yang HL, Zhang J, et al. The Optimized Fabrication of Nanobubbles as Ultrasound Contrast Agents for Tumor Imaging. Sci Rep. 2015;5:13725. | ||
Ristau BT, O’Keefe DS, Bacich DJ. The prostate-specific membrane antigen: lessons and current clinical implications from 20 years of research. Urol Oncol. 2014;32(3):272–279. | ||
Wang L, Li L, Guo Y, et al. Construction and in vitro/in vivo targeting of PSMA-targeted nanoscale microbubbles in prostate cancer. Prostate. 2013;73(11):1147–1158. | ||
Fan X, Wang L, Guo Y, et al. Ultrasonic Nanobubbles Carrying Anti-PSMA Nanobody: Construction and Application in Prostate Cancer-Targeted Imaging. Plos One. 2015;10(6):e0127419. | ||
Pai SS, Ellington AD. Using RNA aptamers and the proximity ligation assay for the detection of cell surface antigens. Methods Mol Biol. 2009;504:385–398. | ||
McNamara JO 2nd, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–1015. | ||
Baek SE, Lee KH, Park YS, et al. RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J Control Release. 2014;196:234–242. | ||
Chu TC, Shieh F, Lavery LA, et al. Labeling tumor cells with fluorescent nanocrystal-aptamer bioconjugates. Biosens Bioelectron. 2006;21(10):1859–1866. | ||
Mathew A, Maekawa T, Sakthikumar D. Aptamers in targeted nanotherapy. Curr Top Med Chem. 2015;15(12):1102–1114. | ||
Dougherty CA, Cai W, Hong H. Applications of aptamers in targeted imaging: state of the art. Curr Top Med Chem. 2015;15(12):1138–1152. | ||
Alibolandi M, Ramezani M, Abnous K, et al. In vitro and in vivo evaluation of therapy targeting epithelial-cell adhesion-molecule aptamers for non-small cell lung cancer. J Control Release. 2015;209:88–100. | ||
Lee IH, An S, Yu MK, Kwon HK, Im SH, Jon S. Targeted chemoimmunotherapy using drug-loaded aptamer-dendrimer bioconjugates. J Control Release. 2011;155(3):435–441. | ||
Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–4033. | ||
Dassie JP, Hernandez LI, Thomas GS, et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther. 2014;22(11):1910–1922. | ||
Rockey WM, Hernandez FJ, Huang SY, et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21(5):299–314. | ||
Wu X, Ding B, Gao J, et al. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int J Nanomedicine. 2011;6:1747–1756. | ||
Kiessling F, Fokong S, Bzyl J, Lederle W, Palmowski M, Lammers T. Recent advances in molecular, multimodal and theranostic ultrasound imaging. Adv Drug Deliv Rev. 2014;72:15–27. | ||
Niu CC, Wang ZG, Zuo GQ, et al. Poly(Lactide-co-glycolide) ultrasonographic microbubbles carrying Sudan black for preoperative and intraoperative localization of lymph nodes. Clin Breast Cancer. 2012;12(3):199–206. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Wibmer AG, Burger IA, Sala E, Hricak H, Weber WA, Vargas HA. Molecular Imaging of Prostate Cancer. Radiographics. 2016;36(1):142–159. | ||
Gupte AA, Hamilton DJ. Molecular Imaging and Precision Medicine. Cardiology. 2016;133(3):178–180. | ||
Kang ST, Yeh CK. Ultrasound microbubble contrast agents for diagnostic and therapeutic applications: current status and future design. Chang Gung Med J. 2012;35(2):125–139. | ||
Shi C, Zhu Y, Xie Z, et al. Visualizing human prostate cancer cells in mouse skeleton using bioconjugated near-infrared fluorescent quantum dots. Urology. 2009;74(2):446–451. | ||
Purohit RS, Shinohara K, Meng MV, Carroll PR. Imaging clinically localized prostate cancer. Urol Clin North Am. 2003;30(2):279–293. | ||
Abou-Elkacem L, Bachawal SV, Willmann JK. Ultrasound molecular imaging: Moving toward clinical translation. Eur J Radiol. 2015;84(9):1685–1693. | ||
Pochon S, Tardy I, Bussat P, et al. BR55: a lipopeptide-based VEGFR2–targeted ultrasound contrast agent for molecular imaging of angiogenesis. Invest Radiol. 2010;45(2):89–95. | ||
Wang CH, Kang ST, Lee YH, Luo YL, Huang YF, Yeh CK. Aptamer-conjugated and drug-loaded acoustic droplets for ultrasound theranosis. Biomaterials. 2012;33(6):1939–1947. | ||
Yang H, Cai W, Xu L, et al. Nanobubble-Affibody: Novel ultrasound contrast agents for targeted molecular ultrasound imaging of tumor. Biomaterials. 2015;37:279–288. | ||
Yin T, Wang P, Zheng R, et al. Nanobubbles for enhanced ultrasound imaging of tumors. Int J Nanomedicine. 2012;7:895–904. | ||
Carson AR, McTiernan CF, Lavery L, et al. Gene therapy of carcinoma using ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2011;37(3):393–402. | ||
Kohler N, Sun C, Wang J, Zhang M. Methotrexate-modified superparamagnetic nanoparticles and their intracellular uptake into human cancer cells. Langmuir. 2005;21(19):8858–8864. | ||
Dassie JP, Liu XY, Thomas GS, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27(9):839–849. | ||
Xu W, Siddiqui IA, Nihal M, et al. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials. 2013;34(21):5244–5253. | ||
Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci U S A. 2011;108(5):1850–1855. | ||
Kim E, Jung Y, Choi H, et al. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials. 2010;31(16):4592–4599. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.