Back to Journals » OncoTargets and Therapy » Volume 13
Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis
Authors Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J
Received 24 December 2019
Accepted for publication 12 March 2020
Published 27 March 2020 Volume 2020:13 Pages 2563—2571
DOI https://doi.org/10.2147/OTT.S243601
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Guobin Zhong,1 Keqiong Wang,1 Jiawei Li,1 Shuzhe Xiao,2 Wei Wei,1 Jianlun Liu3
1Department of Breast Surgery, Guangxi Medical University Cancer Hospital, Nanning, People’s Republic of China; 2Department of Pediatrics, Guangzhou First Municipal People’s Hospital, Guangzhou, People’s Republic of China; 3Department of Breast Surgery, Guangxi Medical University Cancer Hospital, Nanning, China; Department of General Surgery, The Langdong Hospital of Guangxi Medical University, Nanning, People’s Republic of China
Correspondence: Jianlun Liu; Wei Wei Email [email protected]; [email protected]
Purpose: There is an urgent need for new biomarkers for the diagnosis of breast cancer. Exosomes can communicate with cells through transport molecules, including long-chain noncoding RNA (lncRNA), which is considered as a promising noninvasive biomarker. Here, we aimed to determine the potential of long noncoding RNA (lncRNA) H19 in the circulating exosomes for the diagnosis of breast cancer (BC).
Materials and Methods: We measured the levels of lncRNA H19 in serum-derived exosomes from patients with breast cancer (BC) or benign breast disease (BBD) and healthy subjects, using quantitative real-time PCR. H19 levels were also measured for pre-operative and post-operative patients. Receiver operating characteristic curve was constructed, and the area under the curve (AUC) was calculated to determine the applicability of exosomal H19 levels as biomarkers in BC. The relationship between H19 relative expression and clinical features of BC patients was also analyzed.
Results: Exosomal H19 expression levels were upregulated in patients with BC compared to that in patients with BBD and healthy controls (BC vs BBD, P < 0.001; BC vs healthy subjects, P < 0.001). The median serum exosomal H19 levels were significantly lower in post-operative than that in the pre-operative patients (P < 0.001). The AUC for exosomal H19 analysis was 0.870 (95% CI: 0.774– 0.966) with a sensitivity of 87.0% and specificity of 70.6%, which was higher than the AUCs for CA15-3 and CEA, ie, 0.822 and 0.811, respectively. Moreover, exosomal H19 expression levels were associated with lymph node metastasis (P = 0.039), distant metastasis (P = 0.008), TNM stages (P = 0.022), ER (P=0.009), PR (P = 0.018), and Her-2 (P = 0.021).
Conclusion: Our results indicated that serum exosomal H19 acts as a novel biomarker for the diagnosis of BC.
Keywords: biological marker, breast neoplasm, diagnosis, long noncoding RNA
Introduction
Breast cancer (BC) is the most common malignant tumor in women worldwide, accounting for 30% of all new cancer diagnoses in women.1 The most common causes of cancer death in women include lung, breast, and colorectal cancer. The latest data estimated approximately 279,100 new cases and 42,690 deaths due to BC in the United States in 2020.2 It is well known that early diagnosis and treatment can critically benefit the prognosis of BC. However, thick-needle biopsy followed by pathological examination, which is the current gold standard for diagnosis of BC, is uncomfortable and invasive. Although body fluid-based testing contributes notably to cancer diagnosis, the current serum tumor biomarkers used for BC diagnosis such as carbohydrate antigen 15–3 (CA15-3) and carcinoembryonic antigen (CEA) have a low positive rate. Therefore, discovery of alternative novel diagnostic biomarkers with high specificity and high sensitivity for BC diagnosis is urgently required.
Exosomes are a type of membrane vesicles with a diameter of approximately 30–150 nm, which are produced by almost all cells distributed in all body fluids, containing a variety of proteins, RNAs, and lipids and executing the function of cell-to-cell communication by transferring oncogenic molecules, thereby playing crucial roles in tumor growth, progression, and metastasis.3–5 Previous studies have confirmed that long noncoding RNAs (lncRNAs) are non-protein encoding transcripts, having more than 200 nucleotides.6 Disordered regulation of lncRNAs can contribute to human diseases, such as tumor formation and progression.7–9 Based on this, they may be considered feasible as tumor biomarkers. Moreover, several studies have indicated that serum exosomal lncRNAs show increased expression in several human cancers, including colorectal cancer, hepatocellular carcinoma, and gastric cancer.10–13
Emerging evidence indicates H19 is an oncogene.14 The H19 gene encodes a 2.3-kb lncRNA, which is located on human chromosome 11p15.5 and exclusively expressed from the maternal allele.15 H19 expression is widespread during embryonic development but becomes restricted to select tissues such as breast and uterus after birth.16 In cancer, H19 is often overexpressed, and it is associated with many aspects of cancer development.17 Wang’s study showed that circulating exosomal H19 released from bladder cancer cells was valuable for the diagnosis and prognosis of bladder cancer patients in the clinical setting.18 Aberrant H19 expression is observed in solid tumors, including breast cancer. H19 is associated with many biological processes, such as cell proliferation, invasion, and apoptosis in tumors, including breast cancer.19,20 Circulating lncRNA H19 is a reported biomarker for monitoring cancer progression.21 Therefore, we selected serum exosomal lncRNA H19 as a candidate and evaluated its diagnostic value for BC.
In this study, we measured the levels of H19 in serum-derived exosomes, isolated from patients with BC or benign breast disease (BBD) and healthy subjects, using quantitative reverse transcription PCR (qRT-PCR). H19 levels were also measured for the pre-operative and post-operative patients. In addition, we analyzed the applicability of H19 as a novel marker for BC diagnosis and compared its diagnostic potential with that of traditional biomarkers, such as CA15-3 and CEA. The relationship between the relative expression of H19 and the clinical features of BC patients was also analyzed. In summary, the results of this study suggest that exosomal H19 may be a useful tool for the detection of BC.
Materials and Methods
Patient and Sample Collection
A total of 50 patients newly diagnosed with BC were enrolled from Guangxi Medical University Cancer Hospital between May 2018 and December 2018. Inclusion criteria were: 1) female; 2) diagnosed with BC by histopathological examination; 3) lack of prior anti-cancer treatment such as chemotherapy or radiotherapy; 4) absence of other malignancies; 5) available clinical records. Patients with heart disease, organ failure, mental disorder, hypertension, diabetes, traumatic disease, and immune disease were excluded. Blood samples were collected from included patients after diagnosis.
In parallel, serum samples from 50 patients with BBD and 50 healthy participants without BC history were obtained from the same hospital. The diagnosis of BBD was confirmed by pathological examination. The control group was comprised of healthy volunteers, having no health issues that were identified in a routine physical examination. The average age of patients with BC was 49 years (range 41–62). The average age of patients with BBD and healthy volunteers was 44 (range 40–50) and 50 (range 45–65) years, respectively. Blood samples from patients with BC were collected again after day 7 post-operation to avoid the effects of radiation or chemotherapy. After centrifugation at 3000 × g for 10 min, 2 mL of serum was isolated and stored at −80 °C until exosome extraction.
We obtained written informed consent from all subjects, and this study was approved by the ethics committee of Guangxi Medical University Cancer Hospital (approval #:LW2019027). This study was conducted in accordance with the Declaration of Helsinki. Authors had access to information that could identify individual participants during or after data collection.
Exosome Isolation and Identification
Exosomes were extracted using the Exosome Extraction Reagent (RENGEN BIOSCIENCES, SHENYANG, LIAONING, CHINA; http://www.rengenbio.com). Serum samples were thawed in a 25 °C water bath and centrifuged at 3000 × g at 4 °C for 15 min to remove cells and debris. Next, 125 μL exosome extraction reagent was added to 250 μL serum and the mixture was swirled to homogeneity. After incubation at 4 °C for 1 h, the samples were centrifuged at 3000 × g at 4 °C for 30 min. The exosome pellet was diluted in 100 μL phosphate-buffered solution (PBS) (10,010,023, Thermo Fisher Scientific, USA).
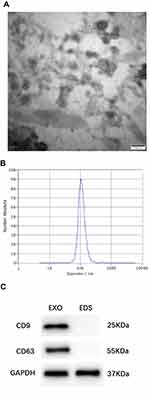
Serum-derived exosomes were identified by transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and Western blotting analysis. Briefly, exosome specimens were fixed in 1% glutaraldehyde solution for 2 h. Thereafter, 20 μL of the diluted mixture was transferred to a clean copper mesh. After staining with 3% phosphotungstic acid solution, an image was captured using transmission electron microscope (TEM; Hitachi, Japan). The exosome specimens were diluted 1000-fold and resuspended in PBS for size distribution analysis using NTA (Malvern Instruments, UK).
For Western blotting analysis, total protein from the exosomes and the exosome-depleted supernatant were extracted with RIPA buffer (Beyotime Biotechnology, China), and the BCA method was performed to determine the protein concentration. The protein fraction (30 µg) was run on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Primary antibodies against CD9 (EXOAB-CD63A-1), CD63 (EXOAB-CD9A-1) were purchased from SBI (System Biosciences, USA), GAPDH (EPR16891) was purchased from Abcam (UK). The secondary antibodies were goat anti-rabbit HRP secondary antibody (EXOAB-VMTN-1) (System Biosciences, USA).
Total RNA Extraction and Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNA was extracted from serum-derived exosomes using RNAiso Blood (Takara, China) according to the manufacturer’s instruction. The extracted RNA was dissolved in 10 μL of RNase-free water. The quality and quantity of RNA were evaluated by a NanoDrop Spectrophotometer (Thermo Fisher Scientific, USA). The total RNAs were reverse-transcribed using a high-capacity cDNA reverse transcription kit (TAKARA) to confirm the expression of lncRNA H19 and internal control. qPCR was performed using Power TB Green (Takara) on the qtower3 G (Analytik Jena, Germany), which was also used for data collection. β-actin was used as an internal control. The primers for lncRNA H19 were (forward) 5′-ACTCAGGAATCGGCTCTGGAAGG-3′ and (reverse) 5′-GATGTGGTGGCTGGTGGTCAAC-3′; primers for β-actin were (forward) 5′-ATCAAGATCATTGCTCCTCCTGA-3′ and (reverse) 5′-CTGCTTGCTGATCCACATCTG-3′. The reaction conditions for RT-qPCR were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C (the optimal annealing temperature) for 31 s, and 72°C for 10 s. The relative expression levels of lncRNA H19 were analyzed by the 2−ΔΔCt method.
Detection of Serum CA15-3 and CEA
Serum CA15-3 and CEA levels were tested by chemiluminescent immunoassay using Roche Cobas E 601 Analyzer (Roche, Switzerland), following the manufacturer’s standard protocol. All methods were performed in accordance with the relevant regulations and guidelines.
Statistical Analysis
SPSS software 22.0 (IBM Corporation, USA) was used for statistical analyses. The data are presented as median and interquartile range. The Kruskal Wallis test with Bonferroni correction was used to compare the differences of H19 in BC group against those in BBD and healthy groups. The Wilcoxon test was used to compare the pre-operative and post-operative serum samples. GraphPad Prism 5.0 (GraphPad Software, USA) was used to prepare the figures. Association of exosomal lncRNA H19 with clinicopathological parameters was examined by the Chi-squared test. Receiver operating characteristic (ROC) curve was constructed, and area under the curve (AUC) was calculated to determine the applicability of exosomal H19 levels as biomarker in BC detection. Two-sided P-value < 0.05 was considered to be statistically significant.
Results
Characterization of Exosomes in Serum
TEM showed the serum-derived exosomes to have a diameter of 50–150 nm with a cup-shaped membrane (Figure 1A). The sizes of exosomes were characterized by an NTA characterization system, and the majority of vesicle particles were between 50–150 nm in diameter (Figure 1B). In addition, CD9 and CD63 were used to identify serum exosomes (Figure 1C).
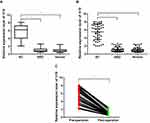
Circulating Exosomal lncRNA H19 Overexpressed in BC Serum
Exosomal H19 expression levels were upregulated in patients with BC compared to that in patients with BBD and in the healthy controls. There were significant differences in the levels of H19 across the groups (BC vs BBD, P < 0.001; BC vs healthy subjects, P < 0.001, Figure 2A and B). Moreover, no significant difference in the expression of exosomal H19 was found among the patients with BBD and healthy subjects. Furthermore, the median serum levels of exosomal H19 were remarkably decreased in post-operative samples compared to that in pre-operative samples (P < 0.001, Figure 2C).
Diagnostic Value of Exosomal H19 for Patients with BC
The value of AUC for exosomal H19 analysis was 0.870 (95% CI: 0.774–0.966, Figure 3A) with a sensitivity of 87.0% and a specificity of 70.6% that was significantly higher than the AUCs for CA15-3 and CEA (0.822 and 0.811, respectively) (Figure 3B and C). We also calculated the combinative diagnostic value of CA15-3 and CEA; AUC was 0.845 (95% CI: 0.702–0.988, Figure 3D). The results indicated that exosomal H19 was superior to the traditional markers in BC diagnosis. Furthermore, AUC for the combination of three markers was 0.914 (95% CI: 0.808–0.983, Figure 3E), thus implying exosomal H19 to be an appropriate diagnostic marker for BC. The diagnostic efficacy of all markers (individual and combinations) for breast cancer are listed in Table 1.
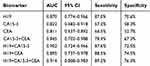 |
Table 1 The Diagnostic Efficacy of All Markers (Individual and Combinations) for Breast Cancer |
Correlation of Serum H19 with Clinicopathological Characteristics
We found exosomal H19 expression levels to be associated with lymph node metastasis (P = 0.039), distant metastasis (P = 0.008), TNM stages (P = 0.022), ER (P=0.009), PR (P = 0.018), and Her-2 (P = 0.021). However, we failed to find any significant correlation between the H19 levels and clinical characteristics in terms of age, tumor size, and Ki67 cells (Table 2).
 |
Table 2 Association Between H19 Relative Expression and Clinical Features of BC Patients |
Discussion
The identification of an effective biomarker for early diagnosis of BC is critical for timely treatment. Prior studies have revealed that circulating exosomes in serum contain protein and nucleic acids that may be indicative of various diseases including cancer, and thus provide opportunities for early cancer diagnosis.22 The analysis of exosomes in plasma or bodily fluids is highly advantageous to pathological biopsy owing to its noninvasiveness and repeatability. Indeed, exosomes are very stable, easy to obtain, and present in almost all body fluids,23,24 allowing for frequent monitoring of malignancies. In our study, we investigated whether lncRNA serum H19 is present in exosomes and whether exosomal H19 could be utilized as a noninvasive diagnostic tool for breast cancer. Our results showed that serum H19 was contained in exosomes, and exosomal H19 was significantly upregulated in the serum of patients with BC compared to those with BBD and NC participants. Moreover, ROC analysis indicated that serum exosomal H19 is a promising diagnostic indicator.
In this study, exosome morphology and size distribution were characterized by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). TEM showed that serum-derived exosomes were 50–150 nm in diameter. Furthermore, exosome size was measured using NTA, and the majority of vesicle particles were 50–150 nm in diameter. CD9 and CD63 are known exosomal protein markers routinely used for the determination of serum exosomes25–27. These results demonstrated that we have successfully extracted exosomes from serum. There was a difference in total exosome content between N versus T serum.28,29 The exosome pellet was washed with PBS to ensure that there was no serum residue. Therefore, H19 was measured in serum exosomes rather than in the serum. Recently, Hon et al have identified 19,175 potentially functional lncRNAs in the human genome that are also regulated by transcription factors and epigenetic modifications.30 They reported that most lncRNAs are similar to mRNA: they are transcribed by RNA polymerase II, are 5′ capped, 3′ polyadenylated, and can be spliced.31,32 Shao et al proved that lncRNA CASC9 may be used as a novel therapeutic target and as a potential diagnostic marker for BC.33 Huang et al identified a panel of serum noncoding RNAs (ncRNAs) as potential diagnostic and prognostic biomarkers for BC that included the ncRNAs let-7a, miR-155, miR-574-5p, and MALAT1.34 Based on our experiments, we found that exosomal H19 could effectively distinguish BC patients from BBD and NC subjects with a significantly high AUC value of 0.870 as well as a sensitivity of 87.0% and a specificity of 70.6%. β-actin was consistently highly expressed in our experiments, showing that ACTB expression in serum exosomes is not affected in breast cancer. The following groups have also chosen β -actin as an internal control for normalizing of RNA levels in exosomes.35,36 For the first time, its potential in the diagnosis of BC was proved. Moreover, the clinical diagnostic value of H19 was shown to be significantly higher than that of CEA and CA15-3. Therefore, the best option would be to combine H19, CEA, and CA15-3 in such a way that the diagnostic efficiency is as high as 0.914, which is much better than that of any of the markers alone. Therefore, we believe that detection of a combination of exosomal H19, CEA, and CA15-3 may be a viable complement to current BC detection strategies. Our study also showed that exosomal H19 levels were associated with poor clinical outcome, including positive lymph node metastasis and positive distant metastasis, which indicated that high levels of exosomal H19 may be correlated with an advanced BC. For patients undergoing modified radical mastectomy, a remarkable decrease in exosomal H19 may indicate the release of exosomal H19 from tumor cells into the bloodstream.
H19 is beneficial in the development of BC, probably by the following mechanisms. First, the H19-derived miR-675 gives rise to two functional microRNAs, namely miR-675-5p and miR-675-3p.37 Vennin et al had identified that the overexpression of miR-675-5p in BC cell lines induces the downregulation of c-Cbl and Cbl-b proteins, and promotes the proliferation and metastasis of tumor cells.38 Matouk et al found that H19 upregulates Slug expression, resulting in the suppression of E-cadherin through mir-675 to regulate epithelial-mesenchymal transition of BC and promote invasion and metastasis of BC cells.39 Second, Li et al revealed that H19 upregulates DNA methyltransferase DNMT1 by sponging miR-152, thereby promoting BC cell proliferation and invasion.40 Last, but not the least, Barsyte-Lovejoy et al demonstrated H19 as a Myc-up-regulated gene that potentiates the tumorigenic phenotype of BC cells.41
Conclusions
In conclusion, our data show that exosomal H19 expression levels are upregulated in patients with BC. Moreover, the ROC results show that the diagnostic accuracy of exosomal H19 was superior to the traditional markers in BC diagnosis. Our results indicate that serum exosomal H19 acts as a novel biomarker for the diagnosis of BC. These findings warrant further investigation. Certainly, more studies are required for further investigation into the diagnostic potential of serum exosomal H19 for BC treatment and prognosis.
Abbreviations
AUC, area under the curve; lncRNA, long noncoding RNA; BC, breast cancer; BBD, benign breast disease; ROC, receiver operating characteristic; qPCR, quantitative real-time PCR; TEM, transmission electron microscopy; NTA, nanoparticle tracking analysis; PBS, phosphate-buffered solution.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author (Jianlun Liu) on reasonable request.
Acknowledgments
This study was supported by a grant (no. 81760481) awarded by the National Natural Science Foundation of China.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Li M, Zeringer E, Barta T, Schageman J, Cheng A, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc London Ser B. 2014;369(1652):20130502. doi:10.1098/rstb.2013.0502
4. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8(1):83. doi:10.1186/s13045-015-0181-x
5. Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi:10.1016/j.cell.2016.01.043
6. Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi:10.1038/nature10398
7. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
8. Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi:10.1038/nature12451
9. Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. doi:10.1007/s10142-016-0524-x
10. Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–85563. doi:10.18632/oncotarget.v7i51
11. Sun L, Su Y, Liu X, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9(15):2631–2639. doi:10.7150/jca.24978
12. Lin LY, Yang L, Zeng Q, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17(1):84. doi:10.1186/s12943-018-0834-9
13. Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17(1):68. doi:10.1186/s12943-018-0817-x
14. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165.
15. Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi:10.1002/bies.200900170
16. Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113(1–4):188–193. doi:10.1159/000090831
17. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14(1):184. doi:10.1186/s12943-015-0458-2
18. Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal h19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi:10.12659/MSM.912018
19. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi:10.1016/j.canlet.2013.01.033
20. Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Et Biophys Acta Mol Cell Res. 2017;1864(10):1887–1899. doi:10.1016/j.bbamcr.2017.08.001
21. Zhou W, Ye XL, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(483):eaak9557. doi:10.1126/scisignal.aak9557
22. Mizutani K, Terazawa R, Kameyama K, et al. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34(7):3419–3423.
23. Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of Hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27(6):710–716. doi:10.1158/1055-9965.EPI-17-0770
24. Khurana R, Ranches G, Schafferer S, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. Rna. 2017;23(2):142–152. doi:10.1261/rna.058834.116
25. Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–1405. doi:10.1111/jcmm.2019.23.issue-2
26. Dai J, Escara-Wilke J, Keller JM, et al. Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J Exp Med. 2019;216(12):2883–2899. doi:10.1084/jem.20190158
27. Zhang H, Liu J, Qu D, et al. Serum exosomes mediate delivery of arginase 1 as a novel mechanism for endothelial dysfunction in diabetes. Proc Natl Acad Sci U S A. 2018;115(29):E6927–E6936. doi:10.1073/pnas.1721521115
28. Liu W, Hu J, Zhou K, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–3851. doi:10.2147/OTT.S140062
29. Li W, Lu Y, Yu X, Yong M, Ma D, Gao Q. Detection of exosomal tyrosine receptor kinase B as a potential biomarker in ovarian cancer. J Cell Biochem. 2019;120(4):6361–6369. doi:10.1002/jcb.v120.4
30. Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5ʹ ends. Nature. 2017;543(7644):199–204. doi:10.1038/nature21374
31. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
32. Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33(8):540–552. doi:10.1016/j.tig.2017.05.004
33. Shao G, Wang M, Fan X, et al. lncRNA CASC9 positively regulates CHK1 to promote breast cancer cell proliferation and survival through sponging the miR195/497 cluster. Int J Oncol. 2019. doi:10.3892/ijo.2019.4734
34. Huang S-K, Luo Q, Peng H, et al. A panel of serum noncoding RNAs for the diagnosis and monitoring of response to therapy in patients with breast cancer. Med Sci Monit. 2018;24:2476–2488. doi:10.12659/MSM.909453
35. Liu L, Zuo L, Yang J, et al. Exosomal cyclophilin A as a novel noninvasive biomarker for Epstein-Barr virus associated nasopharyngeal carcinoma. Cancer Med. 2019;8(6):3142–3151. doi:10.1002/cam4.2185
36. Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234(11):20721–20727. doi:10.1002/jcp.v234.11
37. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna. 2007;13(3):313–316. doi:10.1261/rna.351707
38. Vennin C, Spruyt N, Dahmani F, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6(30):29209–29223. doi:10.18632/oncotarget.4976
39. Matouk IJ, Raveh E, Abu-lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014;1843(7):1414–1426. doi:10.1016/j.bbamcr.2014.03.023
40. Li Z, Li Y, Li Y, et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017;31(9):e21933. doi:10.1002/jbt.2017.31.issue-9
41. Barsyte-Lovejoy D, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–5337. doi:10.1158/0008-5472.CAN-06-0037
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.