Back to Journals » Lung Cancer: Targets and Therapy » Volume 8
Detection of ROS1 rearrangement in non-small cell lung cancer: current and future perspectives
Authors Rossi G , Jocollé G, Conti A, Tiseo M, Zito Marino F, Donati G, Franco R , Bono F, Barbisan F, Facchinetti F
Received 6 March 2017
Accepted for publication 9 May 2017
Published 7 July 2017 Volume 2017:8 Pages 45—55
DOI https://doi.org/10.2147/LCTT.S120172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Pan-Chyr Yang
Giulio Rossi,1 Genny Jocollé,2 Antonia Conti,3 Marcello Tiseo,4 Federica Zito Marino,5,6 Giovanni Donati,7 Renato Franco,5,6 Francesca Bono,8 Francesca Barbisan,9 Francesco Facchinetti4,10
1Pathology Unit, 2Oncology Unit, Azienda USL Valle d’Aosta, Regional Hospital “Parini”, Aosta, 3Medical Illustrator, Riccione, 4Medical Oncology Unit, University Hospital of Parma, Parma, 5Pathology Unit, Istituto Nazionale Tumori Fondazione G. Pascale, 6Pathology Unit, Luigi Vanvitelli University of Campania, Naples, 7Unit of Thoracic and Senology Surgery, Azienda USL Valle d’Aosta, Regional Hospital “Parini”, Aosta, 8Unit of Pathologic Anatomy, San Gerardo Hospital, IRCCS, Monza, 9Pathology Unit, University Hospital, Azienda Ospedali Riuniti, Ancona, Italy; 10INSERM, U981, Gustave Roussy Cancer Campus, Villejuif, France
Abstract: ROS1 rearrangement characterizes a small subset (1%–2%) of non-small cell lung cancer and is associated with slight/never smoking patients and adenocarcinoma histology. Identification of ROS1 rearrangement is mandatory to permit targeted therapy with specific inhibitors, demonstrating a significantly better survival when compared with conventional chemotherapy. Detection of ROS1 rearrangement is based on in situ (immunohistochemistry, fluorescence in situ hybridization) and extractive non-in situ assays. While fluorescence in situ hybridization still represents the gold standard in clinical trials, this technique may fail to recognize rearrangements of ROS1 with some gene fusion partner. On the other hand, immunohistochemistry is the most cost-effective screening technique, but it seems to be characterized by low specificity. Extractive molecular assays are expensive and laborious methods, but they specifically recognize almost all ROS1 fusions using a limited amount of mRNA even from formalin-fixed, paraffin-embedded tumor tissues. This review is a discussion on the present and futuristic diagnostic scenario of ROS1 identification in lung cancer.
Keywords: lung, adenocarcinoma, ROS1, FISH, immunohistochemistry, NGS, rearrangement
Introduction
ROS1 is a gene encoding a receptor tyrosine kinase; it is closely related to ALK and LTK and identified in several human tumors, including non-small cell lung cancer (NSCLC).1–8
Recently, the US Food and Drug Administration approved the use of crizotinib, (Xalkori®, Pfizer Inc., New York, NY, USA) a specific small molecule inhibitor, in the therapy of ROS1 rearranged NSCLC.9–11
Detection of ROS1 rearrangement is then a critical step in the treatment of NSCLC and may be performed using different techniques, including immunohistochemistry (IHC), fluorescence in situ hybridization (FISH) and molecular extractive methods (e.g., reverse transcription-polymerase chain reaction [RT-PCR]).
Since ROS1 protein is absent in normal lung tissue and the prevalence of ROS1 rearrangement in NSCLC ranges from 0.5% to 2%, IHC appears a cost-effective screening assay, thus permitting rapid results with less cost.12–17
All the different methodologies adopted to identify ROS1 rearrangement have some advantages as well as limitations when compared to each other. In the current clinical practice, FISH represents the gold standard in light of its use in determining ROS1 positivity in clinical trials. However, several studies comparing the sensitivity and specificity of other techniques with FISH results have been published.18–36
While coordinated use of IHC and FISH testing does represent the routine practice in real-life laboratories, emerging molecular assays, including mRNA expression of the 3′ region over 5′ region of ROS1 gene (NanoString assay) and next-generation sequencing (NGS), could become an appealing and futuristic standard, permitting simultaneous tests for several “druggable” drivers using limited amount of tumor tissue or liquid biopsies.37–40
The ROS1 oncogene
ROS1 gene is located at chromosome 6q22 and encodes for a receptor tyrosine kinase belonging to the insulin receptor family (Figure 1). The rearrangement of ROS1 gene leads to a constitutively activated downstream signaling with oncogenic properties. ROS1 rearrangement was firstly detected in a glioblastoma cell line,41 but was also reported in cholangiocarcinoma, gastric adenocarcinoma, ovarian serous carcinoma, colonic adenocarcinoma, inflammatory myofibroblastic tumor, angiosarcoma, epithelioid hemangioendothelioma and spitzoid melanocytic tumors.2–7,34 The fusion gene partners of ROS1 comprise several genes, including CD74, EZR, FIG1, CCD6, KDELR2, LRI3, SDC4, SLC34A2, TPM3 and TPD52L1 (Figure 1). All rearrangements involve the 3′ region of the kinase domain of ROS1 to the 5′ region of the partner gene. ROS1 rearrangements characterize about 0.5%–2% of unselected NSCLCs.
The seminal work by Rikova et al42 first identified ROS1 and ALK fusion genes in NSCLC using a phosphoproteomic approach, characterizing tyrosine kinase signaling in tumor cell lines and samples.
Although not every single fusion transcript has been evaluated for its oncogenic potential thus far, the preservation of the entire ROS1 kinase domain, whatever the partner gene may be, should be sufficient to drive carcinogenesis.42 Once constitutively activated, ROS1 signaling mainly rests on extracellular regulated MAP kinase (ERK), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PI3K)/mechanistic target of rapamycin (mTOR) and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) intracellular pathways (Figure 2).1
  | Figure 2 A possible futuristic algorithm for detecting “druggable” oncogenic drivers in NSCLC using non-in situ methods. Abbreviation: NSCLC, non-small cell lung cancer. |
Of note, ROS1 and ALK are evolutionarily conserved and share >80% sequence identity within their ATP-binding sites.9,12 These latter observations are strongly sustained by the clinical activity of crizotinib in both ROS1- and ALK-driven NSCLC and allow to approach them, with special regards to their inhibition by novel compounds, in a parallel way. Nevertheless, biologic differences between the two oncogenes, still not objectified at the cellular level, ostensibly account for diversities observed in preclinical models and in the clinics for ROS1- and ALK-rearranged NSCLC.
The well-defined oncogenic role that ROS1 plays in lung cancer carcinogenesis, as well as the major clinical improvement generated by the ALK/MET/ROS1 inhibitor crizotinib (which, indeed, recently received the US Food and Drug Administration approval for ROS1-rearranged cases too) make the detection of ROS1 rearrangement a crucial task in the field of molecular medicine.11 Moreover, the relatively long survival of ROS1-positive NSCLC patients stimulates researchers and clinicians to find new therapeutic strategies to maintain an active ROS1 deaddiction. As evidenced in EGFR-mutated and ALK-rearranged NSCLC, overcoming resistance to first-generation targeted treatments with novel molecules is achievable and dramatically fruitful, at least in significant quotes of patients.
This review is focused on the current status of the methodologies used to detect ROS1 oncogene rearrangement in NSCLC. Presently, in situ (IHC and FISH) and extractive, non-in situ assays are available worldwide with important differences concerning advantages and limitations. In particular, a summary of the studies comparing IHC and FISH results is reported and the future scenario of ROS1 detection in routine practice along with the advent of promising non-in situ techniques is reported. A present and futurist algorithmic approach to ROS1 rearrangement in NSCLC is briefly discussed.
Clinical implications of ROS1 detection
The identification of ROS1 rearrangement is of crucial interest in NSCLC patients due to the therapeutic consequences it generates.
Cytotoxic treatment with pemetrexed–platin doublets shows special activity in ROS1-positive patients,43 as seen for lung tumors driven by either ALK or RET rearrangements.44,45 Nevertheless, ROS1 positivity appears to be the best biomarker of pemetrexed activity in terms of both response rate and long-term outcomes, with estimations of median progression-free survivals of around 7 months.46–48
After having demonstrated significant activity in ALK-driven diseases,49 the tyrosine kinase inhibitor crizotinib, firstly developed as an anti-MET molecule, was shown to harbor relevant activity in ROS1-rearranged NSCLC too.9 As seen for the first- and second-generation EGFR inhibitors gefitinib, erlotinib and afatinib, the biologic and clinical behavior of crizotinib in ROS1- (and ALK-) positive patients configures the scenario of the disruption of oncogene addiction. Being strictly dependent on ROS1 signaling for survival, growth and progression, its pharmacologic inhibition engenders apoptosis of tumor cells, therefore translating in clinical benefit. Similar to the other oncogene-addicted models in NSCLC, tumor responses are observed in the vast majority of patients, making disease progressions rare events being often explained by diagnostic, pharmacokinetics or molecular caveats.9,10 Albeit ROS1 rearrangement does not seem to be an intrinsic prognostic factor, long-term disease control exerted by crizotinib is mostly relevant, almost doubling the one obtained in EGFR- and ALK-driven tumors undergoing specific treatment. According to the most recent updates, more than 19 months of progression-free survival are achievable with crizotinib in ROS1-postive cancers,50 compared to the 9–12 months observed with the cited oncogenes.9,51,52
Nevertheless, as the intrinsic nature of advanced NSCLC implies, almost unequivocally, every ROS1-positive patient will undergo disease progression while receiving crizotinib. Already assimilated for EGFR and ALK models, beyond-progression strategies, together with local treatments in terms of oligo-progressive diseases involving or not the central nervous system,53–55 are ostensibly applicable in ROS1-rearranged tumors too. Sooner or later anyway, the molecular escapes that cancer cells find out to slip away crizotinib inhibition need to face novel-generation inhibitors. As seen for the mentioned oncogene-addicted NSCLC, mutations occurring in ROS1 kinase domains preclude crizotinib activity. Since their first report,56 a few of them have been reported to be clinically meaningful.57 Given the homology in their respective kinase domains, ALK and ROS1 share a spectrum of active inhibitors beyond crizotinib (e.g., ceritinib, lorlatinib, entrectinib), developed in order to overcome the resistance to the first-generation molecule. Several clinical trials, series and single cases reported the activity of these molecules, sometimes with regard to the precise reversion of the molecular event leading to crizotinib resistance.57 Administration of such drugs after crizotinib exhaustion appears to be of major importance, engendering new responses and positive long-term outcomes, similar to what is observed in ALK-rearranged tumors57 and in EGFR-mutated ones harboring the resistance mutation T790M, undergoing osimertinib treatment.58
Such evidence witnesses the dramatic relevance of the correct detection of ROS1 rearrangements in patients whose cancers lack other genetic abnormalities. The clinical benefit originating from ROS1 inhibition entails that not even a single ROS1-positive patient should be undiagnosed.
Molecular diagnostics
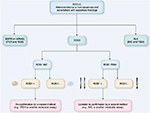
The identification of ROS1 gene rearrangement is mandatory to treat the patients with ROS1-positive NSCLC (Figure 3).
It is generally observed that ROS1 gene rearrangements more often occur in younger and never/light smokers with adenocarcinoma. In a recent study on 727 lung adenocarcinomas from patients with stage IV disease, ROS1 fusions were independently associated with female sex, younger age at diagnosis and absence of smoking history.59 Compared with ALK-positive adenocarcinoma, the ROS-1 positive counterpart is more significantly associated with a peripheral location.60–62
Despite all these data, the clinicopathologic features cannot be robustly used to recognize ROS1-positive patients. In addition, since this genetic alteration occurs in about 0.5%–2% of all NSCLCs, a screening test is necessary in terms of cost-effectiveness.19
ROS1 rearrangement is generally detected using in situ methods, namely FISH and IHC (Figure 4).
Since the clinical trials demonstrating crizotinib efficacy in ROS1-positive patients have substantially adopted FISH testing, this method is considered the “gold standard” for determining ROS1 positivity. Nevertheless, several experiences have evidenced a fair-to-perfect agreement between FISH and IHC tests using the ROS1 rabbit primary antibody D4D6.19–36
At the same time, validated extractive technologies (RT-PCR, NGS, nCounter platform) represent new promising tools in detecting multiple actionable fusions (ALK, ROS1, RET, NTRK) using small amount of tumor RNA.33,63–65
FISH and IHC
FISH using a dual color “break-apart” probes approach is considered the “gold standard” in detecting ROS1 gene rearrangement. If tumor cells are rearranged, the two ends of ROS1 are separated and the portion containing the tyrosine kinase domain is fused with another partner to create an ROS1 fusion gene.
The probes label the 3′ (centromeric) part of the fusion breakpoint with green fluorochrome and the 5′ (telomeric) part with orange fluorochrome. The criteria for ROS1 FISH identification in NSCLC are identical to those proposed for ALK rearrangement, with two main patterns as follows: 1) the break-apart pattern (“conventional” pattern) with one fusion signal and two separated 3′ and 5′ signals and 2) an atypical pattern showing an isolated 3′ signal (usually one fusion signal and one isolated 3′ green signal without the corresponding 5′ signal; Figure 5).19 The green fluorochrome is the part containing the kinase domain of ROS1 gene. FISH testing for ROS1 is applicable either on biopsy or in cytologic specimens, and the cut-off of rearranged signals to quote ROS1 positivity is based on detection of 15% or more among 50 neoplastic nuclei.66,67
Different probes have been used in literature to detect ROS1 rearrangement (Table 1), and it is important to note that some break-apart FISH assays for ROS1 fusion cannot detect intrachromosomal deletion, as GOPC–ROS1 rearrangement.68
IHC is an effective screening tool to detect ROS1-positive NSCLC, since either sensitivity or specificity may reach over 90% when compared with FISH results (Table 1).18–36 Indeed, screening of NSCLC (all nonsmokers and smokers with nonsquamous NSCLC) by IHC may prevent unnecessary FISH analysis in ROS1-negative cases and, thus, dramatically reduce the costs of testing.69
Data obtained from IHC are based on the use of the rabbit primary monoclonal antibody D4D6 (Cell Signaling Technology, Danvers, MA, USA) applied at different dilutions (usually from 1:50 to 1:500) with different amplification kits and detection systems in various automated immune stainers (Table 1).
ROS1 protein is basically absent in normal human lung tissue, but in IHC, its expression may be observed in reactive alveolar type II pneumocytes and macrophages (Figure 6). Therefore, it is mandatory to insert a positive external control or a cell block obtained from ROS1-rearranged cell line, preferentially on the same slide of the tumor to be tested or on a separate slide to be included in the same batch.
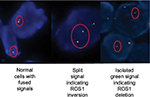  | Figure 6 FISH testing may lead to normal setup with fused signals (on the left) or alterations of signals with the classic split signal (in the middle) or deletion signal (on the right). |
Pathologists should also be aware that in bone biopsies performed in metastatic NSCLC, it is possible to observe osteoclasts revealing a moderate-to-strong granular cytoplasmic staining.
Among the different scoring systems to detect ROS1 rearrangement by IHC, a semi-quantitative method using the expression intensity (negative [score 0], weak [1+], moderate [2+] or strong staining [3+]) was considered. Some authors adopted an h-score (multiplying the percentage of positive tumor cells and the intensity of staining from 0 to 3+, ranging from 0 to 300), with optimal threshold for ROS1 positivity defined as >100.20 ROS1-rearranged tumors usually show diffuse immunohistochemical expression (>75% of tumor cells) with a 2+/3+ staining intensity, basically corresponding to an h-score >100.
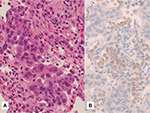
ROS1 expression at IHC level typically reveals finely granular cytoplasmic and/or membranous staining (Figure 7), although the staining pattern may depend on the different gene fusion partners.19 The same IHC protocols used in formalin-fixed, paraffin-embedded (FFPE) tissue specimens may be employed on cell blocks obtained from fine-needle aspirates and pleural/pericardial effusions Although ROS1 IHC works on conventional alcohol-fixed cytology, FISH technique is superior to IHC on nonbloody cytologic smears and cytospin slides.65,66 Conversely, IHC should be preferred in FFPE small biopsy and cell block containing few tumor cells (Figure 8). Some authors suggested to consider IHC ROS1 expression only when at least 20 tumor cells are clearly identifiable in the samples to be analyzed,22 but, in the absence of a clear-cut IHC cut-off, even a positive result on a few neoplastic cells should be considered appropriate and sufficient to quote a tumor as positive/rearranged.
  | Figure 8 A surgically resected lung adenocarcinoma. Notes: (A) Hematoxylin–eosin stain, strongly expressing ROS1. (B) Immunohistochemistry, clone D4D6, score 3+. Magnification: ×200. |
When using clone D4D6 together with highly sensitive amplification kits, the correlation between IHC and FISH is good, although presently, IHC seems to be less specific than FISH when compared with the more robust similar diagnostic scenario already established for ALK rearrangement. Several other promising anti-ROS1 primary monoclonal and polyclonal antibodies are commercially available, although no overt data have been published so far. Another promising primary rabbit monoclonal antibody (EPMGHR2; Abcam, Cambridge, UK) is now commercially available.
Extractive technologies
Extractive assays are based on RT-PCR and massive parallel NGS with kits including several fusion genes (ROS1, ALK, RET, NTRK).70 While the sensitivity and specificity of RT-PCR are quite good, the presence of numerous ROS1 fusion partners identified (e.g., CD74, FIG, SLC34A2, TPM3, SDC4, EZR, LRIG3, GOPC, MSN, KDELR2 and CCDC6)1 or still unknown in the absence of a clear predictive value and the availability of good-quality RNA obtained from FFPE small specimens could limit the use of RT-PCR in routine practice.
nCounter platform (NanoString assay) is a multiplex mRNA-based promising test showing high specificity and sensitivity with referenced tests in detecting ROS1.70 This method may detect several gene fusions by direct profiling using a limited amount of RNA.
Reguart et al33 recently demonstrated the identification of ALK, RET and ROS1 fusions using nCounter technology in FFPE samples of advanced stage NSCLC. Agreement in detecting ROS1 was 87.2% and 86% with IHC and FISH, respectively.
A combined assay involving a pan-receptor tyrosine kinase immunohistochemical cocktail of antibodies (NTRK1, NTRK2, NTRK3, ROS1 and ALK) followed by an RNA-based anchored multiplex PCR NGS assay has been recently proposed.71
The promising results of extractive methodologies applied in clinical practice to determine ROS1 rearrangement seem to candidate non-in situ methods as the near-standalone assays, thus limiting the role of IHC and FISH.
ROS1 rearrangement is commonly mutually exclusive with other alterations involving driver oncogenes in lung cancer. According to this view, a recent study on 62 patients with ROS1-rearranged NSCLC failed to evidence concurrent ALK and EGFR gene alterations, while concomitant KRAS mutations were detected in two cases (3.2%). No concurrent mutations in BRAF, ERBB2, PIK3CA, AKT1 or MAP2K1 were detected. While no concomitant ALK and ROS1 rearrangements were identified, exceedingly rare mutations in EGFR and KRAS were observed.72
Of note, the occurrence of KRAS mutation in ROS1-rearranged lung adenocarcinoma seems to lead to crizotinib resistance in preclinical studies.73
By contrast, a recent work evidenced frequent (36%) concomitant oncogenic driver mutations involving EGFR (6 cases), KRAS (2 cases), PIK3CA and BRAF among 25 lung adenocarcinomas showing ROS1 positivity by IHC.74
Similarly, among 15 cases of ROS1-positive lung adenocarcinomas, 2 patients had TP53 mutations, 1 patient showed an R248L mutation co-occurring with a MAP2K1 missense mutation (K57N) and 1 had an EGFR mutation in exon 21 (P848L). Two out of 17 patients analyzed with NGS or by Sanger sequencing had BRAF mutations and 1 patient showed c-MET mutation (R988C). Overall, 66.7% of patients with ROS1-positive adenocarcinoma analyzed by NGS had further genetic aberrations.75
ROS1 testing into a practical algorithm
The availability of specific inhibitors of ROS1 should lead to test ROS1 rearrangement simultaneously with EGFR mutation and ALK rearrangement in all advanced stage never/light smokers with squamous cell carcinoma and nonsquamous NSCLC.19,38,76
Optimization of tumor tissue with simultaneous preparation of extra blank sections in small biopsy and cell block is another possibility aimed at maximizing the yield of molecular testing, including ROS1 rearrangement.37
ROS1 rearrangement is generally mutually exclusive with other genetic alterations in NSCLC, but a subset may concurrently harbor EGFR or KRAS mutations.
The role of KRAS mutations in an algorithm of predictive biomarkers in routine practice is still debatable. However, when considered as the most common oncogenic driver occurring in about one-third of adenocarcinomas in the Caucasian population, its value as a negative selecting biomarker precluding useless search of further rare targetable gene alterations seems at least reasonable.
According to the recent proposal by an European Board of Pathologists, a routine clinical practice algorithm based on IHC screening with further confirmation by ROS1 break-apart FISH assay in IHC-positive and doubtful cases seems entirely appropriate (Figure 9).19 In the near future, the advent of highly sensitive extractive methods using a small amount of tumor RNA in a single-tube assay to simultaneously detect several oncogenic fusions (ALK, RET, ROS1 and NTRK1 gene rearrangements; Figure 10) and mutations, coupled to a decrease in the cost of instruments will be implemented even in routine practice, possibly limiting the use of IHC and FISH methods.33,77,78
  | Figure 10 A practical algorithm commonly used in real-life routine practice. Abbreviation: NSCLC, non-small cell lung cancer. |
Disclosure
The authors report no conflicts of interest in this work.
References
Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19(15):4040–4045. | ||
Aisner DL, Nguyen TT, Paskulin DD, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res. 2014;12(1):111–118. | ||
Gu TL, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 2011;6(1):e15640. | ||
Birch AH, Arcand SL, Oros KK, et al. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumors. PLoS One. 2011;6(12):e28250. | ||
Giacomini CP, Sun S, Varma S, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9(4):e1003464. | ||
Antonescu CR, Suurmeijer AJ, Zhang L, et al. Molecular characterization of inflammatory myofibroblastic tumors with frequent ALK and ROS1 gene fusions and rare novel RET rearrangement. Am J Surg Pathol. 2015;39(7):957–967. | ||
Charest A, Lane K, McMahon K, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer. 2003;37(1):58–71. | ||
Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. | ||
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS-1 rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. | ||
Mazieres J, Zalcman G, Crino L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results fromtheEUROS1cohort. J Clin Oncol. 2015;33(9):992–999. | ||
FDA. FDA expands use of Xalkori to treat rare form of advanced non-small cell lung cancer; 2016. Available from: http://www.fda.gov/newsevents/newsroom/pressannouncements/ucm490329.htm. Accessed March 11, 2016. | ||
Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18(7):865–875. | ||
Kim MH, Shim HS, Kang DR, et al. Clinical and prognostic implications of ALK and ROS1 rearrangements in never-smokers with surgically resected lung adenocarcinoma. Lung Cancer. 2014;83(3):389–395. | ||
Go H, Kim DW, Kim D, et al. Clinicopathologic analysis of ROS1-rearranged non-small-cell lung cancer and proposal of a diagnostic algorithm. J Thorac Oncol. 2013;8(11):1445–1450. | ||
Lee J, Lee SE, Kang SY, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer. 2013;119(9):1627–1635. | ||
Yoshida A, Kohno T, Tsuta K, et al. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol. 2013;37(4):554–562. | ||
Cai W, Li X, Su C, et al. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann Oncol. 2013;24(7):1822–1827. | ||
Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18(17):4570–4579. | ||
Bubendorf L, Büttner R, Al-Dayel F, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Archiv. 2016;469(5):489–503. | ||
Viola P, Maurya M, Croud J, et al. A Validation Study for the Use of ROS1 Immunohistochemical Staining in Screening for ROS1 Translocations in Lung Cancer. J Thorac Oncol. 2016;11(7):1029–1039. | ||
Yoshida A, Tsuta K, Wakai S, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol. 2014;27(5):711–720. | ||
Sholl LM, Sun H, Butaney M, et al. ROS1 imunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol. 2013;37(9):1441–1449. | ||
Mescam-Mancini L Lantuejoul S, Moro-Sibilot D, et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer. 2014;83(2):168–173. | ||
Rimkunas VM, Crosby KE, Li D, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18(16):4449–4457. | ||
Shan L, Lian F, Guo L, et al. Detection of ROS1 gene rearrangement in lung adenocarcinoma: comparison of IHC, FISH and real-time RT-PCR. PLoS One. 2015;10(3):e0120422. | ||
Boyle TA, Masago K, Ellison KE, Yatabe Y, Hirsch FR. ROS1 immunohistochemistry among major genotypes of non-small-cell lung cancer. Clin Lung Cancer. 2015;16(2):106–111. | ||
Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther. 2016;9:131–138. | ||
Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol. 2015;10(4):611–618. | ||
Lee SE, Lee B, Hong M, et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol. 2015;28(4):468–479. | ||
Fu S, Liang Y, Lin YB, et al. The frequency and clinical implication of ROS1 and RET rearrangements in resected stage IIIA-N2 non-small cell lung cancer patients. PLoS One. 2015;10(4):e0124354. | ||
Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One. 2014;9(7):e103333. | ||
Selinger CI, Li BT, Pavlakis N, et al. Screening for ROS1 gene rearrangements in non-small-cell lung cancers using immunohistochemistry with FISH confirmation is an effective method to identify this rare target. Histopathology. 2017;70(3):402–411. | ||
Reguart N, Teixidó C, Giménez-Capitán A, et al. Identification of ALK, ROS1, and RET fusions by a multiplexed mRNA-based assay in formalin-fixed, paraffin-embedded samples from advanced non-small-cell lung cancer patients. Clin Chem. 2017;63(3):751–760. | ||
Su Y, Goncalves T, Dias-Santagata D, Hoang MP. Immunohistochemical Detection of ROS1 Fusion. Am J Clin Pathol. 2017;147(1):77–82. | ||
Zhou J, Zhao J, Zheng J, et al. A prediction model for ROS1-rearranged lung adenocarcinomas based on Histologic Features. PLoS One. 2016;11(9):e0161861. | ||
Warth A, Muley T, Dienemann H, et al. ROS1 expression and translocations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology. 2014;65(2):187–194. | ||
Wu J, Lin Y, He X, et al. Comparison of detection methods and follow-up study on the tyrosine kinase inhibitors therapy in non-small cell lung cancer patients with ROS1 fusion rearrangement. BMC Cancer. 2016;16:599. | ||
Dietel M, Bubendorf L, Dingemans AM, et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): recommendations of the European Expert Group. Thorax. 2016;71(2):177–184. | ||
Kerr KM, Bubendorf L, Edelman MJ, et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol. 2014;25(9):1681–1690. | ||
Suh JH, Johnson A, Albacker L, et al. Comprehensive genomic profiling facilitates implementation of the National comprehensive cancer network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism-driven clinical trials. Oncologist. 2016;21(6):684–691. | ||
Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A. 1987;84(24):9270–9274. | ||
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. | ||
Zou HY, Li Q, Engstrom LD, West M, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A. 2015;112(11):3493–3498. | ||
Kim HR, Lim SM, Kim HJ, et al. The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol. 2013;24(9):2364–2370. | ||
Camidge DR, Kono SA, Lu X, et al. Anaplastic lymphoma kinase gene rearrangements in non-small cell lung cancer are associated with prolonged progression-free survival on pemetrexed. J Thorac Oncol. 2011;6(4):774–780. | ||
Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21(16):3631–3639. | ||
Chen YF, Hsieh MS, Wu SG, et al. Efficacy of pemetrexed-based chemotherapy in patients with ROS1 fusion-positive lung adenocarcinoma compared with in patients harboring other driver mutations in East Asian populations. J Thorac Oncol. 2016;11(7):1140–1152. | ||
Song Z, Su H ZY. Patients with ROS1 rearrangement-positive non-small-cell lung cancer benefit from pemetrexed-based chemotherapy. Cancer Med. 2016;5(10):2688–2693. | ||
Zhang L, Jiang T, Zhao C, et al. Efficacy of crizotinib and pemetrexed-based chemotherapy in Chinese NSCLC patients with ROS1 rearrangement. Oncotarget. 2016;7(46):75145–75154. | ||
Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. | ||
Shaw A, Riley GJ, Bang YJ, et al. Crizotinib in advanced ROS1-rearranged non-small cell lung cancer (NSCLC): updated results from PROFILE 1001. Ann Oncol. 2016;27 (Suppl 6): 1206PD. | ||
Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. | ||
Park K, Tan EH, O’Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17(5):577–589. | ||
Yap TA, Macklin-Doherty A, Popat S. Continuing EGFR inhibition beyond progression in advanced non-small cell lung cancer. Eur J Cancer. 2017;70:12–21. | ||
Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol. 2012;7(12):1807–1814. | ||
Khalifa J, Amini A, Popat S, Gaspar LE, Faivre-Finn C; International Association for the Study of Lung Cancer Advanced Radiation Technology Committee. Brain metastases from NSCLC: radiation therapy in the era of targeted therapies. J Thorac Oncol. 2016;11(10):1627–1643. | ||
Awad MM, Katayama R, McTigue M, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;368:2395–2401. | ||
Facchinetti F, Rossi G, Bria E, et al. Oncogene addiction in non-small cell lung cancer: focus on ROS1 inhibition. Cancer Treat Rev. 2017;55:83–95. | ||
Marchetti A, Barberis M, Di Lorito A, et al. ROS1 Gene fusion in advanced lung cancer in women: a systematic analysis, review of the literature, and diagnostic algorithm. Precis Oncol. Epub 2017 Feb 22. | ||
Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. 2017;376(7):629–640. | ||
Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016;2(3):313–320. | ||
Scarpino S, Rampioni Vinciguerra GL, Di Napoli A, et al. High prevalence of ALK+/ROS1+ cases in pulmonary adenocarcinoma of adoloscents and young adults. Lung Cancer. 2016;97:95–98. | ||
Yoon HJ, Sohn I, Cho JH, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine (Baltimore). 2015;94(41):e1753. | ||
Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. | ||
Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer. 2015;87(2):122–129. | ||
Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. | ||
Bozzetti C, Nizzoli R, Tiseo M, et al. ALK and ROS1 rearrangements tested by fluorescence in situ hybridization in cytological smears from advanced non-small cell lung cancer patients. Diagn Cytopathol. 2015;43(11):941–946. | ||
Savic S, Bubendorf L. Role of fluorescence in situ hybridization in lung cancer cytology. Acta Cytol. 2012;56(6):611–621. | ||
Suehara Y, Arcila M, Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18(24):6599–6608. | ||
Rossi G, Ragazzi M, Tamagnini I, et al. Does immunohistochemistry represent a robust alternative technique in determining drugable predictive gene alterations in non-small cell lung cancer? Curr Drug Targets. 2017;18(1):13–26. | ||
Lira ME, Choi YL, Lim SM, et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn. 2014;16(2):229–243. | ||
Murphy DA, Ely HA, Shoemaker R, et al. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol. Epub 2016 Mar 29. | ||
Lin JJ, Ritterhouse LL, Ali SM, et al. ROS1 fusions rarely overlap with other oncogenic drivers in non-small cell lung cancer. J Thorac Oncol. 2017;12(5):872–877. | ||
Cargnelutti M, Corso S, Pergolizzi M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget. 2015;6(7):5182–5194. | ||
Wiesweg M, Eberhardt WE, Reis H, et al. High prevalence of concomitant oncogene mutations in prospectively identified patients with ROS1-positive metastatic lung cancer. J Thorac Oncol. 2017;12(1):54–64. | ||
Scheffler M, Schultheis A, Teixido C, et al. ROS1 rearrangements in lung adenocarcinoma: prognostic impact, therapeutic options and genetic variability. Oncotarget. 2015;6(12):10577–10585. | ||
Lindeman NI, Cagle PT, Beasley MB, et al; College of American Pathologists International Association for the Study of Lung C, Association for Molecular P. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn. 2013;15(4):415–453. | ||
Zheng Z, LiebersM, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.