Back to Journals » Infection and Drug Resistance » Volume 13
Detection of Novel Gene Mutations Associated with Pyrazinamide Resistance in Multidrug-Resistant Mycobacterium tuberculosis Clinical Isolates in Southern China
Authors Hameed HMA , Tan Y, Islam MM , Lu Z, Chhotaray C, Wang S , Liu Z, Fang C , Tan S , Yew WW, Zhong N, Liu J, Zhang T
Received 12 September 2019
Accepted for publication 20 December 2019
Published 22 January 2020 Volume 2020:13 Pages 217—227
DOI https://doi.org/10.2147/IDR.S230774
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eric Nulens
Video abstract presented by HM Adnan Hameed.
Views: 346
HM Adnan Hameed, 1, 2,* Yaoju Tan, 3,* Md Mahmudul Islam, 1, 2 Zhili Lu, 1 Chiranjibi Chhotaray, 1, 2 Shuai Wang, 1, 2 Zhiyong Liu, 1 Cuiting Fang, 1, 2 Shouyong Tan, 3 Wing Wai Yew, 4 Nanshan Zhong, 5 Jianxiong Liu, 3 Tianyu Zhang 1, 2, 5
1State Key Laboratory of Respiratory Disease, Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (CAS), Guangzhou, People’s Republic of China; 2University of Chinese Academy of Sciences (UCAS), Beijing, People’s Republic of China; 3State Key Laboratory of Respiratory Disease, Guangzhou Chest Hospital, Guangzhou, People’s Republic of China; 4Stanley Ho Centre for Emerging Infectious Diseases, The Chinese University of Hong Kong, Hong Kong, People’s Republic of China; 5National Clinical Research Center for Respiratory Disease, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tianyu Zhang
Guangzhou Institutes of Biomedicine and Health (GIBH), Chinese Academy of Sciences (CAS), Room A132, 190 Kaiyuan Ave, Science Park, Huangpu District, Guangzhou 510530, People’s Republic of China
Tel +86-2032015270
Email [email protected]
Jianxiong Liu
Guangzhou Chest Hospital, 62 Hengzhigang Road, Yuexiu District, Guangzhou 510095, People’s Republic of China
Tel +86-2083595977
Email [email protected]
Objective: Pyrazinamide (PZA) is a cornerstone of modern tuberculosis regimens. This study aimed to investigate the performance of genotypic testing of pncA+ upstream region, rpsA, panD, Rv2783c, and clpC1 genes to add insights for more accurate molecular diagnosis of PZA-resistant (R) Mycobacterium tuberculosis.
Methods: Drug susceptibility testing, sequencing analysis of PZA-related genes including the entire operon of pncA (Rv2044c-pncA-Rv2042c) and PZase assay were performed for 448 M. tuberculosis clinical isolates.
Results: Our data showed that among 448 M. tuberculosis clinical isolates, 113 were MDR, 195 pre-XDR and 70 XDR TB, while the remaining 70 strains had other combinations of drug-resistance. A total of 60.04% (269/448) M. tuberculosis clinical isolates were resistant to PZA, of which 78/113 were MDR, 119/195 pre-XDR and 29/70 XDR TB strains. PZAR isolates have predominance (83.3%) of Beijing genotype. Genotypic characterization of Rv2044c-pncA-Rv2042c revealed novel nonsynonymous mutations in Rv2044c with negative PZase activity which led to confer PZAR. Compared with phenotypic data, 84.38% (227/269) PZAR strains with mutations in pncA+ upstream region exhibited 83.64% sensitivity but the combined evaluation of the mutations in rpsA 2.60% (7/269), panD 1.48% (4/269), Rv2783c 1.11% (3/269) and Rv2044c 0.74% (2/269) increased the sensitivity to 89.59%. Fifty-seven novel mutations were identified in this study. Interestingly, a frameshift deletion (C− 114del) in upstream of pncAwt nullified the effect of A− 11G mutation and induced positive PZase activity, divergent from five PZase negative A− 11G PZAR mutants. Twenty-six PZAR strains having wild-type-sequenced genes with positive or negative PZase suggest the existence of unknown resistance mechanisms.
Conclusion: Our study revealed that PZAR rate in MDR and pre-XDR TB was markedly higher in southern China. The concomitant evaluation of pncA+ UFR, rpsA, panD, Rv2783c, and Rv2044c provides more dependable genotypic results of PZA resistance. Fifty-seven novel mutations/indels in this study may play a vital role as diagnostic markers. The upstream region of pncA and PZase regulation are valuable to explore the unknown mechanism of PZA-resistance.
Keywords: tuberculosis, pyrazinamidase, drug resistance, molecular diagnosis, novel mutations, frameshift deletion
Introduction
Pyrazinamide (PZA), an analog of nicotinamide, is a key component of current and new anti-tuberculosis (TB) regimens for treatment of both drug-susceptible and multidrug-resistant (MDR) TB, because it plays a critical role in shortening the TB therapy from 9 to 12 months to 6 months.1 PZA has a unique sterilizing activity against Mycobacterium tuberculosis persisters that are not killed effectively by other anti-TB drugs, but its mechanism of action is complex and not well understood yet.2
PZA is a prodrug that is converted to its active form, pyrazinoic acid (POA), by an enzyme, pyrazinamidase (PZase), encoded by pncA gene.3 Averagely, 70% to 90% of PZA resistance (R) incidences are due to mutations in the pncA gene.4,5 In recent studies, RpsA which encodes the ribosomal protein S1, involved in translation and trans-translation, has been reported as a target of PZA.6 The mutations in rpsA seem to have a minor role in PZAR in the clinical isolates having no mutation in pncA gene.7 Besides, another gene panD encoding aspartate decarboxylase involved in beta-alanine and pantothenate synthesis was recently identified, whose mutations were also associated with PZAR in M. tuberculosis strains lacking pncA and rpsA mutations.8
In our recent study, we discovered Rv2783 as a new potential target of POA in M. tuberculosis.9 Rv2783, a bifunctional enzyme which was proved to be able to synthesize and hydrolyze (p)ppGpp and to synthesize and hydrolyze ssDNA and ssRNA without any template.9 In addition to the above genes, ClpC1 was reported as another new target of PZA which acts as an unfoldase in concert with the proteases ClpP1 and ClpP2 of the caseinolytic protease complex but the mutations in the coding region of ClpC1 were associated with PZAR.10
The Wayne assay exhibits the association between PZase activity and genetic mutations in PZA-associated genes. Mutations in pncA gene are considered to be responsible for the failure of key-lock recognition mechanism between the enzyme and its substrate. Similarly, when mutation occurs in the upstream flanking region (UFR) of pncA, the RNA polymerase may not be able to transcribe it. Thus, there is no PZase activity and PZA cannot be converted into POA.11 pncA (Rv2043c) is co-transcribed as a polycistron (Rv2044c-pncA-Rv2042c) with its upstream gene Rv2044c and downstream gene Rv2042c.12 However, apart from frequently observed mutations at position −11 in the putative regulatory region of pncA, no reports showed yet that mutations in the instant upstream gene of wild-type (wt) pncAwt may develop PZAR in M. tuberculosis.
The prior studies described the correlation between PZAR and pncA mutations. Whereas no study has been conducted so far for simultaneous characterization of the mutations in pncA+ UFR, rpsA, panD, Rv2783c, and clpC1 for the detection of PZAR in MDR, pre-XDR (MDR with additional resistant to fluoroquinolone or a second-line injectable drug, e.g. kanamycin, amikacin) and XDR (MDR in addition with resistant to both fluoroquinolones and second-line injectable drugs) M. tuberculosis clinical isolates. The currently available phenotypic PZA susceptibility testing methods are complicated and deceptive because of frequent false-resistance and false-susceptible results.13 Genotypic characterization of PZA-related genes has been endorsed to overcome the shortcomings of phenotypic susceptibility testing methods.14
In this study, to evaluate the performance of genotypic testing method of PZA-associated genes for detecting PZAR strains, we performed the phenotypic and genotypic characterization of 448 M. tuberculosis clinical isolates from southern China. Considering the observation that UFR of pncA has an indispensable role in PZase regulation as well as PZAR,11,12 we have additionally evaluated the complete operon of pncA (Rv2044c-pncA-Rv2042c) to identify the possible role of surrounding genes of pncA in PZAR M. tuberculosis.
Materials and Methods
Ethical Approval
The current study was approved by the Ethics Committee of Guangzhou Chest Hospital (GZXK-2016-015) in accordance with the WHO-approved guidelines.
Collection of M. tuberculosis Clinical Isolates
In this study, 448 drug-resistant M. tuberculosis clinical isolates (resistant ≥ one anti-TB drug) were collected during the period from December 2016 to November 2018 from TB patients at Guangzhou Chest Hospital, the biggest TB-specialized hospital in southern China. Ziehl–Neelsen staining and commercial MPB64 monoclonal antibody assay (GENESIS, Hangzhou, China) were performed to confirm the M. tuberculosis species.15
Drug Susceptibility Testing
Drug susceptibility testing (DST) of 448 M. tuberculosis isolates was first assessed by Mycobacterial Growth Indicator Tubes (MGIT) 960 (Becton Dickinson, Franklin Lakes, NJ, USA). The critical concentrations (μg/mL) for DST were consistent with WHO recommendations; isoniazid (INH; 0.1), rifampicin (RIF; 1.0), ethambutol (EMB; 5.0), PZA (100), streptomycin (STR; 1.0), levofloxacin (LVX; 1.0), moxifloxacin (MXF; 0.25) and amikacin (AMK; 1.0).16 In order to measure the extent of phenotypic PZAR by the gold standard MGIT 960 system, the higher concentrations of PZA (300 and 900 μg/mL) were tested particularly for those PZAR (100 μg/mL) strains which showed inconsistent phenotypic and genotypic results. The DST results were also verified via indirect proportion method on Löwenstein–Jensen medium using the recommended concentrations (μg/mL); INH (0.2), RIF (40.0), EMB (2.0), STR (4.0), LVX (2.0), MXF (1.0) and AMK (30.0).16 The growth on a control medium (drug-free medium) was compared with the growth on drug-containing medium and the resistance was determined when 1% or more growth was noticed at the critical concentration of drug in the medium.16
PZase Activity Assay
PZase activity assay was performed with some modifications in a Wayne test8 to determine the PZase regulation. Briefly, 3 to 4 pure and freshly grown M. tuberculosis colonies on LJ medium were scraped off and transferred into 1 mL Middlebrook 7H9 medium supplemented with albumin-dextrose-catalase (ADC) containing PZA (100 and 200 μg/mL) in 1.5 mL Eppendorf tubes. The colonies were incubated at 37°C in a shaker for 3, 5, and 7 days. Later, 15 μL of 2% Fe2+ was added into the bacterial cells and incubated at 4°C for 2–4 hrs for color appearance. M. tuberculosis H37Rvwt (pyrazinamide-susceptible; PZAS) and M. bovis Bacillus Calmette-Guérin (BCG) Tice (PZAR) were used as positive and negative controls, respectively.
PZA-Associated Genes Amplification and Sequencing
Genomic DNA was extracted from freshly grown M. tuberculosis cultures using MagMAX Total Nucleic Acid Isolation Kit (Ambion, Life Technologies, NY, USA) according to the manufacturer’s instructions. All PZA-associated genes (pncA, rpsA, panD, Rv2783c and clpC1) including their 5ʹ upstream (≤ − 200 bp) to 3ʹ downstream (≤ + 200 bp) regions were amplified in all M. tuberculosis isolates using the newly designed primers in this study (Table 1). The complete operon of pncA (Rv2044c-pncA-Rv2042c) was additionally analyzed in PZAR strains. PCR products were examined on agarose gels, purified by PCR purification kit (Qiagen, Hilden, Germany) and sequenced at the BGI (Guangzhou, China). The DNA sequences were compared with the reference sequence of M. tuberculosis H37Rv (Accession number: NC_000962.3) using the software BioEdit version 7.2.6.1.
 |
Table 1 Primers and PCR Products of PZA-Associated Genes with Brief Explanatory Notes |
Detection of Beijing and Non-Beijing Genotypes
Multiplex PCR method was used to distinguish Beijing and non-Beijing genotypes as previously described17 that region spanning genes Rv2816 to Rv2819 including part of Rv2820 is missing in Beijing genotype of M. tuberculosis strain. A set of primers BJ-F: 5ʹ-ACCGAGCTGATCAAACCCG-3ʹ and BJ-R: 5ʹ-ATGGCACGGCCGACCTGAATGAACC-3ʹ amplified the 239 bp PCR product containing region-specific part of Rv2819 and part of Rv2820 in Beijing genotypes. Whereas another pair of primers NBJ-F: 5ʹ-GATCGCTTGTTCTCAGTGCAG-3ʹ and NBJ-R: 5ʹ-CGAAGGAGTACCACGTGGAG-3ʹ is used to detect non-Beijing genotypes by amplification of 539 bp PCR fragment from Rv2819 gene. The PCR products were observed on agarose gels.
Statistical Analysis
Phenotypic PZA susceptibility testing and the PZase assay were used as references. Associations among multiple aspects between PZAS and PZAR strains were analyzed with Pearson Chi-square test. A paired Chi-square test was used via MEDCALC® statistical software (https:/www.medcalc.org/calc/diagnostic_test.php) to measure the sensitivity, specificity, odds ratio (OR), 95% confidence interval (CI) and accuracy of PZA genotypic susceptibility testing method. P-value of <0.05 was considered statistically significant.
Results
Demographic Characteristics
Of the 448 M. tuberculosis isolates, 71.20% (319/448) were from male patients and 28.79% (129/448) were from female patients. Age range was 15 to 89 years with a mean age of ~41.2 years. Majority of TB patients 62.5% (280/448) examined in this study were previously treated cases and 69.86% (313/448) TB patients enrolled in the current study was migrant population.
Drug Susceptibility Profiles of M. tuberculosis Clinical Isolates
Among the 448 M. tuberculosis clinical isolates, 113 were identified as MDR, 195 pre-XDR and 70 XDR TB, while the remaining 70 strains had a random pattern of drug resistance. Total 60.04% (269/448) M. tuberculosis clinical isolates assessed by gold standard MGIT 960 system were PZAR in this study which were categorized as 69.02% (78/113) MDR, 61.02% (119/195) pre-XDR and 41.42% (29/70) XDR M. tuberculosis which collectively covered 59.8% (226/378) of the total (M/pre-X/X-DR) TB strains (Table 2).
 |
Table 2 Prevalence of MDR, Pre-XDR and XDR TB in PZAR Strains |
Genotypes of PZA-Resistant Isolates
Genotyping by multiplex PCR-based method demonstrated that overall 80.58% (361/448) belonged to Beijing genotype and the other 19.42% (87/448) to non-Beijing genotype in total studied isolates. The distribution of these total Beijing and non-Beijing genotypes in PZAR and PZAS strains is given in Table 3. PZAR isolates predominantly (83.3%; 224/269) belonged to the Beijing genotype, whereas, 16.7% (45/269) PZAR isolates were from non-Beijing genotype family of M. tuberculosis strains.
 |
Table 3 Distribution of Beijing and Non-Beijing Genotypes |
Association Between Mutations in pncA + UFR and PZase Enzyme Activity
Among the 84.38% (227/269) PZAR strains detected with pncA + UFR mutations (Table S1), 23.78% (54/227) exhibited insertions/deletions (indels) in pncA coding region, 5.72% (13/227) had stop codon mutations, 59.03% (134/227) carried single nonsynonymous mutation, 0.44% (1/227) had multiple mutations, 0.88% (2/227) showed synonymous mutation and 5.28% (12/227) had mutations both in pncA and its UFR. The amino acid substitutions/indels in pncA were scattered over the gene including the nonsynonymous mutations near the active site or metal-binding site. However, 4.84% (11/227) PZAR strains had mutations/indels only in the UFR of pncAwt with frequently existed substitutions at −11 position. The PZase assay revealed that 3.96% (9/227) PZAR strains had positive or weakly positive PZase activity, though they had indels or synonymous mutation in pncA gene or its UFR while the remaining 96.03% (218/227) were PZase negative (Table 4).
 |
Table 4 Correlation of Genotypic Susceptibility Testing and PZase Activity Assay |
In addition, among the substitutions located at −11 position, five A−11G PZAR mutants were detected with negative PZase activity but a very interesting phenomenon was identified in this study that one A−11G PZAR mutant having a deletion of C nucleotide at position −114 in the UFR of pncA showed positive PZase activity, which suggests that C−114del possibly altered the effect of A−11G mutation and induced the normal regulation of PZase (PZase positive) in this PZAR strain. On the whole, 21.14% (48/227) new nonsynonymous mutations/indels were found in pncA and its UFR in this study.
Mutations in rpsA, panD, Rv2783c and clpC1
In this study, 2.60% (7/269) PZAR strains carried nonsynonymous mutations (Thr29Met; Gln162Arg; Ala412Val) in RpsA (Table S2). Two of them (Gln162Arg, Ala412Val) were identified first time in this study. A synonymous mutation (Arg212Arg) in RpsA was frequently observed in both PZAS and PZAR strains. Further, we found 1.48% (4/269) PZAR strains with Leu132Pro and Pro134Ser mutations in the C-terminus of PanD which possibly alter the protein structure and cause resistance. The mutation Leu132Pro is recognized as a novel mutation in PanD. It is interesting to note that, a new nonsynonymous mutation (Ser149Pro) was detected in Rv2783c of 1.11% (3/269) PZAR strains, whereas no mutation occurred in clpC1 gene or its promoter region.
Characterization of Rv2044c-pncA-Rv2042c in pncAwt PZAR Strains
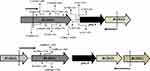
Most importantly, 10.40% (28/269) PZAR strains were found without mutation in the above-sequenced genes. The phenotypic resistance of these strains were reconfirmed by using higher concentration (300 and 900 μg/mL) of PZA and the genotypic assessment of entire operon of pncA (Rv2044c-pncA-Rv2042c) was performed. It is worth noting that among the total PZAR strains, 0.74% (2/269) PZAR strains were identified with nonsynonymous mutations (Tyr70His, Ile71Asn, Trp80Gly) and (Trp80Leu, Ala82Asp) in Rv2044 (the instant upstream gene of pncA) and produced negative PZase activity (Figure 1). Interestingly, 9.7% (26/269) of the total PZAR strains were discovered without any mutation in all known PZA-associated genes. Among these 26 PZAR strains, 34.61% (9/26) produced negative PZase activity and 65.38% (17/26) PZAR strains had positive PZase activity (Table 4) in PZase assay. The genotypic reassessment also confirmed no mutation in all sequenced genes in these PZAR strains. Overall, 23.45% (57/243) novel mutations/indels were identified in pncA + UFR, rpsA, panD, Rv2783c and Rv2044c genes among the total 90.3% (243/269) genetic mutations in PZAR strains.
DNA Sequencing Analysis of Phenotypic PZAS Strains
Of the total 39.95% (179/448) PZAS strains, 1.67% (3/179) harbored synonymous mutations, 0.55% (1/179) insertion and 3.35% (6/179) were noticed with single nonsynonymous mutations only in pncA of phenotypic PZAS strains with positive PZase activity (Table S3), while 94.41% (169/179) PZAS strains were found with wild-type pncA and other sequenced genes along with positive PZase activity.
Prediction of PZAR Isolates Based on DNA Sequencing
To examine the performance of DNA sequencing method for detection of PZAR isolates, the genotypic and phenotypic results of 448 M. tuberculosis isolates were compared. Using the phenotypic results as a reference, the sequencing method exhibited 75.09% sensitivity and 96.09% specificity for the mutations only in the pncA coding region, but the genotypic characterization of pncA accompanied by its UFR improved the sensitivity to 83.64% while specificity remained unchanged. The individual sequencing of rpsA, panD, Rv2783c, and Rv2044c showed low sensitivities, indicating minor diagnostic role of their own. However, the combined evaluation of mutations in pncA + UFR, rpsA, panD, Rv2783c, and Rv2044c genes increased the sensitivity from 75.09% to 89.59% with 92.19% accuracy and 96.09% specificity (Table 5).
 |
Table 5 Evaluation of Sequencing Method and Phenotypic Susceptibility Testing for Detection of PZA Resistance |
Evaluation of Association of PZase Assay with MGIT 960 and DNA Sequencing
Phenotypically PZAR M. tuberculosis strains assessed by MGIT 960 usually show PZase negative activity (PZase -) because of mutation in pncA gene which disrupts the PZase regulation. Whereas PZAR M. tuberculosis strains without pncA mutation possess positive PZase activity (PZase +). The correlation between PZase activity assay and MGIT 960 showed the sensitivity (85.13%, [95% CI: 80.31–89.16]) and specificity (100%, [95% CI: 97.96–100]) with the accuracy of (91.07%, [95% CI: 88.04–93.54]) due to the high number (269/448) of PZAR strains. Besides, the association between genotypic testing and PZase assay exhibited the sensitivity (81.78%, [95% CI: 76.64–86.21]) and specificity (96.09%, [95% CI: 92.11–98.41]) with the accuracy of (87.50%, [95% CI: 84.08–90.42]) because of high incidence of resistance-conferring pncA+ UFR mutations.
Discussion
This study shows the prevalence rates of PZAR in MDR (69%) and pre-XDR (61%) TB in southern China were higher than the MDR TB observed in South Africa (52.1%), Japan (52.8%) and Thailand (49.0%),18,19,20 but in line with another recently published study.21 Beijing genotype was the most dominant genotype 80.58% (361/448) observed in southern China, similar to the studies from Beijing22 and Zhejiang.23 PZA has an imperative role in current and new anti-TB regimens including the promising Pa-824 + Moxifloxacin + PZA, bedaquiline + PZA24 and clofazimine + PZA containing regimens.25 Our study comprehensively investigates the genetic mutations in PZA-related genes (pncA, rpsA, panD, Rv2783c, and clpC1) in addition with the complete operon of pncA (Rv2044c-pncA-Rv2042c) in a wide-range of M. tuberculosis clinical isolates.
The detection of 84.38% (227/269) pncA + UFR mutations in PZAR strains in this study was markedly higher than previous reports from Thailand (75.0%),19 Brazil (45.7%)26 and other parts of China (78.0%).23 Though mutations in pncA scattered all over the gene and its UFR but a certain frequency of pncA mutations has been observed at amino acid residues 3 to 12, 46 to 62, 67 to 85, 94 to 103, and 132 to 142 which were the areas close to the PZase active sites (Asp8, Lys96, and Val139) or metal ion binding sites (Asp49, His51, His57, and His71).23 Our findings are coherent with the previous studies.20,23 So, the absence of “hot spot” regions make it quite challenging to develop rapid diagnostic assays for detection of PZAR isolates based on indel or nonsynonymous mutations in the pncA.13,27
The rpsA gene has been ambiguously discussed in the literature as both supportive and opposing studies have been published regarding the role of RpsA in PZAR strains.12,28,29 However, two novel mutations (Gln162Arg; Ala412Val) are identified in RpsA of PZAR isolates in this study. Moreover, mutations in panD have also been associated with PZAR in clinical isolates lacking mutations in pncA and rpsA.8 The C-terminus of PanD protein spanning amino acid residues 114–139, where all the PanD mutations are usually mapped may affect the binding of the active form of PZA (POA) without abolishing PanD’s enzymatic activity.30 We discerned (Leu132Pro and Pro134Ser) mutations in the C-terminus of PanD in PZAR isolates. Leu132Pro is recognized as a novel mutation in our study. Though another study reported (Leu132Arg) and (Leu136Arg) mutations in PanD31 but these mutations were different from our findings regarding the amino acid substitutions.
Furthermore, Rv2783 plays an important role in the general homeostasis of (p)ppGpp during dormancy; however, POA inhibits its function in wild-type strain, but the Asp67Asn mutation helps in circumvention of POA effects.9 In this study, we identified three PZAR strains with positive PZase activities bearing one novel nonsynonymous mutation (Ser149Pro) in Rv2783, which supports that Rv2783 is a potential target of POA.9 Likewise, clpC1 (Rv3596c) involved in protein degradation by assembling a protease complex through ClpP1 and ClpP2, was proposed to be a new target of PZA and the mutations (Gly99Asp, Lys209Glu) in N-terminal and D1 domain have been interrelated with PZAR strains.10,27 However, there was no any mutation in clpC1of M. tuberculosis clinical isolates in this study. This degradative protease is essential for the viability, survival, and virulence of M. tuberculosis. Therefore, the M. tuberculosis strains containing mutations in ClpC1 may not be able to survive well in the host, so it is hard to get such mutants in clinical isolates.7,32
On the other hand, 10.40% (28/269) PZAR strains were observed without any mutation in pncA, rpsA, panD, Rv2783c, and clpC1 genes. However, to determine the significant role of UFR of pncA and its surrounding genes in PZase regulation,11,12 we sequenced the complete operon of pncA (Rv2044c-pncA-Rv2042c) in these PZAR strains. Notably, two of these PZAR strains were detected with novel missense mutations “Tyr70His, Ile71Asn, Trp80Gly” and “Trp80Leu, Ala82Asp” in Rv2044 first time in this study. Interestingly, these strains exhibited the negative PZase activity in PZase assay which was likely to be the effect of these mutations. However, in-depth impact of these mutations on PZase regulation needs to be investigated in imminent studies. This observation is also supported by another recent study where an important frameshift deletion was identified in Rv2044c that interrupted the stop codon and led to its fusion with pncA which engendered the addition of a novel domain of unknown function (DUF2784) to the PZase enzyme.33 Overall, 23.45% (57/243) novel mutations/indels were detected in PZAR strains in our study.
PZAS strains with missense mutations/insertion in pncA were reconfirmed by phenotypic and genotypic assays. The mutation (T92G→Ile31Ser) has been previously described in both PZAS and PZAR isolates with similar PZase characteristics,19 whereas Thr47Ala was also reported without its involvement in PZAR.34 The mutations in susceptible strains were unable to encounter the threshold of resistance.13 No nonsynonymous mutations/indels were identified in the genes other than pncA in PZAS strains.
Compared to the phenotypic data, the sensitivity for detecting PZAR by DNA sequencing of pncA gene along with its UFR was 83.64% with 96.09% specificity which was consistent with the data from South Korea (84.6%)35 and the United States (84.6%),36 only a little higher than the studies from northern China (77.97%),23 Thailand (75%)19 and Sierra Leone (70%)37 but lower than the findings from Netherlands (96.8%).38 Moreover, the combined assessment of mutations in pncA + UFR, rpsA, panD, Rv2783c, and Rv2044c increased the sensitivity up to 89.59% which strengthens their share for detection of PZAR M. tuberculosis clinical isolates.
In agreement with the several studies, the current study demonstrated that mutations in pncA gene turns the regular enzyme activity into PZase negative; however, nine PZAR strains with synonymous mutations/indels in the pncA or its UFR in our study retained the positive/weakly positive PZase activity and may have the involvement of enigmatic mechanism(s) of resistance against PZA. The PZase activity assay in association with MGIT 960 susceptibility testing showed 85.13% sensitivity. In the same way, the correlation between PZase assay and genotypic testing presented the sensitivity of 81.78% and 96.09% specificity in our study, which is significantly higher than previously published study (68.6%) and (45.7%)26 but slightly lower than another report 88.2% and 88.8%,39 respectively. Similar to previous reports, the mutations in rpsA, panD and Rv2783c were not found to be associated with loss of PZase activity in PZAR strains lacking pncA mutation.6,8,9,26,30
In contrast, the substitution at position −11 of pncA usually possess negative PZase activity in PZAR strains.40 While we identified one A−11G PZAR mutant having a deletion of C nucleotide at position −114 in the UFR of pncA showed PZase positive, that was completely divergent from five other PZase negative A−11G PZAR mutants, which proposes that C−114del might turn the impact of A−11G mutation and produced positive PZase activity in this PZAR strain. This needs to be further studied in the forthcoming studies. Moreover, the consistent results of phenotypic and genotypic reassessment of 34.61% (9/26) PZase negative and 65.38% (17/26) PZase positive PZAR strains without any known mutation in all reported PZAR-related genes direct their linkage with cryptic resistance mechanisms of PZA in M. tuberculosis.
Conclusion
In conclusion, to the best of our knowledge, this is the first study, covering the combined phenotypic and genotypic characterization of all PZAR-associated genes in a wide range of M. tuberculosis clinical isolates. PZAR rate among MDR (69%) and pre-XDR TB (61%) was considerably higher in southern China. Though pncA played a major role (>80%) in PZAR but the simultaneous evaluation of other PZAR related genes increased the sensitivity to ~90%. Considering the difficulty of phenotypic susceptibility testing method, our data suggest that concomitant detection of mutations in pncA+ UFR, rpsA, panD, Rv2783c, and Rv2044c is more dependable. The addition of 57 novel mutations/indels from our study in drug-resistance TB mutation database may increase the reliability of molecular diagnosis. In addition, the substitutions at positions −11 and −12 in pncA UFR caused negative PZase activity but deletion of C nucleotide at −114 altered the effect of A−11G mutation and established the normal regulation of PZase, though the strain was still resistant to PZA. In addition, the loss of PZase activity due to newly identified mutations in Rv2044 led to confer PZAR in M. tuberculosis. Lastly, the nine PZase positive PZAR mutants bearing pncA synonymous mutations/indels and 26 PZAR strains lacking any mutation in all reported PZAR-related genes with discordant PZase activities implicate the existence of unknown resistance mechanisms that need to be uncovered.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572037,81973372), the National Mega-project of China for Main Infectious Diseases (2017ZX10302301-003-002, 2017ZX10302301-003-005) and for Innovative Drugs (2019ZX09721001-003-003), the Chinese Academy of Sciences Grants (154144KYSB20190005, YJKYYQ20170036), the Science and Technology Department of Guangdong Province (2017A020212004, 2019B110233003), the Special Funds for Economic Development of Marine Economy of Guangdong Province (GDME-2018C003) and by the Grant (SKLRD-OP-201919) from the State Key Lab of Respiratory Disease, Guangzhou Institute of Respiratory Diseases, First Affiliated Hospital of Guangzhou Medical University. This work was sponsored by Science and Technology Innovation Leader of Guangdong Province (2016TX03R095 to TZ), the UCAS Ph.D. Fellowship Program (to Hameed HMA) and CAS-TWAS President’s Ph.D. Fellowship Program (to Islam MM and Chhotaray C) for international students.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yadon AN, Maharaj K, Adamson JH, et al. A comprehensive characterization of pncA polymorphisms that confer resistance to pyrazinamide. Nat Commun. 2017;8(1):588. doi:10.1038/s41467-017-00721-2
2. Whitfield MG, Streicher EM, York T, et al. Association between genotypic and phenotypic pyrazinamide resistance in Mycobacterium tuberculosis. Int J Infect Dis. 2014;21:96. doi:10.1016/j.ijid.2014.03.628
3. Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662. doi:10.1038/nm0696-662
4. Islam MM, Hameed HMA, Mugweru J, et al. Drug resistance mechanisms and novel drug targets for tuberculosis therapy. J Genet Genomics. 2017;44(1):21–37. doi:10.1016/j.jgg.2016.10.002
5. Ramirez-Busby SM, Rodwell TC, Fink L, et al. A multinational analysis of mutations and heterogeneity in pZase, rpsA, and panD associated with pyrazinamide resistance in M/XDR Mycobacterium tuberculosis. Sci Rep. 2017;7:3790. doi:10.1038/s41598-017-03452-y
6. Shi W, Zhang X, Jiang X, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–1632. doi:10.1126/science.1208813
7. Shi W, Chen J, Zhang S, Zhang W, Zhang Y. Identification of novel mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, and cyp128 associated with pyrazinoic acid/pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2018;62(7):e00430–18. doi:10.1128/AAC.00430-18
8. Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microb Infect. 2013;2:e34. doi:10.1038/emi.2013.38
9. Njire M, Wang N, Wang B, et al. Pyrazinoic acid inhibits a bifunctional enzyme in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2017;61(7):e00070–17. doi:10.1128/AAC.00070-17
10. Yee M, Gopal P, Dick T. Missense mutations in the unfoldase ClpC1 of the caseinolytic protease complex are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2017;61:2.
11. Barco P, Cardoso RF, Hirata RDC, et al. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis clinical isolates from the southeast region of Brazil. J Antimicrob Chemother. 2006;58(5):930–935. doi:10.1093/jac/dkl363
12. Tan Y, Hu Z, Zhang T, et al. Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol. 2014;52(1):291–297. doi:10.1128/JCM.01903-13
13. Liu W, Chen J, Shen Y, et al. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Hangzhou, China. Clin Microbiol Infect. 2018;24(9):
14. Li D, Hu Y. pncA mutations in Mycobacterium tuberculosis is a strong predictor of poor treatment outcome in the therapy of multidrug resistant tuberculosis. Int J Infect Dis. 2016;45:92–93. doi:10.1016/j.ijid.2016.02.246
15. Pang Y, Dong H, Tan Y, et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci Rep. 2016;6:25330. doi:10.1038/srep25330
16. World Health Organization. Technical manual for drug susceptibility testing of medicines used in the treatment of tuberculosis. 2018.
17. Mustafa S, Javed H, Hashmi J, Jamil N, Tahir Z, Akhtar AM. Emergence of mixed infection of Beijing/Non-Beijing strains among multi-drug resistant Mycobacterium tuberculosis in Pakistan. 3 Biotech. 2016;6(1):108. doi:10.1007/s13205-016-0423-9
18. Ando H, Mitarai S, Kondo Y, et al. Pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis isolates in Japan. Clin Microbiol Infect. 2010;16(8):1164–1168. doi:10.1111/j.1469-0691.2009.03078.x
19. Jonmalung J, Prammananan T, Leechawengwongs M, Chaiprasert A. Surveillance of pyrazinamide susceptibility among multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, Thailand. BMC Microbiol. 2010;10(1):223. doi:10.1186/1471-2180-10-223
20. Mphahlele M, Syre H, Valvatne H, et al. Pyrazinamide resistance among South African multidrug-resistant Mycobacterium tuberculosis isolates. J Clin Microbiol. 2008;46(10):3459–3464. doi:10.1128/JCM.00973-08
21. Islam MM, Tan Y, Hameed HMA, et al. Detection of novel mutations associated with independent and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect. 2019;25(8):
22. Sun Z, Chao Y, Zhang X, et al. Characterization of extensively drug-resistant Mycobacterium tuberculosis clinical isolates in China. J Clin Microbiol. 2008;46(12):4075–4077. doi:10.1128/JCM.00822-08
23. Xia Q, Zhao L-L, Li F, et al. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob Agents Chemother. 2015;59(3):1690–1695. doi:10.1128/AAC.04541-14
24. Diacon AH, Dawson R, von Groote-bidlingmaier F, et al. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet. 2012;380(9846):986–993. doi:10.1016/S0140-6736(12)61080-0
25. Diacon AH, Dawson R, Groote-Bidlingmaier F, et al. Bactericidal activity of pyrazinamide and clofazimine alone and in combinations with pretomanid and bedaquiline. Am J Respir Crit Care Med. 2015;191(8):943–953. doi:10.1164/rccm.201410-1801OC
26. Bhuju S, Fonseca L, Marsico AG, et al. Mycobacterium tuberculosis isolates from Rio de Janeiro reveal unusually low correlation between pyrazinamide resistance and mutations in the pncA gene. Infect Genet Evol. 2013;19:1–6. doi:10.1016/j.meegid.2013.06.008
27. Hameed HMA, Islam MM, Chhotaray C, et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis strains. Front Cell Infect Microbiol. 2018;2018(8):114. doi:10.3389/fcimb.2018.00114
28. Akhmetova A, Kozhamkulov U, Bismilda V, et al. Mutations in the pncA and rpsA genes among 77 Mycobacterium tuberculosis isolates in Kazakhstan. Int J Tuberc Lung Dis. 2015;19(2):179–184. doi:10.5588/ijtld.14.0305
29. Gu Y, Yu X, Jiang G, et al. Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn Microbiol Infect Dis. 2016;84(3):207–211. doi:10.1016/j.diagmicrobio.2015.10.017
30. Shi W, Chen J, Feng J, et al. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg Microb Infect. 2014;3(8):e58. doi:10.1038/emi.2014.61
31. Gopal P, Yee M, Sarathy J, et al. Pyrazinamide resistance is caused by two distinct mechanisms: prevention of coenzyme a depletion and loss of virulence factor synthesis. ACS Infect Dis. 2016;2(9):616–626. doi:10.1021/acsinfecdis.6b00070
32. Raju RM, Jedrychowski MP, Wei J-R, et al. Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathog. 2014;10(3):e1003994. doi:10.1371/journal.ppat.1003994
33. Baddam R, Kumar N, Wieler LH, et al. Analysis of mutations in pncA reveals non-overlapping patterns among various lineages of Mycobacterium tuberculosis. Sci Rep. 2018;8(1):4628. doi:10.1038/s41598-018-22883-9
34. Xu P, Wu J, Yang C, et al. Prevalence and transmission of pyrazinamide resistant Mycobacterium tuberculosis in China. Tuberculosis (Edinb). 2016;98:56–61. doi:10.1016/j.tube.2016.02.008
35. Kim H, Kwak H, Lee J, et al. Patterns of pncA mutations in drug-resistant Mycobacterium tuberculosis isolated from patients in South Korea. Int J Tuberc Lung Dis. 2012;16(1):98–103. doi:10.5588/ijtld.10.0739
36. Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first-and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032–2041. doi:10.1128/AAC.01550-10
37. Feuerriegel S, Oberhauser B, George AG, et al. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 2012;12(1):90. doi:10.1186/1471-2180-12-90
38. Simons S, van der Laan T, Mulder A, et al. Rapid diagnosis of pyrazinamide‐resistant multidrug‐resistant tuberculosis using a molecular‐based diagnostic algorithm. Clin Microbiol Infect. 2014;20(10):1015–1020. doi:10.1111/1469-0691.12696
39. Cui Z, Wang J, Lu J, Huang X, Zheng R, Hu Z. Evaluation of methods for testing the susceptibility of clinical Mycobacterium tuberculosis isolates to pyrazinamide. J Clin Microbiol. 2013;51(5):1374–1380. doi:10.1128/JCM.03197-12
40. Pang Y, Zhu D, Zheng H, et al. Prevalence and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. BMC Infect Dis. 2017;17(1):711. doi:10.1186/s12879-017-2761-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.