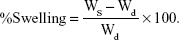Back to Journals » International Journal of Nanomedicine » Volume 14
Designing and enhancing the antifungal activity of corneal specific cell penetrating peptide using gelatin hydrogel delivery system
Authors Amit C, Muralikumar S, Janaki S, Lakshmipathy M, Therese KL, Umashankar V , Padmanabhan P, Narayanan J
Received 22 August 2018
Accepted for publication 8 November 2018
Published 15 January 2019 Volume 2019:14 Pages 605—622
DOI https://doi.org/10.2147/IJN.S184911
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Chatterjee Amit,1,2 Shalini Muralikumar,3 Sargunam Janaki,4 Meena Lakshmipathy,5 Kulandai Lily Therese,4 Vetrivel Umashankar,3 Prema Padmanabhan,5 Janakiraman Narayanan1
1Department of Nanobiotechnology, Vision Research Foundation, Sankara Nethralaya campus, Chennai, Tamil Nadu, India; 2School of Chemical and Biotechnology, SASTRA University, Tanjore, Tamil Nadu, India; 3Centre for Bioinformatics, Vision Research Foundation, Sankara Nethralaya campus, Chennai, Tamil Nadu, India; 4L&T Microbiology Research Centre, Vision Research Foundation, Sankara Nethralaya campus, Chennai, Tamil Nadu, India; 5Department of Cornea, Medical Research Foundation, Sankara Nethralaya campus, Chennai, Tamil Nadu, India
Background: Fungal keratitis is a major cause of corneal blindness accounting for more than one-third of microbiologically proven cases. The management of fungal keratitis is through topical or systemic antifungal medications alone or in combination with surgical treatment. Topical medications such as natamycin and voriconazole pose major challenges due to poor penetration across the corneal epithelium. To address the issue various carrier molecules like nanoparticles, lipid vesicles, and cell penetrating peptides were explored. But the major drawback such as non-specificity and lack of bioavailability remains.
Purpose: In this study, we have attempted to design corneal specific cell penetrating peptide using subtractive proteomic approach from the published literature and tried to improve its bioavailability through gelatin hydrogel delivery system.
Material and Methods: Using subtractive proteomic approach two peptides VRF005 and VRF007 were identified on the basis of solubility, cell permeability and amphipathicity. The peptides were modeled for three-dimensional structure and simulated for membrane penetration. The peptides were characterized using circular dichroism spectroscopy, dynamic light scattering and native polyacrylamide gel electrophoresis. Further uptake studies were performed on primary corneal epithelial cells and the stability was analyzed in corneal epithelial tissue lysates. Insilico prediction of peptides showed it to have antifungal activity which was further validated using colony forming assay and time killing kinetics. The duration of antifungal activity of peptide was improved using gelatin hydrogel through sustained delivery.
Results: VRF005 and VRF007 showed α-helical structure and was within the allowed region of Ramachandran plot. The simulation study showed their membrane penetration. The peptide uptake was found to be specific to corneal epithelial cells and also showed intracellular localization in Candida albicans and Fusarium solani. Peptides were found to be stable up to 2 hours when incubated with corneal epithelial tissue lysate. Dynamic light scattering, and native polyacrylamide gel electrophoresis revealed aggregation of peptides. VRF007 showed antifungal activity up to 24 hour whereas VRF005 showed activity up to 4 hours. Hence gelatin hydrogel-based delivery system was used to improve the activity. Actin staining of corneal epithelial cells showed that the cells were attached on gelatin hydrogel.
Conclusion: We have designed corneal specific cell penetrating peptides using subtractive proteomic approach. Bioavailability and delivery of peptide was enhanced using gelatin hydrogel system.
Keywords: cell-penetrating peptide, gelatin hydrogel, CD spectroscopy, subtractive proteomic approach
Introduction
The corneal epithelium is the outer most layer of the eye and forms an effective barrier against the entry of most pathogens. The tight junctions (TJs) of the corneal epithelium regulate the movement of fluid and macromolecules.1 Microbial invasion typically occurs secondary to ocular trauma resulting in penetration of causal organism in the tissue. The deeper penetration through the stroma and Descemet’s membrane makes the infection more recalcitrant to treatment. Although there are considerable geographic and seasonal variations in the epidemiology of infectious keratitis, fungal infections are a major cause of corneal morbidity in tropical and agricultural countries. Filamentous fungi (Fusarium and Aspergillus) and yeast-like fungi (Candida) are the most frequently isolated organisms in fungal keratitis.2 Currently, three classes of antifungal drugs such as polyenes (eg, natamycin and amphotericin), azoles (derivatives of imidazoles), and pyrimidines (5-fluorocytosine) constitute the medical armamentarium in the management of fungal keratitis.3 Almost all antifungal drugs show poor corneal epithelial penetration resulting in suboptimal bioavailability, making fungal keratitis a potentially devastating condition. This, then, warrants the search for an antifungal agent with good cell penetration and sustained bioavailability.
Although nanoparticles, lipid vesicles, cell-penetrating peptides (CPPs), and CPP-conjugated therapies (CTTs) are now being used for topical drug delivery with high clinical efficacy, they lack cell specificity and bioavailability.4 To address the issue of cell specificity, Heffernan et al5 reported a method to generate glial-specific CPP for multiple sclerosis, from the surface proteins of lymphocytic choriomeningitis virus (LCMV) which infects glial cells. Bioavailability was enhanced by targeting conserved amino acid residues between antimicrobial peptide and CPP,6 but none of them were able to generate tissue-specific peptides.
Semba et al7 published a series of articles on human eye proteome8–10 listing out the set of abundant proteins in different parts of the eye (choroid, retina, lens, iris, ciliary body, etc). Dyrlund et al11 identified and quantified the list of proteins in the layer of the cornea. Hence, in the present study, we attempted to develop corneal-specific CPP by applying subtractive proteomic approach followed by in silico study on the list of proteins published by Semba et al and Dyrlund et al. We successfully identified two CPPs which were validated further experimentally for its structure, cellular uptake, and antifungal activity. Of the two CPPs, one showed antifungal activity for only 4 hours. Hence, we used gelatin hydrogel as a delivery system to improve the duration of its antifungal activity up to 24 hours.
Materials and methods
Ethical approval
The study was reviewed and approved by the local ethics committee at Vision Research Foundation, Sankara Nethralaya, Chennai, India, and the committee deemed that it conformed to the principles of research in accordance with the Declaration of Helsinki (ethics no. 489-2015-P). The study was conducted on the enrolled patients who underwent refractive error correction surgery by obtaining duly signed informed consent forms from the patient/guardian as a part of clinical management.
Peptide designing strategy
Corneal-specific CPPs were designed by creating seven datasets and by consolidating the proteome from the published literature. Dataset 1 consists of common corneal proteins across three regions of cornea (epithelium, endothelium, and stroma).11–13 Similarly, dataset 2 consists of 8,258 proteins expressed in the lens, cornea, choroid, aqueous, retina, tears, and vitreous regions. They were subjected to blast analysis to find conserved retinal-specific proteins.8–10,14 Further, ID match analysis was performed between dataset 1 (609 proteins) and dataset 2 (672 proteins) to identify dataset 3 (338 proteins) consisting of conserved corneal specific proteins. Further, the corresponding sequences were batch retrieved from Uniprot database.
A total of 1,845 CPPs were downloaded from CPP database (http://crdd.osdd.net/raghava/cppsite) and were subjected to redundancy check. These peptides were further analyzed to filter out the ambiguous peptides with undesirable chemical modifications. These 731 nonredundant CPPs made our dataset 4. As a next step, dataset 4 (731 nonredundant CPPs) was subjected to blast analysis with dataset 3 (338 corneal-specific protein sequences) using CLC Genomics Workbench 0.1, set to default parameters, toward identifying the commonly conserved sequences among corneal-specific proteins and CPP dataset. This analysis led us to 218 CPPs which were further filtered with the criteria of >60% sequence identity match and CPP of length 6–16 residues. The peptides surpassing the filtering criteria were further subjected to amphipathicity prediction using the online server AMPHIPASEEK (https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_amphipaseek.html) and also to predict its secondary structure. AMPHIPASEEK provides a score for every residue between a range of 0 and 5 for the given peptide sequences. Higher scores imply high amphipathic nature and vice versa (0= low and 5= high). Hydropathy values were calculated using the online server (https://www.peptide2.com/N_peptide_hydrophobicity_hydrophilicity.php). Following the previously mentioned filtration criteria, dataset 5 consisting of 62 CPPs was obtained.
We downloaded the entire protein dataset of Fusarium solani from the FTP site of NCBI database to design CPP which can penetrate both corneal epithelium and F. solani. This served as our dataset 6. Further comparisons of dataset 4 (731 nonredundant CPPs) vs dataset 6 (F. solani) proteome resulted in the list of 140 CPPs with the abovementioned selection criteria. These 140 CPPs constitute our dataset 7. Finally, to arrive at a CPP which will have the potential to penetrate both corneal epithelium and F. solani, a comparison between dataset 5 vs dataset 7 was done which resulted in 38 CPPs. The 38 CPPs were passed through the criteria of peptide solubility and the cell-penetrating property using Innovagen peptide solubility calculator (https://pepcalc.com/) and CPPpred (http://bioware.ucd.ie/~compass/biowareweb/Server_pages/cpppred.php), respectively. The CPP score is given within the range of 0–1, wherein the peptides with the score of >0.5 are suggestive of better cell penetration. This procedure resulted in four CPPs. Two peptides, VRF005 and VRF007, were selected on the basis of high CPP score, amphiphilicity, and solubility for modeling and experimental validation.
The designed and enriched peptides surpassing all the validation criteria were submitted to I-TASSER server to model the three-dimensional structure. The model having a high confidence score (c-score) signifies higher confidence. Hence high confidence c-score was chosen and further validated for stereochemical properties using Ramachandran plot generated by Procheck. The models having the residues in the outlier regions were optimized by executing the energy minimization script using Modeller 9.15. The models with least DOPE (Discrete Optimized Protein Energy) score were chosen as reliable and were revalidated using Procheck. The designed, modeled, and minimized peptides were further taken for molecular dynamic (MD) simulation studies to study their structural stability. The peptides were preprocessed and optimized using the Schrodinger protein preparation wizard before starting up with an MD production run. The simulation was commenced using Optimized Potential for Liquid Simulations (OPLS) 2005 force field by solvating the cubic system using a Transferable Inter Molecular Potential 3P (TIP3P) water model. The system was neutralized by adding counter ions and energy minimized using OPLS 2005 force field. SHAKE algorithm was used to restrain the geometry of water molecules, bond lengths, and angles of heavy atoms. Periodic boundary conditions were set to simulate a continuous system. The system was further equilibrated using the constant Number, Pressure and Temperature (NPT) ensemble by setting the temperature and pressure parameters to 300 K and 1.0 bar, respectively. Nose–Hoover chain and Martyna–Tobias–Klein were chosen as a coupling algorithm for temperature and pressure, respectively. The equilibrated system was then simulated for a production run of 100 nanoseconds (ns) with a time step of 2 femto seconds (fs), and the trajectories were recorded at every 4.8 pico seconds (ps). The final trajectories were analyzed to perform a collective study on the backbone deviations of the atoms, ie, root mean square deviations convergence, residue fluctuations (root mean square fluctuations), and structure compactness (radius of gyration) throughout the production run of 100 ns to obtain an insight of the secondary structure stability of these modeled peptides.
Membrane-based simulation for shortlisted peptides
The peptides that had been shortlisted from the abovementioned analysis were further subjected to membrane simulations using the Desmond module of Schrodinger using OPLS 2005 force field. The peptides were positioned in the phosphatidylcholine (POPC) lipid bilayer membrane system using the OPM server. The area per lipid for POPC lipid bilayer was 65  2 as reported earlier.15–18 The membrane model system prepared with all the three peptides was integrated at the center of the lipid bilayer and water molecules (TIP3P) sparring at a maximum edge distance of 10 Å from solute atoms.
2 as reported earlier.15–18 The membrane model system prepared with all the three peptides was integrated at the center of the lipid bilayer and water molecules (TIP3P) sparring at a maximum edge distance of 10 Å from solute atoms.
Initial minimization of the system was performed with a convergence threshold value of 1.0 kcal mol−1 Å−1 so as to provide the positional equilibration to all the atoms. The next minimization with and without solute restraints was performed using Berendsen thermostat at temperature 10 K for 12 ps (pico seconds). This was followed by the equilibration step with the NPT ensemble using Berendsen barostat at 1 atm pressure for 12 ps. The temperature of the system was gradually increased to 300 K with restraints on solute heavy atoms and without any restrain for next 24 ps using Berendsen NPT ensemble. The PME method used for coulombic consideration with threshold and a cutoff value of 10 Å was used for the estimation of nonbonded solute–solvent and solvent–solvent interactions, with pressure and temperature of the system maintained at 1.013 atm and 300 K, respectively. The Matrix SHAKE algorithm was employed for the reorientation of hydrogen bonds, and the integration step for the calculation was fixed at 2 fs (femto seconds). Finally, simulation was carried out for 500 ns (nano seconds) and the trajectory frames were generated at an interval of 4.7 ps for all the systems and were analyzed using Visual Molecular Dynamics.
Peptide synthesis
Peptides, VRF005 (KKKWFETWFTEWPKKKK) and VRF007 (KDRPIFQLNTSYWEMGA), were synthesized using solid state synthesis and procured from M/s. Gene Script (https://www.genscript.com/) with an HPLC purity of more than 95%. N terminal of the peptide was labeled with fluorescein isothiocyanate (FITC).
Secondary structure stability and surface zeta potential analysis of peptides
Far-ultraviolet (UV) circular dichroism (CD) spectra of peptides, VRF005, VRF007 (500 nM), and gelatin (100 μg in distilled water, pH 7.0), were recorded on a spectropolarimeter (J810; Jasco International Co., Ltd., Tokyo, Japan) using a 0.1 cm path length quartz cuvette at 37°C. Spectra of VRF005 and VRF007 were recorded from 260 nm to 190 nm at two different temperatures (4°C and 37°C) at a scan rate of 50 nm/min). Further the interaction of peptide (VRF005) with gelatin was studied by incubating 500 nM of peptide and 100 mg of gelatin. The CD measurements were recorded in triplicates.
The stability of the peptide was analyzed in corneal epithelial tissue lysate prepared from the corneal epithelial tissues collected from subjects undergoing photorefractive corrections (ethics no. 489-2015-P). Six corneal epithelial tissues were collected from three subjects (oculus sinister and oculus dextrus). Tissues were lysed using RIPA buffer. Proteins were estimated using bicinchoninic acid (BCA) method as mentioned in the manufacturer’s protocol. Briefly, 1 μM of peptides (VRF005 and VRF007) was incubated with 20 μg and 50 μg of corneal epithelial lysate for a period of 2 hours and 24 hours. Peptide incubated with RIPA buffer served as negative control. SDS-PAGE was carried out for analyzing the degradation of FITC-labeled peptides.
Peptide at 1 μM concentration was run on 16% native PAGE for studying peptide aggregation. The aggregation of the peptides (VRF005 and VRF007) at pH 2, 4, 7, and 10.5 was further validated using Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) equipped with a 4 mW He–Ne laser operating at 633 nm. The peptide concentration was kept as 1 μM. Dip cell cuvette with a 1 cm light path was used, and the scattered light intensities were collected at an angle of 173° for surface potential measurement, and clear disposable cells were used for zeta size measurements.
Characterization of primary corneal epithelial cells and peptide uptake in primary corneal epithelial cells and fungal cells
Corneal epithelial tissues were collected from subjects undergoing refractive error correction (ethics no. 489-2015-P) surgery with prior consent and processed for primary human corneal epithelial cells as reported earlier.19 The tissues were mechanically disrupted to form a single cell suspension and were plated in collagen-coated plates in DMEM/F-12 medium with HEPES buffer, containing sodium pyruvate and l-glutamine: 50 U/mL penicillin and 50 μg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). Once the cells became attached, the cell cycle analysis was performed using propidium iodide and flow cytometry as mentioned in the manufacturer’s protocol.
Cells were lysed with RIPA buffer and were separated on an SDS-PAGE at 100 V in electrophoresis buffer (25 mM Tris, 190 mM glycine, and 0.1% SDS). The proteins were separated and transferred to polyvinylidene difluoride (PVDF) membrane (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) using semidry transblot apparatus (Hoefer, Holliston, MA, USA) at 1.50 mA/cm2. After transferring, the membrane was blocked for 1 hour at 25°C in 5% (w/v) nonfat dry milk (NFDM) powder in TBST (20 mM Tris–HCl pH 7.5, 150 mM NaCl, and 0.1% Tween 20). The membrane was washed with TBST and incubated at 4°C overnight with the following antibodies: claudin 1 (Cell signaling, Danvers, MA, USA), alpha catenin (Cell signaling), E-cadherin (Cell signaling), and ZO2 (Cell signaling). Each antibody was 1,000-fold diluted in either 5% (w/v) BSA (Hi Media, Mumbai, India) or NFDM in TBST. After overnight incubation, the membrane was washed thrice for 5 minutes with TBST and further incubated in the corresponding HRP-conjugated anti-rabbit and anti-mouse secondary antibody. The secondary antibodies used were diluted to 10,000-fold in 5% NFDM (w/v) in TBST. After incubation, the membrane was again washed thrice for 5 minutes with TBST. HRP activity was detected using HRP substrate (catalog no. 1705061; Bio-Rad Laboratories Inc., Hercules, CA, USA) in Bio-Rad Gel Documentation system (Protein Simple, Biorad Hercules, CA, USA).
Peptide uptake experiments were performed after the characterization of the corneal epithelial cells. Peptides were dissolved at a stock concentration of 1 mM in distilled water, and 1 μM of VRF005 and VRF007 was incubated with primary corneal epithelial cells to study the intracellular localization. After 1-hour incubation, the cells were washed with PBS and visualized through fluorescence microscopy using FITC filter. Muller glial cells (MIOM1 – a gift from Prof GA Limb; UCL Institute of Ophthalmology, London, UK), MCF7-ATCC (breast epithelial tumor cells), and NCC-Rb51-Riken (retinoblastoma tumor cells) were used as controls for the experiments.
Similarly, Candida albicans and F. solani cultures were grown for 12 hours in yeast nitrogen broth (YNB), and the population was adjusted to 1.5×105 colony-forming unit (CFU)/mL, and 10 μM of VRF005 and VRF007 was incubated with C. albicans and F. solani cultures for a period of 1 hour and was visualized for the intracellular localization using fluorescence microscopy (Axio Zeiss, Carl Zeiss, Oberkochen Germany).
Antimicrobial activity of peptides was predicted using an online server Antimicrobial Peptide Database:3 (http://aps.unmc.edu/AP/prediction/prediction_main.php). The antifungal efficacy of peptides (VRF005 and VRF007) was carried out against standard strains of C. albicans (ATCC-90028) and F. solani. Minimum inhibitory concentration (MIC) of peptides, VRF005 and VRF007, was performed against C. albicans strain at serial onefold dilution in Sabouraud’s dextrose agar (SDA) media. Medium alone was used as control. A volume of 100 μL from each serially diluted cuvette was spread on SD agar plate. The plates were incubated at 37°C for 48 hours. The appropriate dilution factor was multiplied to attain the final viable cell concentration. A total number of colonies were counted, and the graph was plotted as reported earlier.20
Killing kinetics of peptides
Antifungal activity of native VRF007 and VRF005 was performed against fresh culture of C. albicans (1–5×105 cells/mL) and F. solani (0.4–5×105 cells/mL). Treatment concentration of the peptides was 10-fold higher than their respective MIC values. Natamycin (32 μg/mL) was used as positive control. The experiments were performed in duplicate and were repeated at least three times. The plates were incubated at 25°C, and readings were taken at predetermined time intervals: (30 minutes, 1 hour, 2 hours, 4 hours, 24 hours) in spectrophotometer (600 nm). Then, the graph was plotted using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Microdilution plates (24-well plates) coated with soft gel and soft gel cross-linked with VRF005 (10 μM) were used. C. albicans (1–5×106 cells/mL) and F. solani (0.4–5×106 cells/mL) were inoculated on soft gel and VRF005-cross-linked soft gel. The plates were incubated at 25°C. The readings were taken at predetermined time intervals: (30 minutes, 1 hour, 2 hours, 4 hours, 24 hours) in spectrophotometer (600 nm). Then, the graph was plotted using GraphPad Prism 6.0.
Characterization of gelatin hydrogel and CPP delivery
Hydrogels were prepared using type B gelatin (Sigma-Aldrich Co., St Louis, MO, USA). Two different concentrations of gelatin gels were prepared (100 mg/mL and 50 mg/mL) using 0.8 μg/mL of glutaraldehyde as cross-linker and incubated at 4°C for 4 hours for gelation and cross-linking. For preparing different stiffnesses of hydrogel, two different concentrations of gelatin were used. Higher concentration of gel (100 mg/mL) was referred to as hard gel, and lower concentration (50 mg/mL) was referred to as soft gel. The cross-linked gelatin hydrogels were immersed in a 50 mM glycine aqueous solution under agitation for 1 hour to block the residual aldehyde groups of glutaraldehyde, followed by two washes with double-distilled water for 1 hour. Similarly, for the cross-linking of peptide (VRF005), 56.6 μg of peptide was cross-linked with gelatin using glutaraldehyde as cross-linker. The hydrogels were freeze-dried and used for scanning electron microscopy (SEM) analysis using field-emission gun (FEG)-SEM (JSM-7600F; JEOL Ltd., Tokyo, Japan). The pore size of the hydrogel was calculated using ImageJ.
Swelling behavior
The hydrogels were immersed in deionized water at room temperature. The samples were taken out from deionized water at selected time intervals, wiped with wet tissue paper to remove surface droplets, weighed, and placed back in deionized water. The swelling ratio Qm was calculated using the following equation, where Ws is the weight of swollen hydrogels and Wd is the initial weight of the sample.21
|
Fourier transform infrared (FTIR) spectroscopy analysis
Soft gelatin hydrogel with and without peptide VRF005 was freeze-dried. The chemical compositions of the hydrogels were identified with infrared spectroscopy (Tensor 27 FTIR; Bruker Optik, GmbH, Germany). The samples were prepared using potassium bromide (KBr) pellet and placed on sample holder, and the spectrum was recorded in absorbance mode in the region between 400 cm−1 and 4,000 cm−1 using deuterated triglycine sulfate detector.
Peptide release kinetics from gelatin hydrogel
Gelatin hydrogels of two different stiffnesses (soft and hard gels) and with/without glutaraldehyde cross-linker were prepared. The hydrogels were incorporated with 56.6 μg of peptide (VRF005). The standard graph for FITC-labeled peptide was made at 488 nm with different concentrations (1 μg, 5.6 μg, 11.3 μg, 16.9 μg, and 22.6 μg). The release kinetics of peptide from hydrogel was studied in a regular interval, and 500 μL of fresh PBS/deionized water was replaced in the hydrogel. The peptides released into the PBS were quantified using UV–visible spectrophotometer at 488 nm. The amount of peptide released was measured using the peptide standard graph. The antifungal activity of gelatin and gelatin-cross-linked peptides (VRF005) were measured using time-killing kinetics as mentioned earlier.
Analysis of corneal epithelial cell adhesion on hydrogel
Primary corneal epithelial cells were seeded on both hard and soft hydrogels for 24 hours. Then, the cells are fixed with 4% paraformaldehyde for 10 minutes, washed three times with 1× PBS, and permeabilized with 0.1% Triton-X for 5 minutes, followed by 1× PBS wash. Cells were further stained with FITC–phalloidin (Cytoskeleton Inc., Denver, CO, USA), and the cell nucleus was counterstained with Hoechst staining; furthermore, they were imaged with a fluorescence microscope (Axio Observer; Carl Zeiss Meditec AG, Jena, Germany).
Statistical analyses
All data were presented as mean and SD of at least three independent experiments. Statistical analysis was performed using the GraphPad Prism 6.0 statistical program. One-way ANOVA was followed by Bonferroni post hoc test when compared among the groups for the same experimental duration. Significance was defined by P-values of <0.05.
Results
Designing and modeling of CPPs
CPPs were designed from conserved human corneal proteome and F. solani proteome as shown in Figure S1A. We selected two CPPs for experimental validations based on the CPP score, amphipathicity and solubility, out of four CPPs came as results. The net charge and hydrophobicity ratio were calculated and are shown in Figure S1B. Briefly, VRF005 had a net charge of +5, whereas VRF007 had no charge. Higher grand average of hydropathicity (GRAVY) score of VRF007 (−0.782) is relatively more hydrophobic than VRF005 (−2.024). Peptides were modeled using I-TASSER server. VRF005 had a c-score value of −0.37, whereas the c-score was −1.45 for VRF007. The modeled and validated peptides were further taken for MD simulation to study the secondary structure stability. The final trajectories of peptides showed stability as shown in Figure S2A and B. MD analysis of VRF005 peptide exhibited a stable structure between 70 ns and 100 ns. Furthermore, the peptide had an α-helical structure and flexible amino acid residues throughout the simulation conditions (Figure S2A). VRF007 showed a stable structure between 30 ns and 100 ns, and the partial α-helical structure was changed to random coil during this time period (Figure S2B). The Ramachandran plots showed that both peptides were in the allowed region, VRF005 (Figure S3) and VRF007 (Figure S4). The structure of the peptides was further studied using CD spectroscopy.
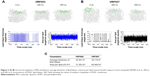
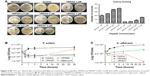
Characterization of peptide using CD spectrophotometer and zeta sizer
Endocytosis and translocation of peptides are dependent on their secondary structure, which are also influenced by temperature. Hence, we studied secondary structure of peptides using CD spectra at two different temperatures, 37°C and 4°C (Figure 1A). VRF007 showed high-intensity negative band at 222 nm and 208 nm and positive intensity band at 200 nm at 37°C. All three vibrations were drastically reduced at 4°C. However, VRF005 showed a negative band at 200 nm which did not vary with changes in temperature. Further, the peptides were characterized using native PAGE and dynamic light scattering for its aggregation property. VRF007 showed three bands in the native PAGE representing different aggregations, whereas VRF005 showed single band in native PAGE (Figure 1B). The predicted molecular weight of the peptide was validated using 16% SDS-PAGE and found to be around 2.5 kDa. The zeta potential of the peptides (VRF005 and VRF007) was +10 mV at both the pH 2 and pH 7, whereas at pH 10.5 it was found to be −40 mV (Figure 1C). The average size of VRF005 aggregation was maintained at 1,200 nm across the entire range of pH (Figure 1D). Interestingly, VRF007 was found to aggregate at different pH (2, 4, 7, and 10.5), and VRF007 exhibited different average size aggregation of 500 nm, 2 μm, and 3 μm at pH 2, 4, and 7, respectively (Figure 1E). At pH 7, VRF007 showed 68% of population within 200 nm size and 29% within 400–600 nm. In an acidic pH, VRF007 had 90% of its population in the size of 400 nm suggesting the effect of pH on the aggregation. The amount of net charge and aggregation determines the crucial property of peptides such as solubility, membrane permeability, and membrane partitioning. Further, the potential of the peptide for cell penetration was predicted using membrane-based simulation followed by cellular uptake studies in corneal epithelial cells.
Molecular simulation of VRF005 and VRF007 for membrane penetration
The average thickness of lipid bilayer in the presence of VRF005 and VRF007 was about 36.067 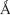 and 38.719
and 38.719 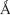 , respectively. It was noted that VRF007 peptide showed higher perturbation for the lipid bilayer during the simulation. The area per lipid value for peptides VRF005 and VRF007 along the bilayer was 73.305
, respectively. It was noted that VRF007 peptide showed higher perturbation for the lipid bilayer during the simulation. The area per lipid value for peptides VRF005 and VRF007 along the bilayer was 73.305 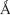 2 and 69.460
2 and 69.460 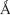 2, respectively (Figure 2A–C). Hence, both the peptides (VRF005 and VRF007) had the ability to penetrate the membrane system. This observation was further validated by uptake studies.
2, respectively (Figure 2A–C). Hence, both the peptides (VRF005 and VRF007) had the ability to penetrate the membrane system. This observation was further validated by uptake studies.
Peptide uptake by corneal epithelial cells and fungal cells
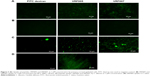
SEM of corneal epithelial tissue confirmed the presence of squamous epithelium, wing cells, and basal cells (Figure S5A), and 98% of the cells were in G0–G1 phase of cell cycle. The predominant presence of cells in this phase is a characteristic feature of corneal epithelial cells (Figure S5B). Expression of corneal epithelial cell markers, claudin 1, alpha catenin, ZO2, and E-cadherin, was observed in cells isolated from the tissues (Figure S5C). The uptake study showed that VRF005 and VRF007 were able to penetrate corneal epithelium and C. albicans, but VRF005 alone showed uptake in F. solani (Figure 3A, C, D).
Evaluation of peptide stability, toxicity, and antifungal activity
Stability of peptides was performed in different protein concentrations (10, 20 and 30 μg) obtained from corneal tissue to understand the time course of degradation inside the cellular environment. Both the peptides were stable for 2 hours at both concentrations (Figure S5D). However, VRF005 was stable in corneal epithelial tissue lysate at 25 μg concentration for 24 hours but got degraded at 50 μg concentration, whereas VRF007 peptide was degraded after 24 hours at both 25 μg and 50 μg concentrations (Figure S5E). The peptides did not show any uptake in retinoblastoma cell line NCC-RB51 (Figure 3B), MIOM1 and MCF7 cells (Figure S6A and B), suggesting the specificity of designed peptides towards corneal epithelial cells.
Primary corneal epithelial cells were treated with different concentrations (1 μM, 5 μM, and 10 μM) of peptides for a period of 24 hours, and cell viability was performed by MTT assay. Our data showed that the peptides were nontoxic and more than 80% of the cells were viable for the period of 24 hours at 1 μM concentration (Figure S5F).
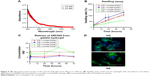
The potential antifungal activity of the peptides was validated through colony-forming assay and time-killing kinetics. VRF007 and VRF005 exhibited antifungal activity at MIC of 1 μg/mL (Figure 4A), whereas the conventional antifungal drug natamycin was effective only at 64 μg/mL. The colony-forming ability of C. albicans decreased substantially when treated with peptides. Killing kinetics of the peptide at 10-fold higher concentration than their respective MIC values showed drastic decrease in the growth of the fungus F. solani and C. albicans for a period of 4 hours by both peptides (Figure 4B and C). However, after 4 hours, the activity of VRF005 was completely lost and increase in the growth of F. solani and C. albicans was observed. Interestingly, VRF007 inhibited the growth of C. albicans and F. solani for 24 hours. The efficacy studies indicated that VRF005 is less effective than the VRF007 even though it was more stable than the later peptide (Figure S5D and S5E). Therefore, hydrogel-mediated delivery could potentially improve the efficacy of VRF005.
Characterization of gelatin hydrogel for peptide delivery
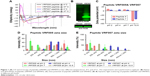
We used gelatin hydrogel delivery system for sustained release and enhanced antifungal activity of VRF005. UV–visible spectrometry results showed that both the gels were optically transparent between 300 nm and 700 nm (Figure 5A). The swelling ratio was considerably altered for the soft gel in the presence of cross-linker where it was not visibly altered for the hard gel in the presence of cross-linker (Figure 5B).
Peptide release kinetics from gelatin hydrogel
The cumulative release of peptide VRF005 from hard and soft gels was determined by UV spectrophotometry (Figure 5C). A total of 54.6 μg and 44 μg of VRF005 were released from hard and soft gels with cross-linker out of 56.6 μg of the peptides loaded on to the gel. Similarly, 36.7 μg and 32.7 μg of peptide were released from hard gel and soft gel without cross-linker, respectively, at 72 hours indicating the effect of cross-linker on peptide release.
Cell adhesion in gelatin hydrogel
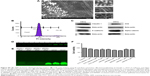
Actin (phalloidin) staining of corneal epithelial cells seeded on both hard and soft gel was analyzed after 24 hours of incubation. The cells grown on soft gel exhibited prominent stress fibers (Figure 5D). Therefore, soft gelatin hydrogel was used for VRF005 peptide delivery. VRF005 was cross-linked with gelatin hydrogel and examined by SEM. The pore size of soft gelatin hydrogel was on average of 120 μm (Figure 6A). The soft hydrogel, after cross-linking with VRF005, the pore size reduced to 50 μm (Figure 6A). The cross-linking of the peptide VRF005 with gelatin hydrogel was analyzed using vibrational spectroscopy (Figure 6B). VRF005 encapsulation on gelatin exhibited intense peaks at 1,600 cm−1 and 900 cm−1 compared to gelatin hydrogel (Figure 6B). CD spectra also showed that gelatin has a strong negative trough around 197 nm and no broad positive band in the n–π* region (Figure 6C). Furthermore, VRF005 peptide addition to soft hydrogels increased the intensity of the negative peak around 200 nm region (Figure 6C) without altering the structure.
Antifungal activity of peptide released from gelatin hydrogel
Soft gel was chosen based on the sustained release kinetics of VRF005 and corneal epithelial cell attachment. Soft gel cross-linked with VRF005 peptide showed antifungal activity against C. albicans and F. solani for the period of 24 hours (Figure 6D and E). Hence, the overall antifungal activity of VRF005 was drastically improved through gelatin hydrogel.
Discussion
Earlier studies have reported the designing of CPP from proteins such as heparin-binding motif of human eosinophil cationic protein, human nuclear body protein, SP140-like protein, and 12 isoforms of annexin, a family of membrane-interacting human proteins.22–24 However, there is no report of using the total proteome to design a CPP which would be tissue specific. In this context, we have attempted to design a corneal-specific CPP using subtractive proteomic approach. The tissue specificity of the peptide was validated by the cellular uptake studies. Both VRF005 and VRF007 showed intracellular localization in primary corneal epithelial cells but not in retinoblastoma cell line, breast cancer cell line, and Muller glial cell line (NCC-RB51, MCF-7, and MIOM1 cells), thus confirming their tissue specificity. Interestingly, peptides also showed localization in C. albicans and F. solani. VRF007’s secondary structure was altered from α-helical structure to random coil, when there was a change in temperature from 37°C to 4°C, whereas VRF005 did not show any structural alterations in secondary structure indicating the stability of VRF005 at both the temperatures. VRF007 showed three bands in native PAGE, whereas VRF005 gave a single band indicating VRF007 aggregation. This was further confirmed by measuring peptide-aggregating sizes in the different ionic strength medium. Antimicrobial peptide has been reported to be influenced by pH, hydrophobicity, and aggregation.25
Peptides, VRF005 and VRF007, at pH 2–7 have a lower zeta potential, whereas at pH 10.5 have higher zeta potential which indicated the high degree of stability. However, low zeta potential for VRF007 peptide indicated peptide aggregation. Aggregating peptide also has the potential for antimicrobial activity.26 Time-killing kinetics showed drastic reduction in fungal growth (C. albicans and F. solani). VRF005 lost the activity after 4 hours of treatment, whereas VRF007 maintained the antifungal activity up to 24 hours. The superior antifungal activity of VRF007 peptide could be attributed to its aggregation property.27 We found that VRF005 was comparatively more stable than VRF007. However, the activity was much lower than VRF007. Hence sustained delivery of the peptide VRF005 was needed to improve the activity. Hence, we cross-linked VRF005 peptide with gelatin hydrogel to sustain the release and improve the activity.
FTIR data indicated the cross-linking of peptides with gelatin. The appearance of 1,100 cm−1 (C–N of amide III), 1,600 cm−1 (NH amide I), and loss of 1,400 cm−1 (aromatic C–C) in VRF005 indicated encapsulation within gelatin hydrogel. The shift in the following wavenumbers clearly indicated the cross-linking of peptides in the gel.21 CD spectra analysis revealed that peptide VRF005 cross-linked with gelatin hydrogel did not induce any changes in the gelatin’s secondary structure. The reduction in pore size of the peptide-cross-linked gelatin hydrogel compared to native gelatin hydrogel indicated that cross-linking of the peptides occurs not only on the surface but also throughout the gel. Corneal epithelial cells showed prominent attachment on soft gelatin hydrogel indicating the preferred stiffness. Soft gel-cross-linked peptide exhibited sustained drug release for 24 hours and improved the activity of VRF005 from 4 hours to 24 hours in both C. albicans and F. solani. Earlier reports have used gelatin hydrogel for small molecule (antifungal) drug delivery.28 To the best of our knowledge, gelatin hydrogel has not been reported for CPP delivery with antifungal activity.
Conclusion
We used novel subtractive proteomic approach to design tissue-specific CPP. In addition, the CPP also had antifungal activity. The duration of the antifungal activity of the peptide was drastically improved by incorporating in to gelatin hydrogel delivery system which may have clinical relevance. This hydrogel-based delivery would also improve the bioavailability of the drug for a longer duration, potentially reducing the frequency of application necessary for therapeutic use.
Acknowledgments
JN and AC acknowledge the Department of Biotechnology (DBT), Government of India, for funding (BT/PR14690/MED/32/496/2015) and fellowship. The authors thank SRM University Nanotechnology Research Center for SEM and Indian Institute of Technology (IIT) Madras for CD spectroscopy. Authors also thank “High performance computing” supported by Science and Engineering Research Board (SERB), Government of India, grant No. YSS/2014/000282. They thank Dr Sailaja Elchuri, Department of Nanobiotechnology, Vision Research Foundation, Sankara Nethralaya campus, for critically reviewing the manuscript and also thank A Samdani, VRF, for proofreading the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Albert DM, Jakobiec FA, Miller JW. Albert & Jakobiec’s Principles and Practice of Ophthalmology. Philadelphia, PA, Edinburgh: Saunders/Elsevier; 2008. | ||
Gopinathan U, Garg P, Fernandes M, Sharma S, Athmanathan S, Rao GN. The epidemiological features and laboratory results of fungal keratitis: a 10-year review at a referral eye care center in South India. Cornea. 2002;21(6):555–559. | ||
Manzouri B, Vafidis GC, Wyse RK. Pharmacotherapy of fungal eye infections. Expert Opin Pharmacother. 2001;2(11):1849–1857. | ||
Desai P, Patlolla RR, Singh M. Interaction of nanoparticles and cell-penetrating peptides with skin for transdermal drug delivery. Mol Membr Biol. 2010;27(7):247–259. | ||
Heffernan C, Sumer H, Guillemin GJ, Manuelpillai U, Verma PJ. Design and screening of a glial cell-specific, cell penetrating peptide for therapeutic applications in multiple sclerosis. PLoS One. 2012;7(9):e45501. | ||
Mishra B, Wang G. The Importance of Amino Acid Composition in Natural AMPs: An Evolutional, Structural, and Functional Perspective. Front Immunol. 2012;3:221. | ||
Semba RD, Enghild JJ, Venkatraman V, Dyrlund TF, van Eyk JE. The Human Eye Proteome Project: perspectives on an emerging proteome. Proteomics. 2013;13(16):2500–2511. | ||
Zhang P, Dufresne C, Turner R, et al. The proteome of human retina. Proteomics. 2015;15(4):836–840. | ||
Zhang P, Karani R, Turner RL, et al. The proteome of normal human retrobulbar optic nerve and sclera. Proteomics. 2016;16(19):2592–2596. | ||
Zhang P, Kirby D, Dufresne C, et al. Defining the proteome of human iris, ciliary body, retinal pigment epithelium, and choroid. Proteomics. 2016;16(7):1146–1153. | ||
Dyrlund TF, Poulsen ET, Scavenius C, et al. Human cornea proteome: identification and quantitation of the proteins of the three main layers including epithelium, stroma, and endothelium. J Proteome Res. 2012;11(8):4231–4239. | ||
Galiacy SD, Froment C, Mouton-Barbosa E, et al. Deeper in the human cornea proteome using nanoLC-Orbitrap MS/MS: An improvement for future studies on cornea homeostasis and pathophysiology. J Proteomics. 2011;75(1):81–92. | ||
Karring H, Thøgersen IB, Klintworth GK, Møller-Pedersen T, Enghild JJ. A dataset of human cornea proteins identified by Peptide mass fingerprinting and tandem mass spectrometry. Mol Cell Proteomics. 2005;4(9):1406–1408. | ||
Nordgaard CL, Berg KM, Kapphahn RJ, et al. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47(3):815–822. | ||
Broemstrup T, Reuter N. Molecular dynamics simulations of mixed acidic/zwitterionic phospholipid bilayers. Biophys J. 2010;99(3):825–833. | ||
Gurtovenko AA, Vattulainen I. Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. J Phys Chem B. 2008;112(7):1953–1962. | ||
Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J. 2005;88(4):2626–2637. | ||
Kučerka N, Nieh M-P, Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta. 1808;2011(11):2761–2771. | ||
Albert R, Veréb Z, Csomós K, et al. Cultivation and characterization of cornea limbal epithelial stem cells on lens capsule in animal material-free medium. PLoS One. 2012;7(10):e47187. | ||
Meletiadis J, Meis JF, Mouton JW, Verweij PE. Analysis of growth characteristics of filamentous fungi in different nutrient media. J Clin Microbiol. 2001;39(2):478–484. | ||
Xing Q, Yates K, Vogt C, Qian Z, Frost MC, Zhao F. Increasing mechanical strength of gelatin hydrogels by divalent metal ion removal. Sci Rep. 2014;4:4706. | ||
Fang SL, Fan TC, Fu HW, et al. A novel cell-penetrating peptide derived from human eosinophil cationic protein. PLoS One. 2013;8(3):e57318. | ||
Wang H, Ma J, Yang Y, Zeng F, Liu C. Highly Efficient Delivery of Functional Cargoes by a Novel Cell-Penetrating Peptide Derived from SP140-Like Protein. Bioconjug Chem. 2016;27(5):1373–1381. | ||
Young Kim H, Young Yum S, Jang G, Ahn DR. Discovery of a non-cationic cell penetrating peptide derived from membrane-interacting human proteins and its potential as a protein delivery carrier. Sci Rep. 2015;5:11719. | ||
Malik E, Dennison SR, Harris F, Phoenix DA. pH Dependent Antimicrobial Peptides and Proteins, Their Mechanisms of Action and Potential as Therapeutic Agents. Pharmaceuticals. 2016;9(4):E67. | ||
Sun J, Xia Y, Li D, du Q, Liang D. Relationship between peptide structure and antimicrobial activity as studied by de novo designed peptides. Biochim Biophys Acta. 2014;1838(12):2985–2993. | ||
Mohanram H, Bhattacharjya S. Resurrecting inactive antimicrobial peptides from the lipopolysaccharide trap. Antimicrob Agents Chemother. 2014;58(4):1987–1996. | ||
Winnicka K, Wroblewska M, Wieczorek P, Sacha PT, Tryniszewska E. Hydrogel of ketoconazole and PAMAM dendrimers: formulation and antifungal activity. Molecules. 2012;17(4):4612–4624. |
Supplementary materials
  | Figure S3 (A) Best model of peptide VRF005. (B) Procheck-generated Ramachandran plot (VRF005 named as Peptide _6). |
  | Figure S4 (A) Best model of peptide VRF007. (B) Procheck-generated Ramachandran plot (VRF007 named as Peptide _1). |
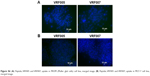  | Figure S6 (A) Peptide, VRF005 and VRF007, uptake in MIOM (Muller glial cells) cell line, merged image. (B) Peptide, VRF005 and VRF007, uptake in MCF-7 cell line, merged image. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.