Back to Journals » Infection and Drug Resistance » Volume 11
Detection of Mycobacterium lepromatosis in patients with leprosy in India
Authors Ahuja M, Lavania M , Singh I, Turankar RP , Chhabra S, Narang T , Dogra S , Sengupta U
Received 20 February 2018
Accepted for publication 15 May 2018
Published 10 October 2018 Volume 2018:11 Pages 1677—1683
DOI https://doi.org/10.2147/IDR.S166035
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
This paper has been retracted.
Madhvi Ahuja,1,* Mallika Lavania,1,* Itu Singh,1 Ravindra P Turankar,1 Seema Chhabra,2 Tarun Narang,3 Sunil Dogra,3 Utpal Sengupta1
1Department of Molecular Biology, Stanley Browne Laboratory, TLM Community Hospital, Delhi, India; 2Department of Immunopathology, Postgraduate Institute of Medical Education and Research, Chandigarh, India; 3Department of Dermatology Venereology and Leprology, Postgraduate Institute of Medical Education and Research, Chandigarh, India
*These authors contributed equally to this work
Introduction: The most commonly noted reactions in leprosy patients are type 1 reactions and erythema nodosum leprosum, with some rare phenomenon of host response known as Lucio phenomenon or leprosy of Lucio and Latapi which is caused by Mycobacterium lepromatosis. So far, no case of M. lepromatosis has been reported from India.
Materials and methods: The main objective of this study was to detect any positive cases of M. lepromatosis in India with such a complication. We screened slit skin smear/biopsy samples from lepromatous leprosy (LL) patients reporting to The Leprosy Mission Community Hospitals across the country. Eighty-eight slit skin smears were collected from leprosy patients in 70% ethanol. DNA was extracted from all these samples. Polymerase chain reaction (PCR) was done for 2 genes; one set was for 16S rRNA and the other set was for coproporphyrinogen III oxidase (hemN) gene. Then, sequencing was done for all positive amplicons. Homology of the sequences was analyzed using the Basic Local Alignment Search Tool at the National Center of Biotechnology Information database.
Results: Among 88 isolates, we found 4 positive cases for M. lepromatosis. All 4 were LL cases with a bacteriological index ranging from 2+ to 4+. On the basis of the National Center of Biotechnology Information Basic Local Alignment Search Tool analysis, the sequenced amplicons of both genes matched with the M. lepromatosis 16S rRNA and phosphofructokinase genes but not with hemN gene of lepromatosis. This is the first report for the presence of M. lepromatosis in LL cases from India.
Conclusion: This new species M. lepromatosis exists beyond Mexico, Singapore and it is the cause of DLL in India also. It may cause dual infections along with M. leprae in endemic areas like India.
Keywords: lepromatous leprosy, phosphofructokinase, M. lepromatosis, coproporphyrinogen III oxidase (hemN) gene, 16S rRNA gene.
Introduction
Mycobacterium leprae has been considered to be the sole causative agent of all known forms of leprosy. The disease manifests with a broad clinicoimmunological spectrum, which has been classified by Ridley and Jopling,1 and has been categorized into tuberculoid leprosy (TT), borderline tuberculoid leprosy, borderline leprosy (BB), borderline lepromatous leprosy, and lepromatous leprosy (LL) types. The clinical manifestation of the patient depends on the host’s own immune response to M. leprae. Leprosy patients often, either during therapy or otherwise, tend to manifest episodes of reactions with exacerbations of existing lesions depending upon their immune status, and these have been most commonly noted as reversal reactions (type 1) and erythema nodosum leprosum reactions.
Another rare phenomenon of host response that has been noted is the Lucio phenomenon. This was first recognized by Lucio and Alvarado in 1852 and further described by Latapi and Chevez-Zamora2 in 1948 in Mexico. It is also known as diffuse leprosy of Lucio and Latapi.3 Later, Han et al,5 while investigating the cause of death of a diffuse lepromatous leprosy (DLL) patient, isolated a Mycobacterium species similar to M. leprae from the freshly frozen autopsied liver sample of the patient and named it Mycobacterium lepromatosis. This mycobacterial species showed 2.1% divergence from the 16S rRNA gene of M. leprae. In addition, comparison of other gene sequences belonging to rpoB, 16S, and hsp65 also confirmed a remarkable level of divergence from the corresponding sequences of M. leprae strains, and thereby it was assigned as a separate species.4 The genome of this species has been recently described by Ang et al,7 and from 126 contigs, which confirms differences at the genome level, both species were found to differ by ~13% in sequence diversity. This is in contrast to <0.005% sequence diversity observed in the global collection of M. leprae strains. This mycobacterial species almost has the same GC content (~57.8%) like M. leprae, which is significantly lower than the GC content of other mycobacterial species (60%–66%). M. lepromatosis is a causative agent for DLL, which carries a higher mortality rate than other forms of leprosy.2,7–10
Earlier, several case reports from India also showed that patients have clinical features suggestive of DLL/Lucio’s Phenomenon.9,11 However, there has not yet been reports of cases of M. lepromatosis, and therefore to search for the existence of M. lepromatosis infection we screened samples isolated from patients visiting the hospitals of The Leprosy Mission, India, from across the country. Thus far, M. lepromatosis has been found in patients with leprosy from Mexico, Canada, Brazil, Singapore, and Myanmar.12–14 Recently, it was found to be a cause of leprosy, along with M. leprae, in red squirrels in the British Isles.15 Thus, M. lepromatosis is present in the Americas, Asia, and Europe.
Materials and methods
Ethical approval
Ethical clearance for this study was provided by The Leprosy Mission Trust India Ethical Committee, under the regulations of the Indian Council of Medical Research. Written informed consent was obtained from all subjects before collection of biological samples.
Materials
In the present study, we analyzed 88 skin slit smear/biopsy samples from MB leprosy patients; the sample was collected in 70% ethanol from patients who visited The Leprosy Mission in different states of India during 2014–2017 years (Table 1).
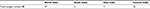  | Table 1 Geographical distribution of patients recruited in this study |
Extraction of DNA from clinical samples
DNA was isolated from all these samples. All samples were processed for lysis in 100 µL Lysis buffer (1 M Tris-EDTA, 0.05% Tween 20, 10 mg/mL proteinase K) at 60°C for 16 hours followed by inactivation of reaction at 97°C for 15 minutes. In case of inhibitors in the lysate, it was passed through a Qiagen column (Qiagen, Hilden, Germany) before using in the polymerase chain reaction (PCR) as a template.
PCR amplification of genes targeting 16Sr RNA gene region and hemN and sequencing
To check the positive cases for M. lepromatosis in our collected samples, PCR was done for 2 sets of genes. One set was for 16S rRNA gene and other set was for coproporphyrinogen III oxidase (hemN) gene. Each PCR reaction contained 5 µL of DNA, 10 µM of each primer, and Hot start Taq polymerase PCR master mix (2×) (Qiagen). The final volume of reaction mixture was made up to 20 µL with nuclease-free water. Details of primers used in this study are listed in Table 2. Amplicons of each reaction were electrophoresed through 3% agarose gel in 1× Tris borate EDTA (TBE) running buffer. Sequencing was done for all positive amplicons. Sequences obtained were analyzed using Basic Local Alignment Search Tool (BLAST) at the National Center of Biotechnology Information database (NCBI, Bethesda, MD, USA).
  | Table 2 List of primers used in this study
|
PCR for M. leprae detection
Each PCR reaction contained 5 µL of DNA, 10 µM of each primer, and Taq polymerase PCR master mix (2×) (Qiagen). The final volume of reaction mixture was made up to 20 µL with nuclease-free water. For M. leprae detection, RLEP gene target was selected; the primer details are listed in Table 2.
PCR targeting rpoB, folP, and gyrA genes to check the drug susceptibility
We also performed PCR sequencing for drug susceptibility testing of 4 samples targeting the genes rpoB, folP, and gyrA. PCR-based gene amplification was done using primers according to the Guidelines of the World Health Organization’s (WHO) “Global Surveillance of Drug Resistance in Leprosy 2008” for detection of mutations in rpoB, gyrA, and folP1 genes in the M. leprae genome.17
Phylogenetic analysis
The gene sequences obtained in this study were analyzed, and additional GenBank accessions (NCBI) for 16S rRNA genes included, EU203590 for M. lepromatosis FJ924, GQ900372 for Sg-1 M. lepromatosis, and GQ900374 for Br-1 M. lepromatosis. Sequence analysis was performed through queries to GenBank using BLAST. Phylogenetic analysis was performed using CLUSTAL W for multialignment.20 The tree construction was done by maximum likelihood method the Tamura–Nei model in MEGA7 software.21
Results
Origin of samples
Out of 88 samples, 47 were from North India followed by 32, 7, and 2 samples from Central, East and South India, respectively (Table 1).
Detection of M. leprae by PCR
Initially, all samples were screened for detection of M. leprae by targeting RLEP. Among these 82 samples, only 3 were negative for RLEP and showed positivity for rpoB gene (Table 3). Out of these 4 RLEP-negative and rpoB cases, 2 belonged to Central India and 1 was from Northern India.
  | Table 3 Geographical distribution of M. leprae and M. lepromatosis detected by PCR Abbreviation: PCR, polymerase chain reaction. |
Detection of M. lepromatosis by PCR
Two gene targets specific for M. lepromatosis were selected – 16S rRNA gene region and hemN gene (Figure 1). Among all samples, we found only 4 cases positive for M. lepromatosis. Positivity of these 4 samples among 88 samples constitutes 4.5% of the total cases, and the clinical details of these 4 patients are summarized in Table 3.
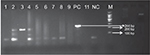  | Figure 1 Gel image for hemN lepromatosis gene-positive samples. Note: Product length: 244 bp. Abbreviation: hemN, coproporphyrinogen III oxidase. |
All 4 presented with LL with infiltration and nodules all over the body. Clinically, they were LL patients with diffuse lesions and a bacteriological index (BI) ranging from 3+ to 4+. All patients were treated according to the WHO regimen of multibacillary leprosy. In our study, we included 2 sets of genes specific for M. lepromatosis as reported by Han et al14 and Singh et al.6 We found amplifications of 142 bp and 244 bp for the 16S rRNA and the hemN genes, respectively. Both the genes were sequenced and found to be matching with M. lepromatosis in the available gene database at the time of analysis on BLAST. But at the time of analysis of hemN gene, we found that BLAST results matched with the gene phosphofructokinase of M. lepromatosis (Figure 2).
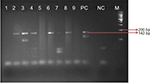  | Figure 2 Gel image for 16S rRNA lepromatosis gene-positive samples. Note: Product length: 142 bp. |
Phylogenetic analysis of samples
The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model.22 The tree with the highest log likelihood (–1,256.25) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pair-wise distances estimated using the Maximum Composite Likelihood approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measuring the number of substitutions per site (next to the branches). The analysis involved 8 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 126 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.21 In figure 3, analysis of phylogenetic tree for 16S rRNA showed that 3 samples CH4, M10 and M6 are closely related to M. lepromatosis, while 2 samples CPH570 and IAI94 were outbranched and formed a different clade with respect to M. lepromatosis. In phylogenetic analysis for hemN gene (Figure 4), no submitted sequence was found in NCBI; but in the analysis, it was clearly seen that CH7 and CPH570 samples branched from the L10 sample, which itself was a positive control of M. lepromatosis (nonsubmitted sequence, gifted from Dr Rahul National Hansen’s Disease Programme Baton Rouge, USA); hence, CH7, CPH570, and L10 all fall into the same clade, confirming the presence of M. lepromatosis in these samples.
  | Figure 3 Molecular phylogenetic analysis by maximum likelihood method for 16S rRNA gene. |
  | Figure 4 Molecular phylogenetic analysis by maximum likelihood method for hemN gene. Abbreviation: hemN, coproporphyrinogen III oxidase. |
Drug susceptibility testing in patients
The drug susceptibility testing was done for the 3 patients positive for M. lepromatosis by 16S rRNA PCR. All patients were classified as having LL based on the Ridley–Jopling scale. Details of these 3 patients are as follows:
Patient 1
This patient (71/F) was from Champa, Chhattisgarh, and registered in 2013. The patient had a BI of 3.3+ and had many raised skin lesions and had undergone diaminodiphenyl sulfone/Dapsone (DDS) monotherapy 40 years ago. She also had active skin lesions with thickened nerves. On analysis, the patient showed resistance to rpoB and folP with mutations at codon positions 411 (Ala–Leu) and 53 (Ala–Thr), respectively. When the sequence was analyzed with BLAST along with the lepromatosis genome, we observed a mutation at codon 54 GGG–CAG (Gln 54 Arg) but no mutation was observed in the gyrA gene (Figure 5).
  | Figure 5 DRS sequence alignment with Mycobacterium species. |
Patient 2
This patient (36/M) was from Pgimer, Chandigarh, and was registered in 2015. The patient presented initially with hypopigmented patches all over the body 7 years ago. The patient presently reported with a new lesion on the face with hypopigmented patches all over the body, back, and on both hands and feet. The patient showed drug sensitivity to all 3 genes.
Patient 3
This patient (45/M) presented with multiple new lesions on face with anesthetic patches and had received multi drug therapy (MDT) for 2 years. He had a BI of 4+ at the time of sample collection. The patient showed drug sensitivity to all 3 genes.
Patient 4
This patient (30/M) was from Champa, Chhattisgarh, and registered in 2017. The patient had a BI of 4+ with infiltration nodules all over the body and had taken multibacillary/multidrug therapy (MB/MDT)/2–3 doses from the government Primary Health Centre (PHC) initially, but then stopped the therapy by himself 7–8 years back after feeling better.
This is the first independent confirmation of the existence of M. lepromatosis in Indian patients.
Discussion
Diffuse Lucio leprosy is characterized by papules, plaques, and necrotizing lesions. It was endemic to Mexico; however, it is not only restricted to Mexico, and recently this phenomenon has been reported in USA, Spain, South and Central America including Brazil, and East and West Asia.25 Lucio phenomenon manifests 3–4 years after onset of disease and is more common in untreated patients or in those receiving inadequate treatment.2,23 Generally, patients with extensive blisters and ulcerations representing type 2 lepra reactions have often been confused and labeled as having Lucio phenomenon.11,24 It was also reported earlier from India that some patients showed clinical features similar to DLL/Lucio’s leprosy.11,9 Latapi and Chevez-Zamora2 and Thappa et al11 also warned about the improper labeling of many cases of nodular LL types as Lucio leprosy with Lucio phenomenon.
In 2008, in Mexico, a new species of Mycobacterium was recognized and named as M. lepromatosis, and this species had led to the death of 2 Mexican DLL patients.7 In addition to Mexico, M. lepromatosis infection has also been reported in Canada, Brazil, Singapore, and Myanmar.12–14 We investigated the prevalence of this newly discovered M. lepromatosis in 82 diffuse LL relapse cases in India. Out of these 82 cases, 3 RLEP-negative rpoB positive cases were from tertiary care hospitals situated in Northern and Central India (Table 1).
According to the published literature, few gene sequences belonging to rpoB, 16SrDNA, and hsp65 of M. lepromatosis show a remarkable level of divergence from the corresponding sequences of M. leprae strains.6 WHO-recommended MDT seems to be effective in treating Lucio phenomenon. As per a case report study in India, it was observed that when MDT for leprosy is introduced at the beginning of Lucio phenomenon outbreak, the prognosis is usually good. Prognosis was noted to be poor even with proper treatment if patient presents with secondary infection and/or anemia with extensive skin involvement (a reported case study from India).25
In the recent past, many studies/case reports have been described with clinical presentations matching very closely those in Lucio phenomenon from other parts of the world, like Brazil and Malaysia, along with confirmation based on molecular findings.18,19 The present study was carried out to assess the prevalence of newly discovered bacteria in DLL patients from India. For the first time, using specific DNA sequences, the presence of M. lepromatosis in diffuse LL has been reported in India. The finding of 4 cases with M. lepromatosis infection from 82 relapsed cases strongly indicates that this mycobacterial species is also prevalent in India. Kai et al19 also reported similar findings after analysis of biopsies obtained from 19 lepromatous cases from Mexico. As the present study was limited to a few selected samples, the data is not suitable to provide data on the actual prevalence of leprosy caused by this mycobacterial species in the country. Therefore, a widely distributed larger study needs to be undertaken to find out the actual prevalence of leprosy caused by M. lepromatosis in India. More cases from various countries need to be investigated to determine whether M. leprae and/or dual infection is the main cause of variations in clinical manifestations in leprosy.
Acknowledgment
We acknowledge the patients and staff from The Leprosy Mission Community Hospital. This work was funded by the Indian Council of Medical Research grant no: 5/8/3(1)/2013-ECD-1. Mycobacterium lepromatosis DNA (not submitted in NCBI) was gifted by Dr Rahul Sharma from National Hansen’s Disease Clinical Center at Baton Rouge, Louisiana (mentioned as M10 and L10 in this study).
Disclosure
The authors report no conflicts of interest in this work.
References
Ridley DS, Jopling WH. Classification of leprosy according to immunity: a 5-group system. Int J Lep. 1966;34(3):255–273. | ||
Latapi F, Chevez-Zamora A. The “spotted” leprosy of Lucio: an introduction to its clinical and histological study. Int J Lep. 1948;16(4):421–437. | ||
Vargas-Ocampo F. Diffuse leprosy of Lucio and Latapí: a histologic study. Lepr Rev. 2007;78(3):248–260. | ||
Han XY, Seo YH, Sizer KC, et al. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol. 2008;130(6):856–864. | ||
Han XY, Sizer KC, Thompson EJ, et al. Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis. Am Soc Mic. 2009;191(19):6067–6074. | ||
Singh P, Benjak A, Schuenemann VJ, et al. Insights into the evolution and origins of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis. Proc Nat Aca Sci U S A. 2015;112(14):4459–4464. | ||
Ang P, Tay YK, Ng SK, Seow CS. Fatal Lucio’s phenomenon in two patients with previously undiagnosed leprosy. J Am Acad Dermatol. 2003;48(6):958–961. | ||
Choon SE, Tey KE. Lucio’s phenomenon: a report of three cases seen in Johor, Malaysia. Int J Dermatol. 2009;48(9):984–988. | ||
Kumari R, Thappa DM, Basu D. A fatal case of Lucio phenomenon from India. Dermatol Online J. 200814:10. | ||
Derbes VJ, Samules M, Williams OP, Walsh JJ. Diffuse leprosy; case in a Louisiana Negro. Arch Dermatol. 1960;81:210–224. | ||
Thappa DM, Karthikeyan K, Kumar BJ. Is it Lucio leprosy with Lucio phenomenon or something else? Indian J Lepr. 2002;74(2):161–166. | ||
Jessamine PG, Desjardins M, Gillis T, et al. Leprosy-like illness in a patient with Mycobacterium lepromatosis from Ontario, Canada. J Drugs in Dermatol. 2012;11(2):229–233. | ||
Han XY, Aung FM, Choon SE, Werner B. Analysis of the leprosy agents M. leprae and M. lepromatosis in four countries. Am J Clin Pathol. 2014;142(4):524–532. | ||
Han XY, Sizer KC, Tan HH. Identification of the leprosy agent Mycobacterium lepromatosis in Singapore. J Drugs Dermatol. 2012;11(2):168–172. | ||
Avanzi C, Del-Pozo J, Benjak A, et al 2016. Red squirrels in the British Isles are infected with leprosy bacilli. Science. 2016;352(6313):744–747. | ||
Donoghue HD, Holton J, Spigelman M. PCR primers that can detect low levels of Mycobacterium leprae DNA. J Med Microbiol. 2001;50(2):177–182. | ||
World Health Organzation. Guidelines for Global Surveillance of Drug Resistance in Leprosy. Geneva: World Health Organization; 2009 | ||
Golchai J, Zargari O, Maboodi A, Maboodi A, Granmayeh S. Lepromatous leprosy with extensive unusual ulcerations and cachexia. Is it the first case of Lucio’s phenomenon from Iran? Int J Lepr Other Mycobact Dis. 2004;72(1):56–59. | ||
Kai M, Morris MF, Miyamoto Y, et al. Mutations in the drug resistance determining region of Mycobacterium lepromatosis isolated from leprosy patients in Mexico. J Dermatol. 2016;43:1345–1349. | ||
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22(22):4673–4680. | ||
Katoh K, Misawa K, Kuma K, Miyata T. MAFT: a novel for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acid Res. 2002;30(14):3059–3066. | ||
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. | ||
Costa IM, Kawano LB, Pereira CP, Nogueira LS Lucio’s phenomenon: a case report and review of the literature. Int J Dermatol. 2005;44(7):566–571. | ||
Saoji V, Salodkar A. Lucio leprosy with Lucio phenomenon. Indian J Lepr. 2001;73(3):267–272. | ||
Kumari R, Thappa DM, Basu D. A fatal case of Lucio phenomenon from India. Dermatol Online J. 2008;14(2):10. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
