Back to Journals » Infection and Drug Resistance » Volume 14
Detection of Plasmid-Mediated Mobile Colistin Resistance Gene (mcr-1) in Enterobacterales Isolates from a University Hospital
Authors Anan MMG, El-Seidi EA, Mostafa MS , Rashed LA, El-Wakil DM
Received 19 May 2021
Accepted for publication 16 July 2021
Published 11 August 2021 Volume 2021:14 Pages 3063—3070
DOI https://doi.org/10.2147/IDR.S318787
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mera Mohammed Galal Anan,1 Eman Ahmed El-Seidi,1 Marwa Salah Mostafa,1 Laila Ahmed Rashed,2 Doaa Mahdy El-Wakil1
1Department of Medical Microbiology and Immunology, Faculty of Medicine, Cairo University, Cairo, Egypt; 2Department of Biochemistry and Molecular Biology, Faculty of Medicine, Cairo University, Cairo, Egypt
Correspondence: Mera Mohammed Galal Anan
Department of Medical Microbiology and Immunology, Faculty of Medicine, Cairo University, Kasr Al-Ainy St., Cairo, 11562, Egypt
Tel +20 1288866285
Email [email protected]
Purpose: Colistin represents one of the last treatment options for infections caused by multi-drug resistant (MDR) Enterobacterales. The emergence of a plasmid-mediated mobile colistin resistance-1 (mcr-1) gene has raised serious concerns about its potential dissemination among bacteria.
Methods: In this study, we evaluated the chromogenic medium, CHROMID® Colistin Resistance (COLR) agar, for the rapid detection of colistin-resistant Enterobacterales using broth microdilution (BMD) as a reference method. We also attempted to detect mcr-1, − 2, − 3, − 4, and − 5 genes, as well as the insertion sequence ISApl1 via polymerase chain reaction (PCR), followed by sequencing of mcr gene(s).
Results: Among the 100 studied Enterobacterales isolates, 53% of them were colistin-resistant, with higher rate among Klebsiella pneumoniae (75%) as compared to Escherichia coli (44.4%). The COLR agar showed 83.2% sensitivity and 97.9% specificity for the detection of colistin resistance. Among colistin-resistant isolates, mcr-1 gene was only detected in four (7.5%) E. coli isolates. The ISApl1 was not found among mcr-1 positive isolates. Sequencing of mcr-1 gene revealed nucleotide sequence homogeneity with the wild-type mcr-1 gene in BLAST.
Conclusion: The COLR agar is a promising phenotypic method for the detection of colistin-resistant Enterobacterales. Multiplex PCR followed by sequencing can be used for mcr genes’ detection and characterization.
Keywords: colistin resistance, Enterobacterales, mcr genes, CHROMID® COLR agar, multiplex PCR, sequencing
Introduction
The ability of Enterobacterales to acquire mobile genetic elements carrying antibiotic resistance has facilitated the development of resistance to multiple antibiotics with subsequent spread of multi-drug resistant (MDR) members of Enterobacterales.1 Colistin resistance has frequently been reported in areas with high rates of carbapenem-resistant Gram-negative bacteria with subsequent widespread use of colistin in clinical practice.2 Polymyxins have gained attention as a last resort antibiotic for the treatment of infections caused by MDR Gram-negative bacteria.3 They have been introduced to the antibiotic armamentarium since 1950s.4 Polymyxin B and polymyxin E (colistin) are used in clinical practice, with almost similar biological activity and differentiated by only a single amino acid.5 These cationic polypeptides target the outer membrane of Gram-negative bacteria,6 therefore destabilizing the lipopolysaccharide (LPS) and consequently increasing the membrane permeability, leading to leakage of the cytoplasmic contents and ultimately causing cell death.7
Various bacterial species are intrinsically resistant to colistin, whereas acquired resistance has been developed due to chromosomal mutations or acquisition of genes carried on plasmids.8 The plasmid-mediated mobile colistin resistance-1 (mcr-1) gene was first recognized in China.5 It encodes the enzyme phosphoethanolamine transferase that modifies the lipid A in the LPS of the bacterial outer membrane, thus decreasing the attachment of colistin, and hence inhibits cell lysis.9
Enterobacterales are the main host of mcr-1 gene which was first located in Escherichia coli then spread to other nosocomial bacterial pathogens; such as Klebsiella pneumoniae to become a source of multiple outbreaks.10 Other Plasmid-mediated colistin resistance genes, including mcr-2, -3, -4, and -5, have been discovered in Enterobacterales strains recovered from human hosts.11 The prevalence of mcr-6, -7, -8, -9, and -10 genes in Enterobacterales is very rare.12,13 It has been shown that mcr-1 sequences are sometimes associated with an upstream and/or downstream ISApl1 copy, an insertion sequence that may play a pivotal role in the mobilization of mcr-1 gene.14,15 However, some mcr-1 sequences lack the insertion sequence ISApl1.16
Detection of colistin-resistant Enterobacterales is usually done by reliable phenotypic techniques, such as the broth microdilution (BMD) method. However, it is quite laborious, time-consuming, and difficult to interpret, and therefore not suitable for most clinical microbiology laboratories.17 Agar dilution is another reference method that can test several strains on the same plate. However, it is time-consuming, and the plates must be used within a week of preparation.6 Disk diffusion is a simple and inexpensive method. However, polymyxins show slow diffusion through the agar which results in small inhibition zones, limiting the predictive accuracy of this method. Therefore, it is not recommended for susceptibility testing of polymyxins.6 Currently, broth disk elution and agar dilution MIC methods are acceptable for testing colistin susceptibility among Enterobacterales, however these methods yield inaccurate results with Acinetobacter spp.18 Alternatively, chromogenic media can be used for screening of colistin resistance among Enterobacterales as they are rapid, accurate, and affordable.19 Meanwhile, genotypic detection of mcr genes can be achieved by the gold standard molecular techniques.20
The aim of the current study was to evaluate the usefulness of CHROMID® Colistin Resistance (COLR) agar as a screening method for the detection of colistin resistance among Enterobacterales as compared to the gold standard BMD method. We also attempted to detect the presence of mcr-1, -2, -3, -4, and -5 genes via multiplex polymerase chain reaction (PCR), to be followed by sequencing of the mcr-1 gene. Finally, the study aimed to evaluate the association of the insertion sequence ISApl1 with mcr-1 gene, if present.
Materials and Methods
This cross-sectional study was conducted during the period from September 2018 through March 2020 and included a total of 100 Enterobacterales isolates (72 E. coli and 28 K. pneumoniae), obtained from the Strain Bank of the Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University, Egypt. The isolates were cultured on MacConkey’s agar plates (Oxoid, UK) and incubated aerobically at 37°C for 24 hours. Although the isolates obtained from the Strain Bank were previously identified, identification was confirmed by conventional microbiological methods including colony morphology, Gram staining, sugar fermentation, and oxidase test. Further identification up to the species level was done by conventional methods including culture on triple sugar iron agar (TSI), motility indole ornithine (MIO), lysine iron agar (LIA), urease test, and citrate test.21,22 These isolates were selected on the basis of being MDR; they constitute 93% ESBL producers, 5% carbapenem-resistant isolates, and 2% AmpC producers. The isolates in the Strain Bank were previously identified as ESBL producers by double disc diffusion23 and by conventional PCR.24 Carbapenem-resistance and AmpC production were detected using the automated Vitek 2 system (bioMérieux, France). The isolates were preserved at −20°C from urine samples collected from hospitalized patients at Kasr Al-Ainy Hospital.25 Informed consent was obtained from each participant. The study protocol was approved by the Ethical Committee of the Medical Microbiology and Immunology Department, Cairo University.
Detection of Colistin Resistance by COLR Agar Medium
Enterobacterales isolates were tested for colistin resistance using COLR medium (bioMérieux, France, cat no. 421170) according to the manufacturer’s instructions.
Detection of Colistin Resistance Using the Broth Microdilution (BMD) Method
The colistin minimum inhibitory concentrations (MICs) of all Enterobacterales isolates were determined by the BMD method according to the Clinical and Laboratory Standard Institute (CLSI) guidelines. Colistin MICs of ≤ 2 µg ∕mL were interpreted as susceptible, whereas MICs of ≥ 4 µg ∕mL were interpreted as resistant.23 The colistin-susceptible E. coli ATCC 25922 strain, with a colistin MIC susceptibility ranging from 0.25–2 µg/mL, was used as a quality control.
Genotypic Detection of Plasmid-Mediated mcr-1, 2, 3, 4, and 5 Genes
Genotypic analysis methods were carried out in the molecular biology unit at the Medical Biochemistry Department, Faculty of Medicine, Cairo University. Genomic DNA was extracted from fresh bacterial isolates using QIAamp DNA Mini Kit (Qiagen, Germany, cat. no. 51306) according to the manufacturer’s instructions. Multiplex PCR was performed for the amplification of mcr-1, -2, -3, -4, and -5 genes using the primers as previously described.11 A total of 35 PCR cycles were done as follows: 94°C for 30 seconds, 58°C for 90 seconds, and 72°C for 60 seconds. An initial denaturation step at 94°C for 2 minutes and a final extension step at 72°C for 10 minutes were performed. Detection of PCR products was done by 2% agarose gel electrophoresis stained with ethidium bromide and visualized under UV illumination using a Biometra T11 Gel documentation system (Biometra, Germany). Visual detection of the expected DNA bands at 320 bp, 715 bp, 929 bp, 1,116 bp, or 1,644 bp was indicative of a positive isolate harboring mcr-1, mcr-2, mcr-3, mcr-4, or mcr-5 gene, respectively.11
Detection of Insertion Sequence ISApl1 in mcr-1 Positive Isolates
Amplification of ISApl1 insertion sequence was done on the extracted DNA from mcr-1 positive isolates using the previously published primers: ISApl1-mcr-F TGGACATTGGGAAGCCGATA and ISApl1-mcr-R GCCACAAGAACAAACGGACT.26 Detection of the amplified PCR product ISApl1 (707 bp length) was done using 2% agarose gel electrophoresis.
DNA Sequencing of mcr-1 Gene
The amplified PCR products from mcr-1 positive isolates were sent to GATC Biotech, Germany for DNA sequencing. Initially, purification of the amplified PCR products was done using the GeneJET PCR Purification Kit (Thermo scientific, USA, cat. no. K0702) according to the manufacturer’s instructions. Then after, genomic DNA sequencing was done by Sanger’s method in the forward direction using the mcr-1 gene forward primer; as described before.11 The obtained sequences were compared to known sequences deposited at the GenBank BLAST program, National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov).
Statistical Analysis
All statistical analyses were performed using the statistical package SPSS (Statistical Package for the Social Sciences) version 25. Mean, standard deviation, median, and interquartile range (IQR) were used for quantitative data, while frequency (count) and relative frequency (percentage) were used for categorical data. Comparisons between quantitative variables were done using the non-parametric Mann Whitney U-test. For evaluation of the chromogenic agar in comparison to the BMD method, standard diagnostic indices including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and diagnostic accuracy were calculated. The chi-square (χ2) test was used to compare categorical data, but the Exact test was used instead when the expected frequency was less than 5. A p-value of ≤ 0.05 was considered as statistically significant.
Results
The present study was carried out on 100 Enterobacterales isolates retrieved from 36 male and 64 female patients; their ages ranged from 2–91 years (mean=47±23 years). The isolates were identified as 72 (72%) E. coli and 28 (28%) K. pneumoniae.
Detection of Colistin Resistance Using COLR Agar
Cultivation of the Enterobacterales isolates on COLR agar after 4‒5 hours incubation in brain heart infusion (BHI) broth containing 10 µg colistin disk revealed that 45/100 (45%) of the isolates showed a positive growth, indicating colistin resistance. Twenty-nine E. coli isolates (29/72, 40.3%) and 16 K. pneumoniae isolates (16/28, 57.1%) were colistin-resistant.
Detection of Colistin Resistance Among Enterobacterales Isolates Using BMD Method
The BMD method showed that out of the 100 studied Enterobacterales isolates, 53 (53%) colistin-resistant isolates were found, among which, 32/72 (44.4%) of E. coli and 21/28 (75%) of K. pneumoniae isolates were colistin-resistant. Interestingly, all of the five carbapenem-resistant isolates included in the study were colistin-resistant. On the other hand, colistin resistance rate among both ESBL and AmpC producers was approximately 50%.
Correlation Between the COLR Agar and BMD Method
Considering the BMD method as the gold standard test for the detection of colistin resistance, growth of Enterobacterales isolates on the COLR agar showed 83.2% sensitivity, 97.9% specificity, 97.8% positive predictive value (PPV), 83.6% negative predictive value (NPV), and 90% accuracy (Table 1). The value of Kappa (statistical measurement of agreement) indicates a significant agreement between the COLR agar and BMD method (p<0.001).
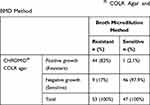 |
Table 1 Correlation Between CHROMID® COLR Agar and BMD Method |
The MIC values were significantly higher in colistin-resistant Enterobacterales isolates grown on COLR agar (median=4; range=2‒32) compared to those that failed to grow (median=2; range=1‒4) (p<0.001). Interestingly, the COLR agar had high sensitivity (100%) in the detection of colistin-resistant Enterobacterales isolates with MIC values ≥8, its sensitivity decreased to 75.7% in isolates with colistin MIC=4. Moreover, 97.8% (46/47) of colistin-susceptible isolates (MIC<4) did not grow on the COLR agar. The distribution of MIC values of colistin is shown in Figure 1.
 |
Figure 1 Colistin MIC values among Enterobacterales in relation to the results of CHROMID® COLR agar. Abbreviations: MIC, minimum inhibitory concentration; COLR, colistin resistance. |
Detection of Plasmid-Mediated mcr-1, 2, 3, 4, and 5 Genes
We found that out of the 53 colistin-resistant isolates detected by BMD method, only four (7.5%) isolates were positive for the presence of mcr-1 gene (Figure S1). All the mcr-1 positive isolates were E. coli, representing 5.6% (4/72) of the total E. coli isolates. Meanwhile, all colistin-resistant isolates were tested negative for the presence of other mcr genes (mcr-2, 3, 4, and 5). Moreover, in the 47 colistin-susceptible isolates, none of the five mcr genes could be detected.
Colistin MICs of the four mcr-1 positive isolates ranged between 4‒16 µg/mL. We also demonstrated significantly higher colistin MICs in mcr-1 positive isolates (median=8, IQR=6‒12) compared to mcr negative isolates (median=4, IQR=2‒4) (p=0.022).
Detection of Insertion Sequence ISApl1 Among mcr-1 Positive Isolates
Conventional PCR was done to detect the presence of ISApl1 among the four mcr-1 positive E. coli isolates. However, ISApl1 could not be detected in any of them.
Sequencing of mcr-1 Gene
The four mcr-1 positive E. coli isolates were further sequenced by an ABI 3730 XL automated DNA sequencer. Three isolates matched the sequence of mcr-1 gene when compared to the prototype sequence of the wild mcr-1 gene in the BLAST system (Figure S2), while only one isolate failed to be read by the sequencer, probably due to product degradation during transportation. Comparing the three chromatograms with the mcr-1 wild gene sequence in BLAST using multiple alignment confirmed the homogeneity of the nucleotide sequence for the mcr-1 gene (Table S1).
Discussion
Massive and inappropriate use of antibiotics generates a selective pressure that is followed by rapid emergence and spread of MDR Enterobacterales.27 Colistin is considered one of the last lines of therapy that was used separately or in combination with other antibiotics to effectively treat serious infections caused by MDR pathogens.28 In the present study, we found that out of 100 Enterobacterales isolates, 53% were colistin-resistant, with a 44.4% (32/72) resistance rate among E. coli isolates and a 75% (21/28) resistance rate among K. pneumoniae isolates. Approximately similar rates of colistin resistance were reported by an earlier study conducted in France that detected a 53.1% resistance rate among E. coli and a 76% resistance rate among K. pneumoniae isolates, using the BMD method.29 On the other hand, another study carried out in India demonstrated that out of 64 Enterobacterales isolates tested for colistin MICs by BMD method, none of them were colistin-resistant.30
The discrepancy in colistin resistance among different studies is explained by the diversity in the type of specimens, number of cases, the general condition of patients, geographical regions, various antibiotic policies, and compliance to the infection control measures. Meanwhile, the high rate of colistin resistance in the current study can be elucidated by the fact that the isolates were previously collected from hospitalized patients with urinary tract infection; where the resistance pattern is more common among hospitalized patients due to extensive antibiotic use, the presence of co-morbidities, and the existence of indwelling catheters.
In the present study, the COLR agar showed that 45 (45%) isolates were colistin-resistant, with MICs ranging from 2‒32 μg/mL. Considering the BMD method as the gold standard for the detection of colistin resistance in the current study, the sensitivity, specificity, PPV, NPV, and accuracy of the COLR agar were 83.2%, 97.9%, 97.8%, 83.6%, and 90%, respectively, with 100% sensitivity in the detection of colistin-resistant Enterobacterales isolates with MIC values ≥8 μg/mL. Failure of detection of low-level resistant isolates in the present study may be attributed to the high concentration (10 μg) of colistin used or other constituents of the COLR agar that may have an antimicrobial effect. The efficacy of the COLR agar in the detection of colistin resistance among Enterobacterales was in agreement with García-Fernández et al,19 who revealed that COLR agar has a sensitivity of 88% and a specificity of 100% when compared with the standard BMD method.
In the current study, the mcr-1 gene was detected in four colistin-resistant E. coli isolates; 4% (4/100) of the total isolates, 7.5% (4/53) of colistin-resistant isolates, and 5.6% (4/72) of E. coli isolates. The remaining four mcr genes (mcr-2, 3, 4, and 5) were not detected in any of the studied isolates. All the four mcr-1 positive isolates demonstrated significantly higher colistin MICs compared to mcr-1 negative isolates (p=0.022). The inability to detect mcr genes among the remaining colistin-resistant isolates could be explained by either the presence of other plasmid or chromosomally mediated resistance mechanisms, extrusion of the drug by efflux pump, decreased permeability of bacterial cell membrane, or enzyme-mediated inactivation.
A closely similar rate was observed in a study done in Hong Kong31 which reported that out of 62 colistin-resistant Enterobacterales isolates of human origin, 6.5% (4/62) of E. coli isolates were mcr-1 positive. Similarly, a study conducted in Oman reported that out of 22 colistin-resistant Enterobacterales isolates, a single (4.5%) E. coli isolate carrying mcr-1 gene was detected, whereas none of them was proved to carry the mcr-2 gene.32 A lower incidence rate of the mcr-1 gene was detected by an Egyptian study33 which revealed that among 241 Gram-negative isolates collected from different hospitals, mcr-1 was detected in only one E. coli (0.4%) isolated from sputum of a patient with bacteremia with no history of traveling abroad. A higher incidence rate of the mcr-1 gene was reported by Rebelo et al,11 who found that out of 42 E. coli isolates of animal origin, 14 (33.3%) isolates carried mcr genes and were distributed as follows: nine (21.4%) E. coli isolates harbored mcr-1 gene, one (2.3%) isolate harbored mcr-3, and one (2.3%) isolate harbored mcr-4. Co-occurrence of mcr-1 and mcr-3 was observed in two (4.7%, 2/42) E. coli isolates, whereas a single (2.3%, 1/42) E. coli isolate harbored both mcr-1 and mcr-4 genes. The high prevalence of mcr-1 in animal isolates compared with human clinical isolates worldwide indicates that animals and their products are possible sources of mcr-1 in humans. Moreover, the misuse of colistin in agriculture and the poultry industry may be the main cause of the high incidence of mcr-1 in bacteria isolated from animals and animal products.34
The mobile colistin resistance-1 (mcr) gene is more commonly isolated from E. coli isolates than from K. pneumoniae, a finding that is supported by our study as well as by previous studies.11,19,34,35 Additionally, a previous Egyptian study found that the eight colistin-resistant K. pneumoniae isolates were negative for mcr-1 gene.17 Another study showed that two E. coli isolates and two K. pneumoniae isolates were colistin-resistant and none of them was positive for mcr-1 gene.36
In the current study, sequencing of the mcr-1 gene of the four positive isolates followed by multiple alignments of the chromatograms revealed homogeneity of the nucleotide sequence for mcr-1 among three of the isolates when compared to the NCBI database. Consistent with our results, Johura et al37 confirmed the homogeneity of the nucleotide sequence of mcr-1 in 13 E. coli strains. Moreover, they reported that their E. coli strains were heterogenous, as confirmed by pulsed-field gel electrophoresis, suggesting horizontal transmission of colistin resistance. It is worth mentioning that several variants of mcr-1 have been identified from E. coli strains isolated from animal, sewage, or human urinary tract samples in various countries.38–40
In the present study, we could not detect the ISApl1 in any of the four mcr-1 positive isolates. This finding was consistent with a study done in Oman by Mohsin et al,32 who demonstrated the lack of ISApl1 in the genetic surrounding of the mcr-1 gene. Additionally, a study from the Czech Republic confirmed the absence of ISApl1 element upstream of mcr-1 gene located on IncI2 plasmids.41 On the contrary, Snesrud et al16 found ISApl1 in 23 (30%) out of 77 mcr-1 positive isolates. However, Wang et al35 did not find the ISApl1 in 56.9% (260/457) of the mcr-1 positive isolates, indicating that the mcr-1 transposon may have been completely stabilized in their genomic background. Failure of the detection of the ISApl1 in our study could be attributed to the small number of mcr-1 positive isolates. Furthermore, the current study did not include other members of Enterobacterales which may have affected the actual prevalence of mcr genes among Enterobacterales.
The emergence of plasmid-mediated colistin resistance in Enterobacterales is currently a crucial issue owing to the high potential of their dissemination in clinical settings. It is important to critically develop proper guidelines against the use of this last-line treatment option, so that the spread of resistance can be limited. For better understanding of the actual status of the global colistin-resistance, development and implementation of rapid procedures to detect colistin resistance in clinical microbiology laboratories should be enhanced.
This study has two limitations. First, our investigation was restricted to E. coli and K. pneumoniae isolates obtained from urine samples only. However, E. coli and K. pneumoniae are the most common members of Enterobacterales recovered from clinical specimens. Moreover, urinary tract infections are the most common type of infections caused by Enterobacterales that are encountered in hospitals.42 Future studies investigating other members of Enterobacterales and different types of samples are required. Second, there is a need for overseas transportation of the isolates for sequencing of PCR products which leads to degradation of one of the isolate sequences and subsequent failure of sequence reading.
Conclusion
In summary, the chromogenic COLR agar medium offers a simple, easy, and inexpensive method for rapid detection of colistin-resistant Enterobacterales. However, it is not recommended as a screening test due to its inadequate sensitivity and accuracy. The BMD method remains the more sensitive standard test for colistin resistance. Multiplex PCR is suggested for the determination of the presence of mcr genes in laboratories with limited resources. However, its value for the detection of mcr genes is debatable due to the low prevalence of mcr genes in our hospital. Therefore, further studies using larger sample size are recommended to elucidate the role of mcr genes in colistin resistance as well as to confirm or exclude the association between mcr-1 gene and the insertion sequence ISApl1. In addition, future investigations will be necessary to detect other mechanisms of colistin resistance among Enterobacterales isolates negative for mcr genes, such as mgrB genes inactivation or the presence of insertion sequence disrupting this chromosomal gene.
Ethics Approval and Informed Consent
There is no ethical concern in this study, and this experiment was approved by the Ethical Committee of the Medical Microbiology and Immunology Department, Cairo University. Written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Acknowledgments
The authors are grateful to Kasr Al-Ainy medical school for allowing this work to be conducted, and specifically the Microbiology and Immunology department to provide the required strains.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019;65(1):34–44. doi:10.1139/cjm-2018-0275
2. Büchler AC, Gehringer C, Widmer AF, Egli A, Tschudin-Sutter S. Risk factors for colistin-resistant Enterobacteriaceae in a low-endemicity setting for carbapenem resistance –a matched case– control study. Euro Surveill. 2018;23(30):1700777. doi:10.2807/1560-7917.ES.2018.23.30.1700777
3. Timmerman R, McGarry R, Yiannoutsos C, et al. Excessive toxicity when treating central tumors in a Phase II study of stereotactic body radiation therapy for medically inoperable early-stage lung cancer. J Clin Oncol. 2006;24(30):4833–4839. doi:10.1200/JCO.2006.07.5937
4. Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of. Escherichia Coli Lancet Infect Dis. 2016;16(3):281. doi:10.1016/S1473-3099(16)00006-2
5. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
6. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi:10.1128/CMR.00064-16
7. Falagas ME, Kasiakou SK, Saravolatz LD. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis. 2005;40(9):1333–1341. doi:10.1086/429323
8. Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat. 2010;13(4–5):132–138. doi:10.1016/j.drup.2010.05.002
9. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. doi:10.1016/S1473-3099(16)00061-X
10. Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 2016;16(3):287–288. doi:10.1016/S1473-3099(16)00056-6
11. Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6):17–00672. doi:10.2807/1560-7917.ES.2018.23.6.17-00672
12. Ngbede EO, Poudel A, Kalalah A, et al. Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int J Antimicrob Agents. 2020;56(3):106108. doi:10.1016/j.ijantimicag.2020.106108
13. Borowiak M, Baumann B, Fischer J, et al. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front Microbiol. 2020;11:80. doi:10.3389/fmicb.2020.00080
14. Zurfluh K, Tasara T, Poirel L, Nordmann P, Stephan R. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc. 2016;4(4):e00796–16.
15. Li R, Xie M, Zhang J, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393–401. doi:10.1093/jac/dkw411
16. Snesrud E, He S, Chandler M, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60(11):6973–6976. doi:10.1128/AAC.01457-16
17. Emara MM, Abd-Elmonsef MM, Abo Elnasr LM, Elfeky AA. Study of mcr-1 gene-mediated colistin resistance in Gram-negative isolates in Egypt. Egypt J Med Microbiol. 2019;28(3):9–16.
18. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
19. García-Fernández S, García-Castillo M, Ruiz-Garbajosa P, et al. Performance of CHROMID® Colistin R agar, a new chromogenic medium for screening of colistin-resistant Enterobacterales. Diagn Microbiol Infect Dis. 2019;93(1):1–4. doi:10.1016/j.diagmicrobio.2018.07.008
20. Nordmann P, Jayol A, Poirel L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis. 2016;22(6):1038–1043. doi:10.3201/eid2206.151840
21. Cricton P. Enterobacteriaceae. In: Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology.
22. Mladenović KG, Muruzović MŽ, Žugić Petrović T, Stefanović OD, Čomić LR. Isolation and identification of Enterobacteriaceae from traditional Serbian cheese and their physiological characteristics. J Food Saf. 2018;38(1):e12387. doi:10.1111/jfs.12387
23. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
24. Yuan XY, Yu DY, Qu XH, et al. Increased resistance rate to ceftazidime among blood culture isolates of ESBL-producing Escherichia coli in a university-affiliated hospital of China. J Antibiot. 2016;69(3):169–172. doi:10.1038/ja.2015.100
25. Collee JG, Marr W. Specimen collection, culture containers and media. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology.
26. Veldman K, van Essen-zandbergen A, Rapallini M, et al. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother. 2016;71(8):2340–2342. doi:10.1093/jac/dkw181
27. Malik B, Bhattacharyya S. Antibiotic drug resistance as a complex system driven by socio-economic growth and antibiotic misuse. Sci Rep. 2019;9(1):1–12.
28. Tran TB, Velkov T, Nation RL, et al. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int J Antimicrob Agents. 2016;48(6):592–597. doi:10.1016/j.ijantimicag.2016.09.010
29. Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect. 2018;24(2):175–179. doi:10.1016/j.cmi.2017.06.002
30. Shaikh S, Pandya H, Arora S, Kamtekar T. Comparison of colistin susceptibility testing by Vitek 2 compact and broth microdilution method for carbapenem-resistant isolates in a tertiary diagnostic center. Int J Intern Med Geriatr. 2019;1(2):54–58.
31. Wong SC, Tse H, Chen JH, Cheng VC, Ho PL, Yuen KY. Colistin-resistant Enterobacteriaceae carrying the mcr-1 gene among patients in Hong Kong. Emerg Infect Dis. 2016;22(9):1667–1669. doi:10.3201/eid2209.160091
32. Mohsin J, Pál T, Petersen JE, et al. Plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli ST10 bloodstream isolate in the Sultanate of Oman. Microb Drug Resist. 2018;24(3):278–282. doi:10.1089/mdr.2017.0131
33. Elnahriry SS, Khalifa HO, Soliman AM, et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016;60(5):3249–3250. doi:10.1128/AAC.00269-16
34. Tanfous FB, Raddaoui A, Chebbi Y, Achour W. Epidemiology and molecular characterization of colistin-resistant Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia. Int J Antimicrob Agents. 2018;52(6):861–865. doi:10.1016/j.ijantimicag.2018.08.022
35. Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi:10.1016/S1473-3099(16)30527-8
36. Liassine N, Assouvie L, Descombes MC, et al. Very low prevalence of mcr-1/mcr-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int J Infect Dis. 2016;51:4–5. doi:10.1016/j.ijid.2016.08.008
37. Johura FT, Tasnim J, Barman I, et al. Colistin-resistant Escherichia coli carrying mcr-1 in food, water, hand rinse, and healthy human gut in Bangladesh. Gut Pathog. 2020;12(1):1–8. doi:10.1186/s13099-020-0345-2
38. Tijet N, Faccone D, Rapoport M, et al. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One. 2017;12(7):e0180347. doi:10.1371/journal.pone.0180347
39. Yang YQ, Li YX, Song T, et al. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother. 2017;61(5):e01204–16. doi:10.1128/AAC.01204-16
40. Zhao F, Feng Y, Lü X, McNally A, Zong Z. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol. 2017;8:2094. doi:10.3389/fmicb.2017.02094
41. Zelendova M, Papagiannitsis CC, Valcek A, et al. Characterization of the complete nucleotide sequences of mcr-1-encoding plasmids from Enterobacterales isolates in retailed raw meat products from the Czech Republic. Front Microbiol. 2021;11:3552. doi:10.3389/fmicb.2020.604067
42. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi:10.1038/nrmicro3432
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
