Back to Journals » Cancer Management and Research » Volume 11
Development and external validation of prognostic nomograms in hepatocellular carcinoma patients: a population based study
Authors Xiao Z , Yan Y , Zhou Q, Liu H, Huang P, Zhou Q, Lai C, Zhang J, Wang J, Mao K
Received 18 October 2018
Accepted for publication 25 February 2019
Published 10 April 2019 Volume 2019:11 Pages 2691—2708
DOI https://doi.org/10.2147/CMAR.S191287
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Kenan Onel
Zhiyu Xiao1,2,*, Yongcong Yan1,2,*, Qianlei Zhou1,2,*, Haohan Liu1,2, Pinbo Huang1,2, Qiming Zhou1,2, Changliang Lai1,2, Jianlong Zhang1,2, Jie Wang1,2, Kai Mao1,2
1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, People’s Republic of China; 2Department of Hepatobiliary Surgery, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, People’s Republic of China
*These authors contributed equally to this work
Background: We attempted to construct and validate novel nomograms to predict overall survival (OS) and cancer-specific survival (CSS) in patients with hepatocellular carcinoma (HCC).
Methods: Models were established using a discovery set (n=10,262) obtained from the Surveillance, Epidemiology, and End Results (SEER) database. Based on univariate and multivariate Cox regression analyses, we identified independent risk factors for OS and CSS. Concordance indexes (c-indexes) and calibration plots were used to evaluate model discrimination. The predictive accuracy and clinical values of the nomograms were measured by decision curve analysis (DCA).
Results: Our OS nomogram with a c-index of 0.753 (95% confidence interval (CI), 0.745–0.761) was based on age, sex, race, marital status, histological grade, TNM stage, tumor size, and surgery performed, and it performed better than TNM stage. Our CSS nomogram had a c-index of 0.748 (95% CI, 0.740–0.756). The calibration curves fit well. DCA showed that the two nomograms provided substantial clinical value. Internal validation produced c-indexes of 0.758 and 0.752 for OS and CSS, respectively, while external validation in the Sun Yat-sen Memorial Hospital (SYMH) cohort produced a c-indexes of 0.702 and 0.686 for OS and CSS, respectively.
Conclusions: We have developed nomograms that enable more accurate individualized predictions of OS and CSS to help doctors better formulate individual treatment and follow-up management strategies.
Keywords: surveillance, epidemiology and end results, overall survival, cancer-specific survival, decision curve analysis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the second most deadly cause of cancer mortality worldwide according to global cancer statistics obtained in 2012, and nearly half of the total number of cases and deaths occur in China.1–3 Improvements in treatment strategies have markedly improved the overall survival (OS) and cancer-specific survival (CSS) of HCC patients, although the long-term survival rate remains low. Many HCC patients die because of disease rather than of other causes. Factors related to prognostic predictions are based on the American Joint Committee on Cancer (AJCC) TNM staging system and the National Comprehensive Cancer Network (NCCN) guidelines.4 However, many factors not included in TNM staging may influence the survival of HCC patients, including patient background characteristics (ie, patient age, sex, race and geographical location), tumor-related factors (tumor size, invasion and histological grade) and treatment received (surgery performed).1,5–7
Nomograms are reliable statistical predictive models that are used to accurately calculate and predict individual survival by combining all risk factors for tumor development.8,9 An increasing number of nomograms are being widely established to provide assistance in formulating individual treatment and follow-up management strategies in several cancers, such as oropharyngeal cancer,10 gastrointestinal stromal tumors,11 adenoid cystic carcinoma,12 bladder cancer,13 and prostate cancer.14 In HCC, many nomograms have been constructed to predict recurrence-free survival and OS after liver resection,6,7,15,16 but these nomograms are all based on a single population or have been unvalidated in an external cohort. More importantly, few studies have focused on nomogram models specific to CSS in HCC patients. To the best of our knowledge, no study has been carried out to predict prognosis using data gathered from HCC patients in the Surveillance, Epidemiology, and End Results (SEER) database.
In the present study, we aimed to establish and validate the first effective and convenient HCC nomogram model to calculate and predict OS and CSS based on clinicopathological risk factors obtained from the SEER database. Moreover, these nomogram models were validated using both an internal SEER dataset and an independent external cohort obtained from our hospital. These nomograms can guide individual treatment and follow-up management in HCC patients.
Materials and methods
Patients and study design
We downloaded clinical data related to all patients under the liver heading (Site Recode ICD-O-3/WHO 2008) in the SEER 18 registry database (1973–2015) using SEER*Stat 8.3.5 software. The flow chart used for data selection is shown in Figure 1. “The International Classification of Diseases for Oncology (ICD-O-3) Hist/behav, malignant” was used to screen HCC cases. The “Year of diagnosis” ranged from 2004 to 2014. “Derived AJCC Stage Group 6th (2004+)”, “CS tumor size (2004+)”, “RX Summ-Surg Prim Site (1998+)”, and “Grading and differentiation codes in ICD-O-2” were used in the present study. “Vital status recode” and “SEER cause-specific death classification” were used to set endpoints for OS and CSS, respectively, while patient survival time was defined using “survival months code”. The inclusion criteria were as follows: diagnostic confirmation achieved based on microscopic analysis and patient background characteristics (ie, age, sex, race and marital status), tumor-related factors (tumor size, invasion and histological grade) and treatment received (surgery performed) were known and available. The exclusion criteria were as follows: death certificate or autopsy only and age <15 years. A total of 15,394 cases in the SEER cohort were included and analyzed in the present study. All HCC patients were randomly divided into a discovery set with N*q samples and an internal testing set with N*(1-q) samples (q=2/3). To further validate our results in a responsible manner, we sought an external testing cohort from Sun Yat-sen Memorial Hospital (SYMH) that included data obtained between January 1, 2009, and December 31, 2012. That dataset included 244 postoperative HCC patients (the SYMH cohort) who were recruited using the above inclusion and exclusion criteria. All diagnoses were confirmed by pathology. The data from the SEER Registry and the Sun Yat-sen Memorial Hospital were rendered anonymous.This retrospective study was reviewed and approved by the ethics committee of Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, and written informed consent was obtained from each patient. The procedure was conducted in accordance with the Declaration of Helsinki.
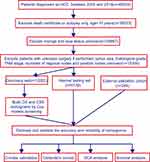 | Figure 1 Study flowchart.Abbreviations: HCC, hepatocellular carcinoma; OS, overall survival; CSS, cancer-specific survival; DCA, decision curve analysis. |
Nomogram construction
We validated that the clinicopathological features of the patients in the discovery and internal testing sets were well balanced (P>0.05) according to the chi-square test. We used a univariate Cox regression analysis to screen for clinicopathological risk factors for OS and CSS in the SEER discovery set. We further performed multivariate Cox regression analysis to screen for independent important factors for HCC patients without violating the PH assumption. All variables were screened using the forward stepwise selection method in a Cox multivariate analysis regression model.17,18 Based on the identified independent important factors, we constructed two nomograms to predict OS and CSS at 1, 3 and 5 years using R software.
Nomogram validation
We calculated concordance indexes (c-indexes) and drew calibration plots for the internal and external validating cohorts, respectively. The c-index quantified the discrimination between two random patients, with a c-index of 0.5 indicating no discrimination and 1 indicating perfect discrimination.19 Calibration plots were constructed to validate the accuracy and reliability of the nomograms for OS and CSS by comparing the nomogram-predicted and actual survival rates determined in a Kaplan-Meier analysis with 1000 bootstrap samples.20
Clinical application value assessment
Decision curve analysis (DCA) was performed to identify and compare the clinical application value between the nomogram model and other clinical features by calculating the net benefits at each risk threshold probability.21,22 The net benefit was determined by subtracting the proportion of all false-positive results from the proportion of true-positive results and weighted by the relative harm caused by giving up treatment compared with the negative consequences of unnecessary treatments.23 Based on the DCA, we further plotted curves to evaluate the clinical impact of the nomogram to help us more intuitively understand its significant value. Accordingly, we evaluated the number of high-risk patients and the number of high-risk patients with outcomes for OS and CSS at different threshold probabilities in a given population.24
Statistical analysis
The chi-square test was used to compare categorical variables. OS was defined as the time from diagnosis to the time of death from any cause or the most recent follow-up, and CSS was calculated from the date of diagnosis to the date of HCC-related death or the most recent follow-up. Kaplan-Meier survival curves were used to compare OS and CSS among different groups, and survival differences were assessed by a two-sided log-rank test in R version 3.3.4 (
Results
Study flowchart and basic clinical characteristics
The study flowchart is presented in Figure 1. A total of 15,394 HCC patients were included in the study and randomly divided into a discovery set (n=10,262) and an internal testing set (n=5,132). Patient clinicopathological features in the discovery and internal testing sets are shown in Table S1. There were no significant differences between the two sets (P>0.05, Table S1). The detailed clinical characteristics of patients in the SYMH cohort (n=244) are shown in Table S1. The median ages (interquartile range) of the patients in the SEER discovery set, the internal testing set, the entire SEER cohort and the SYMH cohort were 63 (56–72), 63 (56–72), 63 (56–72) and 52 (42–59) years, respectively, and the median OS durations were 630, 600, 630 and 1448 days, respectively. The 1-, 3- and 5-year OS rates were 63.99%, 40.91%, and 30.78% and 80.10%, 58.36%, and 45.19% in the SEER and the SYMH cohorts, respectively. The median CSS durations in the above four sets were 1,230, 1,200, 1,200 and 1,701 days, respectively; while the 1-, 3- and 5-year CSS rates were 73.12%, 54.25%, and 45.42% in the SEER cohort. OS and CSS curves are plotted in Figure S1.
 | Table 1 The c-index for the nomogram to predict OS and CSS |
Independent significant factors in the discovery set
To further identify the candidate predictors of OS and CSS, we evaluated all clinicopathological features by Cox proportional hazards regression analysis. Univariate analysis of the Cox regression was performed in 10,262 patients in the discovery set. All clinicopathological features, including age, sex, marital status, race, histological grade, tumor size, surgery performed, and TNM stage, affected OS (Table S2). Multivariate regression analysis was performed on the 8 factors that were shown to significantly affect OS, and all 8 factors were independent prognostic predictors of OS (Figure 2). Similarly, the univariate analysis showed that all evaluated clinical indexes were associated with CSS. In the multivariate analysis, sex, marital status, histological grade, tumor size, surgery performed, and TNM stage were the 6 independent significant factors that predicted CSS (Figure 2). OS and CSS curves stratified by these independent prognostic factors showed significant differences among the groups, as shown in Figures S2 and S3.
Prognostic nomograms for OS and CSS
Based on the independent prognostic factors identified in the multivariate Cox regression analysis, two nomograms were developed to predict 1-, 3-, and 5-year OS (Figure 3A) and CSS (Figure 3B) in HCC patients. The point assignments and prognostic scores for every variable involved in the score models are shown in Table S3.
Performance of the nomograms in the discovery set
All indexes in the nomograms and other single factors in all cohorts are listed in Table 1. The c-index for the OS prediction nomogram was 0.753 (95% CI, 0.745–0.761) in the discovery set, while the c-indexes for TNM stage, histologic grade and tumor size for OS prediction were 0.555 (95% CI, 0.547–0.563), 0.654 (95% CI, 0.646–0.662), and 0.618 (95% CI, 0.612–0.624), respectively, and were much lower than those of the nomogram model. Similarly, the c-index for the CSS prediction nomogram was higher than the c-indexes for TNM stage, grade and tumor size. The calibration plots for the probability of 1-, 3- and 5-year OS demonstrated good concordance between the nomogram prediction and actual observations in the discovery set (Figure 4A); similar results were found for the CSS nomogram (Figure 4B). The discriminatory ability of the OS nomogram was further assessed in a survival analysis. All patients were divided into three groups according to optimal cutoff values determined by X-tile software (for OS, low risk: <133, intermediate risk: 133–214, and high risk: >214; for CSS, low risk: <118, intermediate risk: 118–206, and high risk: >206) according to the nomogram predictions. Kaplan-Meier curves for OS and CSS were plotted for the entire SEER cohort (Figure 5A and B). Based on risk stratification, in the low-, intermediate- and high-risk subgroups, the 1-year OS rates were 85.59%, 52.78%, and 20.57%, the 3-year OS rates were 63.72%, 22.64%, and 4.38%, the 1-year CSS rates were 90.69%, 59.95%, and 23.39%, and the 3 year CSS rates were 75.23%, 31.18%, and 6.88%, respectively.
Validation of the OS and CSS nomograms
The c-index for the OS prediction nomogram was 0.758 (95% CI, 0.746–0.770) in the SEER testing set, which was higher than the c-indexes for the TNM stage, grade and tumor size (Table 1). The c-index for CSS prediction was 0.752 (95% CI, 0.740–0.764) in the SEER testing set, which was higher than the c-index for any of the three single factors (Table 1). Likewise, the c-index for the OS prediction nomogram was 0.702 (95% CI, 0.647–0.757) in the SYMH cohort, which was higher than the c-indexes for TNM stage, grade and tumor size (Table 1). The curves for 1- and 3-year OS and CSS were generally well calibrated in the SEER testing set (Figure 4C and D) and the SYMH cohort (Figure 4E and F). Kaplan-Meier curves for OS and CSS in the low-, intermediate- and high-risk subgroups were plotted for the SYMH cohort (Figure 5C and D), and the results showed that the high-risk subgroups had the worst OS and CSS in the validation cohort.
Assessment of the value of the nomograms as clinical applications
The DCA results of the nomograms and other indexes (ie, histological grade, TNM stage and tumor size) are presented in Figure 5. The results showed that the nomogram indicated a better net benefit after 1 and 3 years than that achieved for the other indexes for predicting OS (Figure 5E and F) and CSS (Figure 5H and I) in the discovery set. Based on these results, we plotted additional curves for the clinical impact of the nomograms to help us more intuitively evaluate its significance. The OS nomogram showed that cost/benefit ratios were lower when the risk threshold was <0.7 (Figure 5G), while the CSS nomogram showed that cost/benefit ratios were lower when the risk threshold was <0.6 (Figure 5J) in the discovery set. Therefore, based on the DCA, the nomograms provided more net benefits for predicting OS (Figure S4A and B) and CSS (Figure S4C and D) in the testing set. Similarly, the nomograms had a stronger clinical impact in all validation sets including the SEER database and SYMH cohorts (Figure S4E and H).
Discussion
HCC is one of the most common cancers and the leading cause of cancer-related deaths worldwide, accounting for more than half of all cases and deaths in China.3,25 Thus, more precise treatment guidelines and follow-up management strategies are urgently needed for HCC patients. Nomogram models are statistical tools that can meet these requirements. Shim et al established and validated predictive nomograms with c-indicex of 0.69 based on data obtained from 1,085 patients with early-stage HCC who underwent curative resection in Asan Liver Center.7 He et al developed and validated a nomogram that predicted survival after recurrence in HCC patients with a high c-index of 0.797.15 However, previous studies on OS and CSS in HCC patients have been limited by a number of factors, such as single-center datasets, small sample sizes, and a lack of external validation.
In the present study, we identified several conventional factors including age, sex, race, marital status, histological grade, TNM stage, tumor size, and surgery performed that significantly affected OS. Histological grade (undifferentiated) (HR=1.920, 95% CI=1.608–2.292, P<0.001) and TNM stage (stage IV) (HR=1.920, 95% CI=1.804–2.378, P<0.001) resulted in higher HRs than did other variables in the multivariate regression analysis. TNM stage has been regarded as the most important factor for predicting OS,4 and the staging system has been considered valuable for predicting survival in patients after liver transplantation.4,26 Interestingly, we found that being married was associated with a better prognosis of HCC (HR=0.9 and 0.822 relative to the OS and CSS of unmarried patients, respectively), which is consistent with the results of many other studies.27,28 Married patients with small intestinal adenocarcinoma have better OS and CSS than unmarried patients. Psychological and economic support from spouses may contribute to improvements in survival in married patients.29 A novel nomogram model that included age, tumor size, margin status and vascular invasion and alpha-fetoprotein (AFP) levels performed well in predicting prognosis in HCC patients after liver resection.30 Another interesting study found that a tumor size greater than 2 cm, multifocal tumors, and vascular invasion were three independent predictors of poor survival in patients with early-stage HCC after surgery.31 However, the effectiveness and accuracy of our OS prediction nomogram was better than that achieved using TNM stages (c-index, 0.753 vs 0.555) in the SEER discovery set. Additionally, the results obtained using our nomograms are more reasonable than those obtained used the universally acknowledged TNM stage and provide a prognostic estimate that can predict individual results. For example, imagine two HCC patients: X and Y. They have the same sex (male; 9 points) and TNM stage (II, 13 points), but different ages and histological grades, with one patients being 45 years old (0 point) and having a well-differentiated tumor (0 point) and the other being 75 years old (26 points) and having an undifferentiated tumor (56 points). However, both patients are stratified into stage II disease based on the TNM staging system, which is associated with specific outcomes. As has been widely acknowledged, these two patients will probably have different prognoses, but the question regarding how to quantify these prognoses remains unresolved. Calculating the total scores of these patients individually is simple according to our OS prediction nomogram: patient X has 22 points, while patient Y has 104 points. The OS nomogram indicates that the 1-, 3-, and 5-year OS probabilities for patient X are nearly 88.5%, 77% and 68%, respectively, while the probabilities are nearly 75%, 49.5% and 35%, respectively, for patient Y. More interestingly, for the CSS nomogram, we identified six clinical factors, namely, sex, marital status, histological grade, TNM stage, tumor size and surgery performed, but not patient age or race, as significant independent prognostic predictors. In contrast, in the OS prediction nomogram, the magnitude of a poor prognosis was found to increase with age, given the same histological grade and TNM stage.
Most of the predictive factors included in our models were the same as those included in other well-accepted models (ie, age, sex, marital status, histological grade, and TNM stage),30,31 but our data included a larger and worldwide sample, which allowed us to estimate the contribution of additional factors and to build independent nomogram models for OS and CSS at cutoff points of 1, 3 and 5 years. However, neither risk stratification nor discrimination could be used associate the clinical consequences of fixed discrimination or calibration. Therefore, using DCA, we evaluated whether nomogram-based medical decisions and strategies could improve patient prognoses to show the clinical value of the nomograms,23,24 even though many other clinical predictive models ignore this factor. We showed that our nomograms add more benefit when used to predict OS and CSS.
Huang et al revealed EXT2, ETV5, and CHODL as independent prognostic factors of HCC by univariate and multivariate Cox analyses from a public database and established a novel nomogram by integrating the three molecular proteins and the TNM staging system, which displayed good performance in predicting long-term prognosis in HCC patients. They focused on the correlation between the expression of molecular markers and patient prognosis. However, we used a large population from the international SEER database to avoid heterogeneity among different medical centers and used the variables that are available and easily obtainable in clinical practice to construct our nomogram models.32 Adhoute et al found that the Barcelona Clinic Liver Cancer (BCLC) nomogram and the NIACE score provided the best prognostic information, but the NIACE could even help treatment strategies because of the low Akaike’s information criterion (AIC) value in a large French HCC cohort. Interestingly, they focused on several current common staging systems and compared their prognostic prediction abilities. In particular, the authors reassigned detailed and accurate scores of risk factors in the BCLC staging system using the nomogram model, whereas our study mainly focused on mining and building a new prognostic staging system based on clinical factors from a worldwide cohort.33
The present study has several merits that should be noted. First, a large population from the international SEER database was used to avoid heterogeneity among different medical centers. The results were also externally validated using the SYMH cohort. Second, the variables included in the nomogram models are available and easily obtainable in routine clinical practice. Third, using DCA, we plotted curves for the clinical impact of the nomograms, and this helped us to more intuitively understand the significant value of the nomograms in a clinical setting. Nevertheless, several potential limitations should also be noted. First, this is a retrospective study. Second, we only analyzed common prognostic factors, while blood and inflammation indicators were not used in multivariable survival analyses. Third, there is a possibility that residual confounding occurred after internal validation as a consequence of overfitting from variables and selection of threshold. However, we performed bootstrapping during internal testing and external validation. Fourth, the BCLC stages were not recorded in the SEER database; thus, we could not compare different outcomes from our nomogram model with those using the BCLC staging system. In the future, we will optimize the model by combining prognostic factors in our study with chronic liver disease condition, other interventions and the severity of liver dysfunction in HCC patients from multicenter cohorts in China.
Conclusions
In conclusion, based on the clinical risk factors identified in a large population-based cohort, we established practical prognostic nomograms that can objectively and accurately predict long-term OS and CSS in HCC around the world. Moreover, the internal and external cohort validation results demonstrate that these nomograms perform very well and have high accuracy and reliability. Our nomograms were demonstrated to be clinically useful based on a DCA and should therefore help clinicians improve individual treatment, make clinical decisions and guide follow-up management strategies in HCC patients.
Abbreviation list
HCC, Hepatocellular carcinoma; OS, overall survival; CSS, cancer-specific survival; AJCC, American Joint Committee on Cancer; SEER, Surveillance, Epidemiology, and End Results; c-index, concordance index; DCA, decision curve analysis; SYMH, Sun Yat-sen Memorial Hospital.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81572407, 81602112, 81672405, 81702404); Key Project of Natural Science Foundation of Guangdong Province, China (No. 4210016041); Science and Technology Program of Guangzhou, China (No.2015A030313096, 2016A030313184, 2017A030313536); Natural Science Foundation of Guangdong Province, China (No. 4250016043); Grant [2013] 163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology; Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors have declared that no conflict of interest exists in this work.
References
1. Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–855. doi:10.1136/gutjnl-2013-306627
2. Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. doi:10.1016/j.suronc.2016.03.002
3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
4. Benson AB
5. Ganne-Carrie N, Layese R, Bourcier V, et al. Nomogram for individualized prediction of hepatocellular carcinoma occurrence in hepatitis C virus cirrhosis (ANRS CO12 CirVir). Hepatology. 2016;64(4):1136–1147. doi:10.1002/hep.28702
6. Kao WY, Su CW, Chiou YY, et al. Hepatocellular carcinoma: nomograms based on the albumin-bilirubin grade to assess the outcomes of radiofrequency ablation. Radiology. 2017;285(2):670–680. doi:10.1148/radiol.2017162382
7. Shim JH, Jun MJ, Han S, et al. Prognostic nomograms for prediction of recurrence and survival after curative liver resection for hepatocellular carcinoma. Ann Surg. 2015;261(5):939–946. doi:10.1097/SLA.0000000000000747
8. Kluth LA, Black PC, Bochner BH, et al. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. Eur Urol. 2015;68(2):238–253. doi:10.1016/j.eururo.2015.01.032
9. Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–180. doi:10.1016/S1470-2045(14)71116-7
10. Fakhry C, Zhang Q, Nguyen-Tan PF, et al. Development and validation of nomograms predictive of overall and progression-free survival in patients with oropharyngeal cancer. J Clin Oncol. 2017;35(36):4057–4065. doi:10.1200/JCO.2016.72.0748
11. Lee CK, Goldstein D, Gibbs E, et al. Development and validation of prognostic nomograms for metastatic gastrointestinal stromal tumour treated with imatinib. Eur J Cancer. 2015;51(7):852–860. doi:10.1016/j.ejca.2015.02.015
12. Ganly I, Amit M, Kou L, et al. Nomograms for predicting survival and recurrence in patients with adenoid cystic carcinoma. An international collaborative study. Eur J Cancer. 2015;51(18):2768–2776. doi:10.1016/j.ejca.2015.09.004
13. Mitra AP, Lam LL, Ghadessi M, et al. Discovery and validation of novel expression signature for postcystectomy recurrence in high-risk bladder cancer. J Natl Cancer Inst. 2014;106:11. doi:10.1093/jnci/dju061
14. Gravis G, Boher JM, Fizazi K, et al. Prognostic factors for survival in noncastrate metastatic prostate cancer: validation of the glass model and development of a novel simplified prognostic model. Eur Urol. 2015;68(2):196–204. doi:10.1016/j.eururo.2014.09.022
15. He W, Peng B, Tang Y, et al. Nomogram to predict survival of patients with recurrence of hepatocellular carcinoma after surgery. Clin Gastroenterol Hepatol. 2018;16(5) :756–764 e710. doi:10.1016/j.cgh.2017.12.002
16. Xu L, Peng ZW, Chen MS, et al. Prognostic nomogram for patients with unresectable hepatocellular carcinoma after transcatheter arterial chemoembolization. J Hepatol. 2015;63(1):122–130. doi:10.1016/j.jhep.2015.02.034
17. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi:10.1136/bmj.g7594
18. Sauerbrei W, Boulesteix AL, Binder H. Stability investigations of multivariable regression models derived from low- and high-dimensional data. J Biopharm Stat. 2011;21(6):1206–1231. doi:10.1080/10543406.2011.629890
19. Mayr A, Schmid M. Boosting the concordance index for survival data–a unified framework to derive and evaluate biomarker combinations. PLoS One. 2014;9(1):e84483. doi:10.1371/journal.pone.0084483
20. Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35(9):2052–2056. doi:10.1097/01.CCM.0000275267.64078.B0
21. Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi:10.1186/1472-6947-8-53
22. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi:10.1177/0272989X06295361
23. Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi:10.1200/JCO.2015.65.9128
24. Kerr KF, Brown MD, Zhu K, Janes H. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol. 2016;34(21):2534–2540. doi:10.1200/JCO.2015.65.5654
25. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
26. Vauthey JN, Ribero D, Abdalla EK, et al. Outcomes of liver transplantation in 490 patients with hepatocellular carcinoma: validation of a uniform staging after surgical treatment. J Am Coll Surg. 2007;204(5) :1016–1027. discussion 1027–1018. doi:10.1016/j.jamcollsurg.2006.12.043
27. Dai D, Jin H, Wang X. Nomogram for predicting survival in triple-negative breast cancer patients with histology of infiltrating duct carcinoma: a population-based study. Am J Cancer Res. 2018;8(8):1576–1585.
28. Aizer AA, Chen MH, McCarthy EP, et al. Marital status and survival in patients with cancer. J Clin Oncol. 2013;31(31):3869–3876. doi:10.1200/JCO.2013.49.6489
29. Chen Z, Cui J, Dai W, Yang H, He Y, Song X. Influence of marital status on small intestinal adenocarcinoma survival: an analysis of the Surveillance, Epidemiology, and End Results (SEER) database. Cancer Manag Res. 2018;10:5667–5676. doi:10.2147/CMAR.S177430
30. Cho CS, Gonen M, Shia J, et al. A novel prognostic nomogram is more accurate than conventional staging systems for predicting survival after resection of hepatocellular carcinoma. J Am Coll Surg. 2008;206(2):281–291. doi:10.1016/j.jamcollsurg.2007.07.031
31. Nathan H, Schulick RD, Choti MA, Pawlik TM. Predictors of survival after resection of early hepatocellular carcinoma. Ann Surg. 2009;249(5):799–805. doi:10.1097/SLA.0b013e3181a38eb5
32. Huang XT, Chen LH, Huang CS. et al. Establishment of a nomogram by integrating molecular markers and tumor-node-metastasis staging system for predicting the prognosis of hepatocellular carcinoma. Dig Surg;2018. 1–7. doi:10.1159/000494219
33. Adhoute X, Penaranda G, Raoul JL, et al. Barcelona clinic liver cancer nomogram and others staging/scoring systems in a French hepatocellular carcinoma cohort. World J Gastroenterol. 2017;23(14):2545–2555. doi:10.3748/wjg.v23.i14.2545
Supplementary materials
 | Figure S1 OS and CSS in the entire SEER cohort. |
 | Figure S2 Curves for OS in subgroups within the entire SEER cohort. Sex (A), marital status (B), age (C), race (D), surgery performed (E), grade (F), TNM stage (G), and tumor size (H). |
 | Figure S3 Curves for CSS in subgroups within the entire SEER cohort. Sex (A), marital status (B), surgery performed (C), grade (D), TNM stage (E), and tumor size (F). |
 | Table S1 Demographic and clinical characteristics of HCC patients in the SEER and SYMH cohorts. |
 | Table S2 Univariate Cox regression analyses of clinicopathological parameters in the SEER discovery set. |
 | Table S3 Point assignments and prognostic scores for each variable in the nomogram models |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





