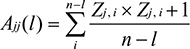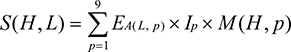Back to Journals » Biologics: Targets and Therapy » Volume 12
Designing of CD8+ and CD8+-overlapped CD4+ epitope vaccine by targeting late and early proteins of human papillomavirus
Authors Kaliamurthi S , Selvaraj G , Kaushik AC, Gu KR , Wei DQ
Received 21 June 2018
Accepted for publication 1 August 2018
Published 2 October 2018 Volume 2018:12 Pages 107—125
DOI https://doi.org/10.2147/BTT.S177901
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Doris Benbrook
Satyavani Kaliamurthi,1 Gurudeeban Selvaraj,1 Aman Chandra Kaushik,2 Ke-Ren Gu,1,3 Dong-Qing Wei1,2
1Centre of Interdisciplinary Science – Computational Life Sciences, College of Food Science and Engineering, Henan University of Technology, Zhengzhou, China; 2The State Key Laboratory of Microbial Metabolism, College of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China; 3College of Chemistry, Chemical Engineering and Environment, Henan University of Technology, Zhengzhou, China
Background and aim: Human papillomavirus (HPV) is an oncogenic agent that causes over 90% of cases of cervical cancer in the world. Currently available prophylactic vaccines are type specific and have less therapeutic efficiency. Therefore, we aimed to predict the potential species-specific and therapeutic epitopes from the protein sequences of HPV45 by using different immunoinformatics tools.
Methods: Initially, we determined the antigenic potential of late (L1 and L2) and early (E1, E2, E4, E5, E6, and E7) proteins. Then, major histocompatibility complex class I-restricted CD8+ T-cell epitopes were selected based on their immunogenicity. In addition, epitope conservancy, population coverage (PC), and target receptor-binding affinity of the immunogenic epitopes were determined. Moreover, we predicted the possible CD8+, nested interferon gamma (IFN-γ)-producing CD4+, and linear B-cell epitopes. Further, antigenicity, allergenicity, immunogenicity, and system biology-based virtual pathway associated with cervical cancer were predicted to confirm the therapeutic efficiency of overlapped epitopes.
Results: Twenty-seven immunogenic epitopes were found to exhibit cross-protection (≥55%) against the 15 high-risk HPV strains (16, 18, 31, 33, 35, 39, 51, 52, 56, 58, 59, 68, 69, 73, and 82). The highest PC was observed in Europe (96.30%), North America (93.98%), West Indies (90.34%), North Africa (90.14%), and East Asia (89.47%). Binding affinities of 79 docked complexes observed as global energy ranged from -10.80 to -86.71 kcal/mol. In addition, CD8+ epitope-overlapped segments in CD4+ and B-cell epitopes demonstrated that immunogenicity and IFN-γ-producing efficiency ranged from 0.0483 to 0.5941 and 0.046 to 18, respectively. Further, time core simulation revealed the overlapped epitopes involved in pRb, p53, COX-2, NF-X1, and HPV45 infection signaling pathways.
Conclusion: Even though the results of this study need to be confirmed by further experimental peptide sensitization studies, the findings on immunogenic and IFN-γ-producing CD8+ and overlapped epitopes provide new insights into HPV vaccine development.
Keywords: human leukocyte antigen, killer cells, overlapped epitopes, time course simulation
Introduction
Human papillomavirus (HPV) is an oncogenic, mucosa-infecting, and etiologic agent that causes over 90% of cases of cervical cancer in the world.1,2 The genome of HPV consists of three regions, viz, noncoding region (extended control), and nonstructural (E1–E7) and structural regions (L1, L2) of the open reading frames.3,4 Briefly, E1 protein is the ATP-dependent DNA helicase which is involved in replication. E2 protein acts as a regulator of viral gene transcription and genome replication. E4 and E5 proteins are involved in the early and late phases of the virus life cycle and immune modulation. E6 and E7 proteins are involved in cell differentiation and targeting numerous negative regulators of the cell cycle (tumor suppressor genes p105Rb and p53). The assemblage of L1 and L2 proteins in capsomers results in the formation of icosahedral capsids around the viral genetic material during formation of progeny.5,6 Identification of CD8+ antigenic determinants is crucial in vaccine development.7 Cytotoxic T lymphocytes (CTLs) are also known as killer cells or CD8 cells matured from the thymus of a specific primary lymphoid organ of the human immune system. CD8+ T cells play a vital role in cell-mediated immune responses against invading intracellular foreign substances like viruses.8 In addition to CD8+ T cells, the peptides of CD4+ T cells are presented on major histocompatibility complex (MHC) class II, which are located in the antigen-presenting cells (APCs), to trigger immune response. Therefore, CD4+ cells are also known as T helper (Th) cells. Initially, APC identifies the pathogen-associated molecular pattern by toll-like receptors (TLRs) which results in the cleavage of antigen into small fragments. Then, MHC class II peptides complex activates the naive CD4+ Th cells and triggers CD8+-mediated cellular immune response or B cell-mediated humoral immune response. Interestingly, when the antigen complexes with MHC class I directly, it results in CD8+-mediated cellular immune response.9,10 The detection of CTL epitopes and the stimulation of immune response play a critical role in an individual’s immune system.
Currently available prophylactic vaccines such as Gardasil (HPV6, 11, 16, and 18) and Gardasil 9 (HPV31, 33, 45, 52, and 58) could provide protection to women by inducing significant immunity, but these vaccines are limited to those aged between 9 and 26 years.11 However, these prophylactic vaccines demonstrated no therapeutic benefits in patients who already had HPV infections.12,13 The US Food and Drug Administration (FDA) and the Centers for Disease Control and Prevention (CDCP) have recommended that HPV vaccines are safe but can cause more pain, uneasiness, soreness, and Guillain–Barre syndrome in patients.14,15
However, to control the prevalence rate of high-grade HPV infections, species-specific screening and vaccination will be needed.16 Out of 150 types of HPV, the 13 high-risk HPV (hrHPV) types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) which induce uncontrolled cell growth in the cervix uteri region have been identified.17,18 Among these, HPV16 and 18 were reported as the major strains to cause cervical cancers (about 67.4%) around the world.19–21 In fact, the contributions of other hrHPV types are limited due to their geographical distribution. HPV45 is a member of HPV18 alpha papillomaviridae family and a species of α7.22–24 Moreover, the prevalence rate of HPV45 in cases of cervical cancer was found to be high varying from 1.6% to 37.4% which was recorded in different geographical regions such as Ghana, Portugal, Serbia, Spain, Pakistan, USA, Japan, Burkina Faso, Northern parts of China, Southeast regions of Brazil, and Northeast of India.18,25–32 Difference in the host genetic framework and oncogenicity of the circulating variants might play a critical role in geographical distribution of HPV45. Together with HPV16 and 18, HPV45 is highly recorded in cases of squamous cell carcinoma (75%) and adenocarcinoma (94%), and in cases of invasive cervical cancer and cervical intraepithelial neoplasia grade 3.2,16,33 Based on the level of enrichment in cervical cancer cases compared to women with normal cytological features, HPV45 has been suggested as the third highest oncogenic agent next to HPV16 and 18.1,30,34,35 In comparison to other hrHPV types, limited reports exist about the peptide research in HPV45.
Immunogenic peptides are one of the excellent resources to develop therapeutic vaccines against hrHPV strains. Instant accessibility of open databases and informatic tools suggest efficient methods to develop vaccines with reduced time, labor, and experimental cost.36,37 Several studies reported that virtual peptides of HPV16 and 18 oncoproteins (E6 and E7) efficiently induce immune response in cervical cancer patients as revealed by their peripheral blood mononuclear cell samples.38–42 However, cross-protection against all the hrHPV strains could not be ignored.40 The cross-reactivity of CTL peptide is suspected to play an essential role in producing immune response against multiple strains.43 Selection of immunogenic epitopes from the whole genome of hrHPV strains may enable designing of novel CTL vaccines. Therefore, we identified the potential immunogenic CD8+, CD8+-overlapped CD4+, and B-cell epitopes from the antigenic proteins of HPV45 and anticipated an epitopes pool with the capability to demonstrate immunogenic responses against all 15 hrHPV strains. The workflow of the study is illustrated in Figure 1.
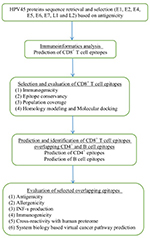  | Figure 1 Flowchart of the overall study. Abbreviations: HPV, human papillomavirus; IFN-γ, interferon gamma. |
Materials and methods
Sequence retrieval and antigenicity-based selection
The whole amino acid sequence of HPV45 genome was retrieved from UniProt Knowledgebase (UniProtKB) database. The amino acid sequences of complete list of nonstructural and structural proteins of HPV45, which included sequences of replication initiation protein E1, regulatory proteins E2, E4, E5, E6, and E7, major capsid protein L1, and minor capsid protein L2, were obtained in October 2017.44 In total, sequences of 139 proteins were extracted which included E1 (11), E2 (12), E4 (04), E5 (03), E6 (27), E7 (26), L1 (32), and L2 (24). Alignment-independent VaxiJen 2.0 tool was employed to determine the antigenicity of the complete list of selected sequences. The protective antigenic score was calculated based on the auto cross-covariance (ACC), which is the transformation of protein sequences into normal vectors with essential amino acid properties.45
|
|
where Ajj(l) denotes ACC, n is the number of amino acids in a sequence, i is the amino acid position, and l is the lag; index j (1–3) was used for the Z scales. Antigenic score of each protein was predicted with the respective threshold value (>0.4). In addition, the same method was followed for the overlapped epitopes.
Immunoinformatics analysis
Prediction of CD8+ CTL epitopes
NetCTL 1.2 server was employed to predict CTL epitopes from the input sequence. The integrated prediction (PI) scores of the CTL epitopes with the presence of MHC ligand were obtained as a result. The threshold value 0.75 with a sensitivity of 0.80 and a specificity of 0.970 was set for CTL epitope prediction.46 The Immune Epitope Database (IEDB) combined tool was used to predict the processing of antigen by MHC class I antigen processing and presentation pathway. NetMHC Artificial Neural Network (ANN) 3.4 was used to find out the natural potential (proteasomal cleavage, transporter associated with antigen processing (TAP) transport, and MHC class I binding) of being a CTL epitope.47 Fifty-one frequently occurring MHC class I alleles, including human leukocyte antigen (HLA)-A (16), HLA-B (23), HLA-C (10), and HLA-E (2), were selected as input. In the TAP transport predictions, the peptide length (9mer) and 0.2 alpha factors were defined as threshold. The MHC class I alleles showing a binding affinity of IC50 <200 nm were selected and used for further study.
Selection and evaluation of CD8+ T-cell epitopes
Determination of immunogenicity
The immunogenicity of the optimized MHC class I peptides was determined using T-cell class I pMHC immunogenicity prediction tool. Based on the properties and position of amino acids within the peptide, the immunogenicity of an MHC peptide was predicted using the following formula.48
|
|
where S denotes the immunogenicity score of a peptide ligand (L) presented on an HLA molecule (H). For every position (p) in the ligand (L), the log enrichment score (E) for the amino acid at that position A (L, p) weighted by the importance of that position Ip was summed. M is the masking of anchor positions on the HLA. In the present analysis, the first, second, and C-terminal amino acids were masked. The same method was followed for the overlapped epitopes.
Analysis of epitopes conservancy
Epitopes conservancy analysis tools were used to find out the degree of conservancy of an epitope within a set of given protein sequences at a given identity level.49 A set of sequences of epitopes (e) and proteins {p} were given as input. The degree of conservation percentage of e within p was estimated as the fraction of {p} that matched the aligned e above a chosen identity level. The conservancy analysis was performed for structural and nonstructural protein sequences of 15 hrHPV strains (HPV16, 18, 31, 33, 35, 39, 51, 52, 56, 58, 59, 68, 69, 73, and 82).
Analysis of population coverage (PC)
The IEDB PC tool was used to calculate the PC of predicted CTL epitopes. It calculates the fraction of individuals predicted to respond to an epitope set based on the HLA genotype frequencies and MHC binding and T-cell restriction data.50 It provided allele frequencies for 21 different ethnicities in 115 countries that assembled into 16 different geographical areas. The number of epitope sets, epitopes with MHC-restricted alleles, MHC class selection, and the population’s area were selected as per the user’s choice during the analysis.
Molecular docking studies
Prediction of peptide structure and homology modeling
PEP-FOLD3 was used to predict the chemical structure of the selected antigenic proteins.51 Hidden Markov model suboptimal conformation sampling approach was applied to predict the structure from the input peptide sequence. For confirmation, 200 simulation runs were selected and the different cluster models were sorted by the sOPEP energy.52 Phyre2 and Iterative Threading ASSEmbly Refinement (I-TASSER) were used to predict the three-dimensional (3D) structure of target receptors. The Phyre2 is a protein modeling and analysis engine which uses highly innovative remote homology detection methods to accurately construct the 3D structure of targets within a duration of 30 minutes to 2 hours.53 I-TASSER is another vital protein structure prediction tool that identifies the respective structural templates from the Protein Data Bank (PDB) through local meta-threading server (LOMETS) with full-length atomic models created by the repetitive template fragment simulations.54 The sequences of MHC class I antigenic allotypes including HLA-A*01:01, HLA-A*02:06, HLA-A*11:01, HLA-A*23:01, HLA-A*26:01, HLA-A*29:02, HLA-A*30:02, HLA-A*32:01, HLA-A*68:01, HLA-B*08:01, HLA-B*15:01, HLA-B*35:01, HLA-B*39:01, HLA-B*58:01, HLA-C*03:03, HLA-C*07:02, HLA-C*08:02, HLA-C*12:03, HLA-C*14:02, HLA-C*15:02, and HLA-C*05:01 were retrieved from the PDB and UniProtKB database. FASTA sequences were given as an input. Scoring percentage of the most favored regions of Ramachandran plot, energy minimization, and root mean square deviation values were used to validate the modeled structure and select a highly precise model.
Molecular docking of predicted peptides’ with target receptors
PatchDock tool was employed for shape representation, surface patch matching, and filtering and scoring.55 Based on the ranking, the elite peptide models were selected for docking with targets. Further, the prepared HLA molecules were used for docking with the predicted epitopes. Then, the results were refined by FireDock server according to the energy function. In the final refinement, a full interface side chain was optimized with an atomic radius of 0.85 to facilitate decreasing the number of conflicts.56 The binding interactions of the docked structures were evaluated using UCSF Chimera 1.11.2, a highly extensible program for interactive visualization (data not shown).57
Prediction and identification of CD8+ epitopes overlapping CD4+ and B-cell epitopes
Prediction of interferon-gamma (IFN-γ)-producing CD4+ T-cell epitopes
IEDB consensus method was used to predict CD4+ T-cell epitopes by combining the following: neural network alignment (Net MHCII 2.2), stabilization matrix method alignment, and CombLib or Sturniolo method. IEDB is a comprehensive dataset consisting of over 10,000 unpublished MHC peptide binding affinities, 29 crystal structures, and 664 experimentally proven CD4+ peptides. The major population in the tool is HLA-DRB locus, which consists of 51 alleles. HLA-DRB1, HLA-DRB3, HLA-DRB4, and HLA-DRB5 subtypes were selected for the analysis. As a result, the lowest percentile rank peptides exhibited higher binding affinity.58 The antigen was presented by the APCs to the CD4+ receptor of Th cells, and then Th cells producing IFN-γ were activated. Moreover, the IFN-γ-producing CD4+ MHC class II-restricted epitopes were analyzed using IFN epitope tool. The prediction program was used with the motif-based model or support vector machine or hybrid algorithms. It showed an accuracy of 81.39% for the prediction of IFN-γ-producing epitopes.59 The dataset consists of 3,705 IFN-γ-inducing and 6,728 non-IFN-γ-inducing MHC class II binders.
Prediction of B-cell epitopes
The B-cell epitopes of the selected antigenic sequences were predicted using ABCpred tool. The results were predicted based on the experimentally trained B-cell epitope datasets. ABCpred is the first server created based on recurrent neural network (ie, Jordan network, a machine-based technique) using fixed-length patterns with an accuracy of 65.93%.60
Evaluation of selected overlapping epitopes
Determination of allergenicity
AllergenFP v.1.0 is an essential tool to predict allergenicity of peptide sequences based on novel descriptor fingerprint approach with an accuracy of 87%, a specificity of 89%, and a sensitivity of 86%. It contains 2,427 allergenic and 2,427 non-allergenic amino acid segments.61
Analysis of cross-reactivity with human proteomes
The similarity analysis was performed to find out the epitopes that match with the human proteome by using the integrated open database, Protein Information Resource, with peptide-matching program.62,63
Systems biology-based virtual cancer pathway prediction and kinetics
A literature survey was performed to collect the required information about the selected protein sequences to design and predict the peptides involved in biochemical pathways.64–74A mathematical and computational model of the entire HPV45 infection signaling pathway interacting with specific genes in the presence of peptides was developed and was visualized using CellDesigner v4.4 modeling tool.75 Nodes in the pathway represented the entities, and edges represented connectivity of one node to another, which was closely related to each other.76 The inhibitory concentration (1 µg) of proposed peptides was collected from the published experimental data and pharmacokinetic studies.64 Mass action kinetics (V=KΠi S) was applied for pharmacokinetic study. Time course and kinetics simulation was performed to concern transition time of cancer and complete biochemical pathway in the presence of targets with peptide using COPASI tool.77
Results
Antigenic proteins
The sequences of proteins E1 (UniProt ID: T2A7A1), E2 (UniProt ID: A9XFM5), E4 (UniProt ID: A9XFL8), E6 (UniProt ID: T2A578), E7 (UniProt ID: M4H3T6), L1 (UniProt ID: I6SCV2), and L2 (UniProt ID: T2A7B7) attained the highest antigenic scores of 0.5027, 0.4353, 0.4668, 0.5795, 0.7251, 0.6404, and 0.5943, respectively. The antigenic scores of all the query proteins are illustrated in Table S1.
CD8+ CTL epitopes
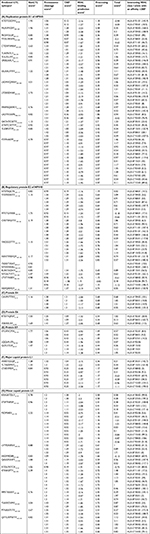
In total, 80 CTL epitopes were predicted from the early proteins E1 (19), E2 (17), E4 (01), E6 (05), and E7 (14), and late proteins L1 (07) and L2 (17) of HPV45 based on MHC class I supertype by NetCTL 1.2 (threshold value >0.75). Among the 80 epitopes, only 50 CTL epitopes showed a higher binding affinity (IC50 <200 nm) for MHC class I alleles as found using NetMHC ANN 3.4. Table 1 depicts the proteasome score, TAP score, MHC score, processing score, total score, and binding affinity of predicted epitopes for different alleles as determined using NetMHC ANN 3.4. According to the results, 17 out of 19 epitopes of E1 protein, such as ATDTGSDMV40–48, MLAVFKDIY200–208, FKDIYGLSF204–212, FTDLVRNFK212–220, CTDWVMAIF226–234, TLIKPATLY246–254, VLILALLRY266–274, LRSSLAALY305–313, SSLAALYWY307–315, LSDMVQWAF350–358, LTDESDMAF362–370, MNMSQWIKY413–421, LADTKVAML506–514, DATHTCWTY516–524, ATHTCWTYF517–525, YLESRVTVF571–579, and FTFPHAFPF579–587, showed binding affinity for more than one HLA (HLA-A, HLA-B, or HLA-C). Among them, epitope FTFPHAFPF interacted with 13 HLA alleles including HLA-A*(02:06, 29:02, 32:01, 68:02, 68:01, 26:01), HLA-B*(15:01, 35:01), and HLA-C*(03:03, 12:03, 07:02, 14:02, 15:02) (Table 1A). Moreover, 12 out of 17 epitopes of E2 protein, including NTEPSQCFK109–117, YVVWDSIYY137–145, MTETGIWEK146–154, CVSYWGVYY158–166, YIKDGDTTY166–174, NSNTWEVQY187–195, TSDDTVSAT209–217, NTHVHNSLL265–273, RLRKYADHY309–317, NTGILTVTY332–340, LTVTYNSEV336–344, and NSVQISVGY187–195, were predicted to have binding affinity for MHC class I alleles. Among these, the CVSYWGVYY exhibited interactions with six alleles, viz, HLA-A*(29:02, 30:02, 11:01, 01:01, 68:01), and HLA-B*35: 01 (Table 1B). In addition, the CAVPVTTRY4–12 epitope of E4 protein was predicted to have binding affinity for HLA-(A*29, A*30, B*58, B*35) allotypes (Table 1C). Among the five epitopes of E6 protein, RTEVYQFAF41–49 and YSRIRELRY72–80 were predicted to have binding affinity for HLA-A*(32:01, 29:02) and HLA-B*58: 01 MHC class I alleles (Table 1D). Out of 13 MHC-binding E7 peptides (Table 1E), STLSFVCPW93–101 (HLA-A*32:01, HLA-B*58:01, HLA-B*57:01), LQQLFLSTL87–95 (HLA-A*02:06), and QLFLSTLSF89–97 (HLA-B*15:02, HLA-B*15:01, HLA-A*32:01) were predicted to have a binding affinity of IC50 <200 nm. Furthermore, three out of seven epitopes of L1 protein, such as HVEEYDLQF51–59, TLTAEVMSY67–75, and LTAEVMSYI68–76, exhibited significant binding affinity for the HLA alleles (Table 1F). Among these, the LTAEVMSYI68–76 demonstrated interactions with six alleles, viz, HLA-A*(68:02, 02:06, 02:01), HLA-B*58: 01, and HLA-C*(12:03, 15:02). In addition, 12 out of 17 epitopes of L2 protein, KRASATDLY10–18, STSFTNPAF154–162, FSDPSIIEV162–170, LVTFDNPAY248–256, HSDFMDIIR273–281, QIGGRVHFY312–320, ATDLYKTCK14–22, STINKSFTY362–370, MPSTAASSY377–385, TSAWDVPIY393–401, PTNAATSTY420–428, and QYYLWPWYY435–443, exhibited significant binding affinity for the HLA alleles. Among these, epitope FSDPSIIEV162–170 demonstrated interactions with nine alleles, viz, HLA-A*(02:06, 02:01, 01:01, 68:02) and HLA-C*(12:03, 05:01, 15:02, 08:02, 03:03) (Table 1G).
Immunogenicity
In the present findings, 27 out of 50 epitopes were found to be immunogenic. A list of immunogenic epitopes from structural and nonstructural proteins is illustrated in Table S2.
Epitopes conservancy
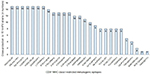
While designing epitope-based vaccines, the use of conserved epitopes may be helpful to provide broader protection across multiple strains and to develop cross-reactive vaccines. Table S3 shows the detailed results of conservancy (≥50%) analysis of the 27 immunogenic epitopes and their positions, and protein sub-sequences and their respective cross-protection against hrHPV strains. The conservation of immunogenic epitopes across the 15 hrHPV strains is shown in Figure 2.
PC
The highest PC was observed in Europe (96.30%), North America (93.98%), West Indies (90.34%), North Africa (90.14%), East Asia (89.47%), Northeast Asia (89.68%), West Africa (88.82%), South Asia (87.57%), Southwest Asia (86.37%), Southeast Asia (86.33%), Oceania (84.47%), East Africa (84.29%), Central Africa (80.16%), and South America (80.95%). About 93.68% of cumulative PC was achieved around the world (Figure 3).
Molecular docking studies
Table 2 depicts the docking interaction of CD8+ MHC class I-restricted epitopes with different HLA alleles. All the epitopes were docked with structurally defined targets. Totally, 79 docked complexes exhibited a range of binding affinities in terms of global energy (-10.80 to -86.71 kcal/mol), attractive van der Waals energy (-5.24 to -36.74 kcal/mol), and repulsive van der Waals energy (0.01–9.72 kcal/mol).
MHC class II-restricted CD4+ epitopes and IFN-γ production
The high-ranked MHC class II CD4+ epitopes with lower percentile value <1% and their IFN-γ-producing capacities are illustrated in Table S4. Among them, the high-ranked CD4+ epitopes and the overlapped potential immunogenic CD8+ T-cell epitope regions are highlighted in bold.
B-cell epitopes
In total, 197 B-cell epitopes (16mer amino acid length) were predicted from N- and C-terminal regions of the E1 (61), E2 (36), E4 (07), E5 (05), E6 (15), E7 (09), L1 (14), and L2 (50) amino acid sequences of HPV45. The predicted B-cell epitopes were selected according to their scores (threshold 0.5) determined by trained recurrent neural network. The B-cell peptides showing the highest score >0.5 exhibited the highest probability to be as epitope. The predicted 16mer B-cell epitopes start position and B-cell epitopes containing the immunogenic CD8+ T-cell epitopes-overlapped regions are shown in Table S5.
Among them, G(VLILALLRY)KCGKNR265–280 (0.89), R(SSLAALYWY)RTGISN306–321 (0.78), V(FTFPHAFPF)DKNGNP578–593 (0.76) and A(MLAVFKDIY)GLSFTD199–214 (0.67), (CVSYWGVYY)IKDGDTT158–173 (0.86), K(NTGILTVTY)NSEVQR331–346 (0.85), DSIYY(MTETGIWEK)TA141–156 (0.82), HKCIDF(YSRIRELRY)Y66–81 (0.83), TLE(RTEVYQFAF)KDLC38–53 (0.67), R(HVEEYDLQF)IFQLCT50–65 (0.87), GK(QIGGRVHFY)HDISP310–325 (0.93), LTRPSS(LVTFDNPAY)E242–257 (0.88), SVSIS(STSFTNPAF)SD149–164 (0.79), NVTVPL(TSAWDVPIY)T387–402 (0.77), SFTNPA(FSDPSIIEV)P156–167 (0.72), and (LVTFDNPAY)EPLDTTL248–263 (0.69) from E1, E2, E6, L1, and L2 amino acid sequences overlapped the potential immunogenic CD8+ T-cell epitopes.
Overlapping epitopes
Table 3 illustrates the epitope sequence, position in the selected protein, allergenicity, antigenicity, immunogenicity, and IFN-γ-producing efficiency of 23 CD8+ T-cell epitopes overlapping CD4+ and B-cell epitopes. All the selected epitopes were found to be immunogenic. Among the 23 epitopes, 19 were identified as non-allergenic and 15 were antigenic with a threshold value of 0.4. In addition, 19 epitopes had the efficiency to induce IFN-γ production. The similarity level zero indicated no unique matches of query epitopes found against the Homo sapiens (Taxonomy ID: 9606). Moreover, these peptides would eliminate the risk of inducing autoimmune disorders.
Virtual biochemical pathways and kinetics
The predicted peptides of E1, E2, E6, and L1 inhibit or trigger the P13K/AKT signaling pathway, MAPK pathway, overexpression of COX-2, downregulation of tumor suppressor p53, transcriptional repressor NF-X1 gene, multifunctional regulator gene (MYC), tumor suppressor pRb, higher expression of minichromosome maintenance proteins, proliferating cell nuclear antigen, P16 protein, antigen Ki-67, alternate reading frame protein (P14ARF), and MDM gene which are involved in uncontrolled cell division, virus immortalization, MAPK pathway, apoptosis, notch pathway, transcription, and telomerase reverse transcription (Figure 4A). These genes and pathways are actively involved in the development of HPV causing cervical cancers including cervical intraepithelial neoplasia and invasive cervical carcinoma. The results demonstrate that the predicted peptides from HPV45 would efficiently inhibit the development of cervical cancer.
The pharmacokinetic simulations are shown in Figure 4B, which indicate the concentration of entities and time in different colors. After 30 seconds of time course simulation, all entities were constant. The results indicated that the proposed peptides of HPV45 could work as best anticancer peptides through the inhibition of various pathways involved in the cervical cancer development. The kinetics simulation was also performed using different concentrations of HPV45 peptides for cross-validation. The results obtained at the concentration of 0.0300 µm support the literature information. Time course and kinetics simulation prediction demonstrated the biological behavior of HPV45 in the presence of peptides where HPV45 interacts with different molecules, which play an important role in cervical cancer inhibition.
Discussion
HPV infection is solely responsible for 99.7% of cervical cancer cases. Fitzmaurice et al78 reported the age-standardized incidence and death rates of cervical cancer in the developing and developed countries from 1990 to 2013. Exploitation of bioinformatic techniques has led to an emerging concept in advanced vaccine design against different diseases.79 Immunoinformatics approaches have played a vital role in the qualitative screening of multiple genomes to predict target epitopes, which provides a rational design to enhance the T-cell immune responses in a cost-effective manner.42,80,81 The possibility of antigen identification at the molecular level has enabled sensible designing of peptide vaccines. An advantage of the epitope-based vaccines is that they have the potential to induce high dosage of immunogenicity at a lower cost. Therefore, the present study aimed at the prediction of potential immunogenic epitopes from the whole genome of hrHPV45 strain for designing epitope-based vaccines.
VaxiJen is the first server developed for the prediction of antigenicity of proteins of viruses, bacteria, fungi, parasites, and tumor cells.45 The potential antigen classification is completely based on the physicochemical properties of proteins and not related to sequence alignment. Therefore, we utilized VaxiJen tool in the present study and predicted potential antigenic proteins with 87% accuracy. The reliable predictions of CTL epitopes are vital for consistent drug development.82,83 The CD8+ receptors consist of single CD8α and CD8β chains. Furthermore, the CD8+ CTLs identify the epitopes which are presented by the MHC class I established on all the nucleated cells. This process is facilitated by the binding of CD8+ to α3 conserved region of MHC class I. Recently, elution studies by Bassani-Sternberg et al84 proved that the 9mer peptides are naturally optimized by the antigen processing process. This result supports the present study.
MHC class I-presenting peptides are synthesized from endogenous processing pathway-derived proteins.85 We predicted 50 potential CD8+ MHC class I-restricted 9mer epitopes based on endogenous processing pathway (proteasomal cleavage, TAP transport, and MHC class I binding algorithm) by using combined methods in the IEDB. The α-chain of MHC class I contains the peptide-binding domain, immunoglobulin-like domain, and a transmembrane region. The heavy chain of the MHC class I molecule is coded by the genes at HLA (A, B, and C) loci. The MHC genes are highly polymorphic in nature; it means the residues in the N- and C-terminal regions of MHC proteins could generate clefts for specific peptide binding. MHC class I alleles exhibit broader binding specificity for peptides between 1,000 and 10,000 amino acids with high promiscuity.86 Neural networks based on computational approaches have played a vital role in the prediction of the binding affinity of a peptide for specific MHC molecules.87 In the present study, the FTFPHAFPF, CVSYWGVYY, LTAEVMSYI, and FSDPSIIEV antigenic epitopes were found to interact with more number of HLA alleles. This specific high-binding affinity is absolutely desired because the efficiency of an epitope in vaccine development greatly depends on the extent of its interaction with HLA alleles. Moreover, activated CD8+ cells kill the targets through three crucial mechanisms including secretion of antitumor and antiviral (IFN-γ) substances,88 membrane disruption by granulysin, and Fas-mediated killing.89
Recent studies reported the prevalence rate of multiple hrHPV genotypes in cervical cancer90 and the poor survival rate associated with invasive cervical carcinomas. Moreover, the hrHPV strains co-infection has been shown to increase the death rate of HIV patients.91 Conserved epitopes provide powerful immunization for prolonged period. Sabah et al92 found that the single 9mer CTL epitope ETSVHEIEL was conserved (100%) in all E6 proteins of HPV58 isolates. While designing an epitope-based vaccine, the use of conserved epitopes may be helpful to provide broader cross-protection across multiple strains. The present study demonstrated that 14 immunogenic CD8+ epitopes showed cross-protection against more than 12 hrHPVs.
Clifford et al93 reported that the HPV45-positive invasive cervical cancer rate was higher in Africa (15.5%) and Europe (6.3%). The highest grade of cervical cancer lesions was also recorded in Asia (2.8%) and Latin America (8.1%). Moreover, cervical cancer is the second fatal cancer contributing to 14% of cancers in Indian women.94 The American Cancer Society estimated that there were 12,820 cases of invasive uterine cervical cancer in 2017.95 The age-standardized incidence rate of cervical cancer in the overall geographical region of China is around 98.9%.96 The present findings showed that the highest PC of the pooled epitopes was recorded in Europe, North America, West Indies, West Indies, North Africa, East Asia, Northeast Asia, West Africa, South Asia, Southwest Asia, Southeast Asia, Oceania, East Africa, Central Africa, South America, Central America, and South Africa. These PC findings showed that the peptide vaccine from HPV45 would be effective for immunizing a huge population over an extensive geographical region.
There is a lack of scientific reports on binding cleft interactions of CD8+ epitopes of HPV45 with HLA molecules. Considering this fact, the binding affinity of the potential 27 immunogenic CD4+ epitopes for receptors such as HLA-A*(02:06, 0201, 11:01, 68), HLA-B*(35:01, 58:01), and HLA-C*05:01 was analyzed using docking tools. It is notable that the prophylactic vaccines from HPV L1 protein are used to treat HPV infections in more than 100 countries.97 In the present study, we found strong binding affinity between the predicted L1 epitope LTAEVMSYI and three different MHC class I alleles HLA-A*(02:06, 02:01, 68, 58:01). Moreover, studies reported that multi-epitope HPV16 E6/E7 vaccine enhanced the clearance of HPV-positive cancer in the mouse model.98,99 TA-CIN vaccine was synthesized from a single fusion protein of HPV16 (L2, E6, and E7), and the clinical Phase I study in healthy volunteers showed significant production of IFN-γ which confirmed T-cell immune response in patients.100 In the present study, minor capsid protein FSDPSIIEV epitope showed affinity for a peptide-binding groove with allele-specific pockets in MHC class I HLA-A*02:01, HLA-A*01:01, HLA-A*68:02, and HLA-C*15:02 alleles. In the case of E6 CD8+ epitopes, RTEVYQFAF showed potential binding interactions with HLA-A*32:01 and HLA-B*58:01 with the highest global binding energy of -61.05 and -58.52 kcal/mol, respectively. Moreover, E2 is the violent regulator of extra-chromosomal replication and E1 protein expression of the viruses. In addition, the E2 proteins from hrHPV cause apoptosis in HPV-transformed cells compared to E2 from low-risk HPV strains. From the present findings, the E1 epitope FTFPHAFPF exhibited excellent affinity for a peptide-binding groove with allele-specific pockets in MHC class I HLA-A*(32:01, 26:01, 68:02), HLA-B*15:01, and HLA-C*(07:02, 14:02) alleles. The E2 CD8+ epitope CVSYWGVYY exhibited excellent affinity for MHC class I HLA-A*(29:02, 30:02, 11:01, 68:01) alleles.
Both MHC class I and II molecules share some superficial similarities and bind to the 9mer peptides.101 However, vital differences exist between the MHC class I and II molecules. The capped nature of the MHC class I peptide-binding groove with allele-specific pockets does not allow variation in the length of peptides (9mer), which is called register shifting. This is because the peptide-binding groove of MHC class I molecules is closed at each end. In contrast, the peptide-binding groove of MHC class II molecules is open at both ends which allows for binding of more extended peptides (ranging from >15 to 24 AA in length). Sercarz and Maverakis102 reported that longer peptides have peptide-flanking residues that lie outside of the peptide-binding groove in MHC class II molecules and might interact with peptides in another distal location. MHC class II proteins primarily present peptides derived from endocytosis of extracellular proteins (exogenous processing pathway). In the present study, promising 15mer CD4+ epitopes were predicted by consensus approaches from the late and early proteins of HPV45. The predicted CD4+ epitopes possessed good binding affinity (<1% lowest percentile rank) for MHC class II alleles. CD4+ immune responses are associated with the production of IFN-γ or IL-2. Furthermore, the IFN-γ-producing CD4+ epitopes were found by using machine learning hybrid method. B cell-mediated humoral immunity involves the recognition of antigens circulating in the body fluid. Recently, Paolini et al103 reported the production of CD8+ and CD4+ lymphocytes induced by anticancer pVAX-E5CP and pVAX-E5MultiCP E5 vaccine (carrying the whole E5 gene or multi-epitope) in the preclinical cancer model. Bristo et al104 reported that immunization with a single peptide that contained both the CD4+ and CD8+ CTL epitopes (ie, 9mer CD8+ MHC class I-restricted peptides nested within the 13mer CD4+ epitopes) elicited the response and production of T cells in an animal model. We also predicted the CD8+-overlapped (9mer) epitopes within the ideal B-cell (16mer) and CD4+ cell (15mer) epitope regions. The results suggested that the CD8+, nested CD4+, or B-cell epitopes may elicit CTL-induced cellular immunity or T cell-induced adaptive immunity or B cell-mediated humoral immunity. These identified epitopes from HPV45 may have substantial implications in the peptide vaccine-based immunotherapy.
Conclusion
Each HPV type is immunologically distinct, which is necessary to find suitable target for the development of therapeutic vaccines. We conclude that the predicted CD8+, nested IFN-γ-producing CD4+, and linear B-cell epitopes can possibly induce both cell-mediated and humoral immunity by CTLs, Th1 cells, and B cells in the virus-infected host system. Moreover, the CD8+ epitopes from HPV45 proteins have the potential to be effectively used for the development of unique cross-protective peptide vaccines against hrHPV-attributed cervical cancer. However, further experimental peptide sensitization studies are needed to confirm our findings on humoral and cell-mediated immune response of the overlapped epitopes. This may enable designing of novel peptide-based therapeutic vaccines with improved efficiency for better protection.
Acknowledgments
The authors are grateful for The Key Research Area Grant 2016YFA0501703 from the Ministry of Science and Technology of China, Henan Natural Science Grant 162300410060, grants from the State Key Lab on Microbial Metabolism, and Joint Research Funds for Medical and Engineering & Scientific Research at Shanghai Jiao Tong University awarded to D-QW. The authors GS and SK are grateful for the Postdoctoral Research Grants 21450003 and 21450004 from Henan University of Technology, Henan Province, China. KG was supported by the research grant 2013BAB11B02 from the Ministry of Science and Technology of China.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Ohlenschläger O, Seiboth T, Zengerling H, et al. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene. 2006;25(44):5953–5959. | ||
de Sanjose S, Quint WGV, Alemany L, et al; Retrospective International Survey and HPV Time Trends Study Group. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11(11):1048–1056. | ||
Mcbride AA. The Papillomavirus E2 proteins. Virology. 2013; 445(1–2):57–79. | ||
Bogani G, Leone Roberti Maggiore U, Signorelli M, et al. The role of human papillomavirus vaccines in cervical cancer: Prevention and treatment. Crit Rev Oncol Hematol. 2018;122:92–97. | ||
Mighty KK, Laimins LA. The role of human papillomaviruses in oncogenesis. Recent Results Cancer Res. 2014;193:135–148. | ||
Graham SV. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5(10):1493–1506. | ||
Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annu Rev Immunol. 2000;18(1):275–308. | ||
Gao GF, Jakobsen BK. Molecular interactions of coreceptor CD8 and MHC class I: the molecular basis for functional coordination with the T-cell receptor. Immunol Today. 2000;21(12):630–636. | ||
Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15(4):203–216. | ||
Shcharansky M, Toth I, Peptide-based synthetic vaccines . Chem Sci. 2016;7(2):842–854. | ||
Petrosky E, Bocchini JA Jr, Hariri S, et al; Centers for Disease Control and Prevention (CDC). Use of 9-valent human papillomavirus (HPV) vaccine: updated HPV vaccination recommendations of the advisory committee on immunization practices. MMWR Morb Mortal Wkly Rep. 2015;64(11):300–304. | ||
Wain G. The human papillomavirus (HPV) vaccine, HPV related diseases and cervical cancer in the post-reproductive years. Maturitas. 2010;65(3):205–209. | ||
Pandhi D, Sonthalia S. Human papilloma virus vaccines: Current scenario. Indian J Sex Transm Dis. 2011;32(2):75–85. | ||
Wakerley BR, Yuki N. Pharyngeal-cervical-brachial variant of Guillain-Barre syndrome. J Neurol Neurosurg Psychiatry. 2014;85(3):339–344. | ||
Unger Z, Maitra A, Kohn J, Devaskar S, Stern L, Patel A. Knowledge of HPV and HPV Vaccine among Women Ages 19 to 26. Women’s Health Issues. 2015;25(5):458–462. | ||
Tjalma WA, Depuydt CE. Don’t forget HPV-45 in cervical cancer screening. Am J Clin Pathol. 2012;137(1):161–162. | ||
Muñoz N, Bosch FX, de Sanjosé S, et al; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. | ||
Sasagawa T, Maehama T, Ideta K, Irie T; Fujiko Itoh J-HERS Study Group. Population-based study for human papillomavirus (HPV) infection in young women in Japan: A multicenter study by the Japanese human papillomavirus disease education research survey group (J-HERS). J Med Virol. 2016;88(2):324–335. | ||
Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121(3):621–632. | ||
Félix A, Alemany L, Tous S, de Sanjosé S, Bosch FX. HPV distribution in cervical cancer in Portugal. A retrospective study from 1928 to 2005. Papillomavirus Res. 2016;2:41–45. | ||
Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55(4):244–265. | ||
de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. | ||
Naghashfar ZS, Rosenshein NB, Lorincz AT, Buscema J, Shah KV. Characterization of human papillomavirus type 45, a new type 18-related virus of the genital tract. J Gen Virol. 1987;68(Pt 12):3073–3079. | ||
Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One. 2013;8(8):e72565. | ||
Awua AK, Sackey ST, Osei YD, Asmah RH, Wiredu EK. Prevalence of human papillomavirus genotypes among women with cervical cancer in Ghana. Infect Agent Cancer. 2016;11(1):4. | ||
Stamenkovic´ M, Kneževic´ A, Kneževic´ I, et al. High-risk human papilloma virus genotypes in cervical carcinoma of Serbian women: Distribution and association with pathohistological findings. Biologicals. 2016;44(5):412–416. | ||
Godínez JM, Heideman DA, Gheit T, et al. Differential presence of Papillomavirus variants in cervical cancer: an analysis for HPV33, HPV45 and HPV58. Infect Genet Evol. 2013;13:96–104. | ||
Aziz H, Iqbal H, Mahmood H, et al. Human papillomavirus infection in females with normal cervical cytology: Genotyping and phylogenetic analysis among women in Punjab, Pakistan. Int J Infect Dis. 2018;66:83–89. | ||
Djigma FW, Ouédraogo C, Karou DS, et al. Prevalence and genotype characterization of Human Papillomaviruses among HIV-seropositive in Ouagadougou, Burkina Faso. Acta Trop. 2011;117(3):202–206. | ||
Hh X, Wang K, Feng XJ, et al. Prevalence of human papillomavirus genotypes and relative risk of cervical cancer in China: a systematic review and meta-analysis. Oncotarget. 2018;9(20):15386–15397. | ||
Basto DL, Vidal JP, Pontes VB, et al. Genetic diversity of human papillomavirus types 35, 45 and 58 in cervical cancer in Brazil. Arch Virol. 2017;162(9):2855–2860. | ||
Das D, Rai AK, Kataki AC, et al. Nested multiplex PCR based detection of human papillomavirus in cervical carcinoma patients of North- East India. Asian Pac J Cancer Prev. 2013;14(2):785–790. | ||
Amaro-Filho SM, Pereira Chaves CB, Felix SP, Basto DL, de Almeida LM, Moreira MAM. HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer. Papillomavirus Res. 2018;5:172–179. | ||
Awua AK, Adanu RMK, Wiredu EK, et al. Unique LCR variations among lineages of HPV16, 18 and 45 isolates from women with normal cervical cytology in Ghana. Virol J. 2017;14(1):85. | ||
Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131(10):2349–2359. | ||
Usman Mirza M, Rafique S, Ali A, et al. Towards peptide vaccines against Zika virus: Immunoinformatics combined with molecular dynamics simulations to predict antigenic epitopes of Zika viral proteins. Sci Rep. 2016;6(1):37313. | ||
Patronov A, Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3(1):120139. | ||
Kather A, Ferrara A, Nonn M, et al. Identification of a naturally processed HLA-A*0201 HPV18 E7 T cell epitope by tumor cell mediated in vitro vaccination. Int J Cancer. 2003;104(3):345–353. | ||
Kim S, Chung HW, Lee K-R, Lim JB. Identification of novel epitopes from human papillomavirus type 18 E7 that can sensitize PBMCs of multiple HLA class I against human cervical cancer. J Transl Med. 2014;12(1):229. | ||
Singh KP, Verma N, Akhoon BA, et al. Sequence-based approach for rapid identification of cross-clade CD8+ T-cell vaccine candidates from all high-risk HPV strains. 3 Biotech. 2016;6(1):39. | ||
Chenzhang Y, Wen Q, Ding X, et al. Identification of the impact on T- and B- cell epitopes of human papillomavirus type-16 E6 and E7 variant in Southwest China. Immunol Lett. 2017;181:26–30. | ||
Khan A, Junaid M, Kaushik AC, et al. Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PLoS One. 2018;13(5):e0196484. | ||
Petrosky E, Bocchini JA Jr, Hariri S, et al; Centers for Disease Control and Prevention (CDC). Use of 9-valent human papillomavirus (HPV) vaccine: updated HPV vaccination recommendations of the advisory committee on immunization practices. MMWR Morb Mortal Wkly Rep. 2015;64(11):300–304. | ||
UniprotConsortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46(5):2699. | ||
Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8(1):4. | ||
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8(1):424. | ||
Tenzer S, Peters B, Bulik S, et al. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci. 2005;62(9):1025–1037. | ||
Calis JJ, Maybeno M, Greenbaum JA, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9(10):e1003266. | ||
Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics. 2007;8(1):361. | ||
Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7(1):153. | ||
Kaur H, Garg A, Raghava GP. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett. 2007;14(7):626–631. | ||
Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J Chem Theory Comput. 2014;10(10):4745–4758. | ||
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. | ||
Pandit SB, Zhang Y, Skolnick J. TASSER-Lite: an automated tool for protein comparative modeling. Biophys J. 2006;91(11):4180–4190. | ||
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server):W363–W367. | ||
Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36(Web Server):W229–W232. | ||
Huang CC, Meng EC, Morris JH, Pettersen EF, Ferrin TE. Enhancing UCSF Chimera through web services. Nucleic Acids Res. 2014;42(W1):W478–W484. | ||
Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. | ||
Dhanda SK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8(1):30. | ||
Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. | ||
Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. | ||
Lucchese G, Stufano A, Kanduc D. Proteome-guided search for influenza A B-cell epitopes. FEMS Immunol Med Microbiol. 2009;57(1):88–92. | ||
Chen C, Li Z, Huang H, Suzek BE, Wu CH; UniProt Consortium. A fast Peptide Match service for UniProt Knowledgebase. Bioinformatics. 2013;29(21):2808–2809. | ||
Nilges K, Höhn H, Pilch H, et al. Human papillomavirus type 16 E7 peptide-directed CD8+ T cells from patients with cervical cancer are cross-reactive with the coronavirus NS2 protein. J Virol. 2003;77(9):5464–5474. | ||
Romanczuk H, Howley PM. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci. 1992;89(7):3159–3163. | ||
Gao LJ, Gu PQ, Zhao W, et al. The role of globular heads of the C1q receptor in HPV 16 E2-induced human cervical squamous carcinoma cell apoptosis is associated with p38 MAPK/JNK activation. J Transl Med. 2013;11(1):118. | ||
Zhang E, Feng X, Liu F, Zhang P, Liang J, Tang X. Roles of PI3K/Akt and c-Jun signaling pathways in human papillomavirus type 16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis in non-small cell lung cancer cells. PLoS One. 2014;9(7):e103440. | ||
Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 2007;67(8):3976–3985. | ||
Zhou Y, Wei Y, Zhu J, et al. GRIM-19 disrupts E6/E6AP complex to rescue p53 and induce apoptosis in cervical cancers. PLoS One. 2011;6(7):e22065. | ||
Xu B, Chotewutmontri S, Wolf S, et al. Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PLoS One. 2013;8(6):e66693. | ||
Doorbar J, Quint W, Banks L, et al. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine. 2012;30(5):F55–F70. | ||
Sun J, Xiong J, Zhen Y, Chen ZL, Zhang H. P53 and PCNA is Positively Correlated with HPV Infection in Laryngeal Epitheliopapillomatous Lesions in Patiets with Different Ethnic Backgrounds in Xinjiang. Asian Pac J Cancer Prev. 2012;13(11):5439–5444. | ||
Vazquez-Vega S, Sanchez-Suarez LP, Andrade-Cruz R, et al. Regulation of p14ARF expression by HPV-18 E6 variants. J Med Virol. 2013;85(7):1215–1221. | ||
Kim SM, Lee JU, Lee DW, Kim MJ, Lee HN. The prognostic signifi cance of p16, Ki-67, p63, and CK17 expression determined by immunohistochemical staining in cervical intraepithelial neoplasia 1. Korean J Obstet Gynecol. 2011;54(4):184–191. | ||
Funahashi A, Morohashi M, Kitano H, Tanimura N. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO. 2003;1(5):159–162. | ||
Kaushik AC, Kumar A, Dwivedi VD, et al. Deciphering the Biochemical Pathway and Pharmacokinetic Study of Amyloid βeta-42 with Superparamagnetic Iron Oxide Nanoparticles (SPIONs) Using Systems Biology Approach. Mol Neurobiol. 2018;55(4):3224–3236. | ||
Hoops S, Sahle S, Gauges R, et al. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22(24):3067–3074. | ||
Fitzmaurice C, Dicker D, Pain A, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–527. | ||
Zhao M, Wei D-Q. Rare diseases: Drug discovery and informatics resource. Interdiscip Sci. 2018;10(1):195–204. | ||
Kamthania M, Sharma DK. Screening and structure-based modeling of T-cell epitopes of Nipah virus proteome: an immunoinformatic approach for designing peptide-based vaccine. 3 Biotech. 2015;5(6):877–882. | ||
Moosavi F, Mohabatkar H, Mohsenzadeh S. Computer-aided analysis of structural properties and epitopes of Iranian HPV-16 E7 oncoprotein. Interdiscip Sci. 2010;2(4):367–372. | ||
Muderspach L, Wilczynski S, Roman L, et al. A phase I trial of a human papillomavirus (HPV) peptide vaccine for women with high-grade cervical and vulvar intraepithelial neoplasia who are HPV 16 positive. Clin Cancer Res. 2000;6(9):3406–3416. | ||
Elemans M, Florins A, Willems L, Asquith B. Rates of CTL killing in persistent viral infection in vivo. PLoS Comput Biol. 2014;10(4):e1003534. | ||
Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics. 2015;14(3):658–673. | ||
Lorente E, García R, López D. Allele-dependent processing pathways generate the endogenous human leukocyte antigen (HLA) class I peptide repertoire in transporters associated with antigen processing (TAP)-deficient cells. J Biol Chem. 2011;286(44):38054–38059. | ||
Brusic V, Bajic VB, Petrovsky N. Computational methods for prediction of T-cell epitopes – a framework for modelling, testing, and applications. Methods. 2004;34(4):436–443. | ||
Meydan C, Otu HH, Sezerman OU. Prediction of peptides binding to MHC class I and II alleles by temporal motif mining. BMC Bioinformatics. 2013;14(Suppl 2):S13. | ||
Slifka MK, Whitton JL. Antigen-Specific Regulation of T Cell–Mediated Cytokine Production. Immunity. 2000;12(5):451–457. | ||
Ghanekar SA, Nomura LE, Suni MA, Picker LJ, Maecker HT, Maino VC. Gamma Interferon Expression in CD8+ T Cells Is a Marker for Circulating Cytotoxic T Lymphocytes That Recognize an HLA A2-Restricted Epitope of Human Cytomegalovirus Phosphoprotein pp65. Clin Diagn Lab Immunol. 2001;8(3):628–631. | ||
Kaliff M, Sorbe B, Mordhorst LB, Helenius G, Karlsson MG, Lillsunde-Larsson G. Findings of multiple HPV genotypes in cervical carcinoma are associated with poor cancer-specific survival in a Swedish cohort of cervical cancer primarily treated with radiotherapy. Oncotarget. 2018;9(27):18786–18796. | ||
Badial RM, Dias MC, Stuqui B, et al. Detection and genotyping of human papillomavirus (HPV) in HIV-infected women and its relationship with HPV/HIV co-infection. Medicine (Baltimore). 2018;97(14):e9545. | ||
Sabah SN, Gazi MA, Sthity RA, et al. Designing of Epitope-Focused Vaccine by Targeting E6 and E7 Conserved Protein Sequences: An Immuno-Informatics Approach in Human Papillomavirus 58 Isolates. Interdiscip Sci. 2018;10(2):251–260. | ||
Clifford GM, Tully S, Franceschi S. Carcinogenicity of human papillomavirus (HPV) types in HIV-positive women: A meta-analysis from HPV infection to cervical cancer. Clin Infect Dis. 2017;64(9):1228–1235. | ||
Chatterjee S, Chattopadhyay A, Samanta L, Panigrahi P. HPV and Cervical Cancer Epidemiology - Current Status of HPV Vaccination in India. Asian Pac J Cancer Prev. 2016;17(8):3663–3673. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Monie A, Hung CF, Roden R, Wu TC. Cervarix: a vaccine for the prevention of HPV 16, 18-associated cervical cancer. Biologics. 2008;2(1):97–105. | ||
Greenbaum JA, Andersen PH, Blythe M, et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J Mol Recognit. 2007;20(2):75–82. | ||
de Oliveira LMF, Morale MG, Chaves AA, et al. Design, immune responses and anti-tumor potential of an HPV16 E6E7 multi-epitope vaccine. PLoS One. 2015;10(9):e0138686. | ||
de Jong A, O’Neill T, Khan AY, et al. Enhancement of human papillomavirus (HPV) type 16 E6 and E7-specific T-cell immunity in healthy volunteers through vaccination with TA-CIN, an HPV16 L2E7E6 fusion protein vaccine. Vaccine. 2002;20(29–30):3456–3464. | ||
Jones EY. MHC class I and class II structures. Curr Opin Immunol. 1997;9(1):75–79. | ||
Sercarz EE, Maverakis E. Mhc-guided processing: binding of large antigen fragments. Nat Rev Immunol. 2003;3(8):621–629. | ||
Paolini F, Curzio G, Cordeiro MN, et al. HPV 16 E5 oncoprotein is expressed in early stage carcinogenesis and can be a target of immunotherapy. Hum Vaccin Immunother. 2017;13(2):291–297. | ||
Bristol JA, Orsini C, Lindinger P, Thalhamer J, Abrams SI. Identification of a ras oncogene peptide that contains both CD4(+) and CD8(+) T cell epitopes in a nested configuration and elicits both T cell subset responses by peptide or DNA immunization. Cell Immunol. 2000;205(2):73–83. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.