Back to Journals » Infection and Drug Resistance » Volume 14
Design of Rapid Detection System for Five Major Carbapenemase Families (blaKPC, blaNDM, blaVIM, blaIMP and blaOXA-48-Like) by Colorimetric Loop-Mediated Isothermal Amplification
Authors Feng W, Niu S , Chang Y, Jia X, Huang S , Yang P
Received 13 January 2021
Accepted for publication 7 May 2021
Published 24 May 2021 Volume 2021:14 Pages 1865—1874
DOI https://doi.org/10.2147/IDR.S301757
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wenjuan Feng,* Siqiang Niu,* Yanbin Chang, Xiaojiong Jia, Shifeng Huang, Ping Yang
Department of Clinical Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shifeng Huang; Ping Yang
Department of Clinical Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, No. 1 Friendship Road, Yuzhong District, Chongqing, People’s Republic of China
Tel +86-18623027077; +86-13637993035
Fax +86-23-89012513; +86-23-89012513
Email [email protected]; [email protected]
Purpose: Carbapenemase-producing Enterobacteriaceae (CPE) infection constitutes a public health threat. Timely and efficient diagnosis is of paramount importance for prompt and effective therapy. In order to quickly and comprehensively detect the five major families of carbapenemases (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48-like), colorimetric loop-mediated isothermal amplification (LAMP) was employed.
Materials and Methods: Five sets of LAMP primers were designed, each of which can, respectively, amplify all the carbapenemase subtypes described in this work. Twenty whole genome sequencing-verified-“standard strains”, including 1 blaNDM-1, 1 blaNDM-5, 1 blaNDM-6, 1 blaNDM-7, 2 blaIMP-4, 1 blaIMP-8, 2 blaKPC-2, 1 blaKPC-3, 1 blaKPC-4, 1 blaKPC-5, 1 blaKPC-6, 1 blaKPC-7, 1 blaOXA-48 and 1 blaOXA-181 carrier, and 1 blaVIM and blaOXA-244, 1 blaKPC-2 and blaIMP-4, 1 blaKPC-2 and blaVIM-1 and 1 blaKPC-2 and blaNDM-1-co-carriers, were used to establish a 25-microliter visual LAMP reaction system (kept at 65°C for 30 minutes in water bath). Color change from bright pink to yellow indicated positive amplification. In addition, 126 pre-verified clinical carbapenem-resistant Enterobacteriaceae (CRE) isolates, including 65 CPE (23 blaNDM, 2 blaOXA-48-like, 1 blaKPC and blaVIM, 2 blaIMP, and 37 blaKPC carriers) and 61 non-CPE, were also detected.
Results: With the lowest detection limit of 10 colony forming units (CFU) per reaction for LAMP and 103 CFU per reaction for PCR, the LAMP system demonstrated dramatically higher sensitivity while retaining the same specificity. Furthermore, we demonstrated concordant results between the two methods for the 126 clinical isolates.
Conclusion: Therefore, LAMP could be used for rapid identification of the five major carbapenemase gene families in routine clinical laboratories.
Keywords: carbapenemase-producing Enterobacteriaceae, loop-mediated isothermal amplification, blaNDM, blaKPC, blaVIM, blaIMP, blaOXA-48-like
Introduction
Antibiotic resistance is one of the most threatening global health challenges.1–3 Carbapenems are considered as the last line of defense against the infection of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) and AmpC cephalosporinases (AmpCs).4,5 However, due to the wide spread of ESBLs-producing Enterobacteriaceae in recent years, the extensive use of carbapenems has led to the quick emergence of carbapenem-resistant Enterobacteriaceae (CRE). Carbapenem resistance in Enterobacteriaceae is mainly caused by the production of carbapenemases, especially Class A Klebsiella pneumoniae carbapenemase (KPC), Class B New Delhi metallo-β-lactamase (NDM), Imipenemase (IMP), Verona Integron-encoded Metallo-β-lactamase (VIM), and Class D oxacillinase-48-like (OXA-48-like).4,6 Several methods have been employed to detect carbapenemase activities, such as the Modified-Hodge test (MHT),7,8 the Ethylenediaminetetraacetic Acid (EDTA) inhibition test,8 MALDI-TOF MS,9 Carba NP test (CNPt),10,11 and the carbapenem inhibition test (CIM).12,13 Yet, some defects, such as low specificity, low sensitivity, lack of rapidity and convenience, still remain. As the standard method for identifying carbapenemase genotypes, polymerase chain reaction (PCR) depends on both expensive instruments and specialized technicians. Therefore, an easier, more convenient, rapid and accurate carbapenemase genotyping method, which can easily be carried out in primary-level laboratories, is essential.
Figure 1 Continued.
Loop-mediated isothermal amplification (LAMP), invented by Notomi et al in 2000,14 is a type of in vitro amplification technique under constant temperature condition without the need of skilled operators and expensive instruments, which has the characteristics of isothermal, rapid, high specificity and sensitivity. The reaction can be completed by setting up 3 pairs of specific primers (FIP, BIP, F3, B3, LF, and LB) for target gene and amplified at 65°C for 30 minutes; moreover, the amplified products can be observed by colorimetric change or visually measured by the presence of turbidity of the byproduct (magnesium pyrophosphate). The technique has been widely used for point-of-care diagnosis and detection of viruses, bacteria and parasites involved in recurrent and emerging infectious diseases.15–18 Recently, LAMP has been used to detect carbapenemase genes in hydroxynaphthol blue dye (HNB), gel electrophoresis, turbidity, SYBR green or calcein for the measurement of the by-products.19–22 However, those previously reported LAMP systems could not detect all the variants that have been reported from each carbapenemase gene family. To more rapidly and comprehensively detect the most common carbapenemase gene families in Enterobacteriaceae (blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48-like), we tried to develop a new colorimetric LAMP system.
In this study, we described the design of three sets of primers and optimized the LAMP assay for detection of each of the five carbapenemase gene families. Of note, extremely strict primer screening criteria were followed, such as TM value, primer terminal stability, GC content and primer spacing. Most importantly, we designed five sets of LAMP primers for each carbapenemase gene family, ensuring no spanning of as many as possible the mutation sites among all the different variants of each carbapenemase family. Theoretically, each set of the LAMP primers can, respectively, detect the carbapenemase subtypes as described in this work. Also, the specificity and sensitivity of the LAMP reactions were determined. Finally, this LAMP system was used for rapid identification of carbapenemase genes in 126 pre-verified clinical CRE isolates.
Materials and Methods
Bacterial Isolates
Twenty whole-genome sequencing-verified “standard strains” from the Microbiology Laboratory of the first affiliated Hospital of Chongqing Medical University, including 1 blaNDM-1, 1 blaNDM-5, 1 blaNDM-6, 1 blaNDM-7, 2 blaIMP-4, 1 blaIMP-8, 2 blaKPC-2, 1 blaKPC-3, 1 blaKPC-4, 1 blaKPC-5, 1 blaKPC-6, 1 blaKPC-7, 1 blaOXA-48 and 1 blaOXA-181 carriers, and 1 blaVIM and blaOXA-244, 1 blaKPC-2 and blaIMP-4, 1 blaKPC-2 and blaNDM-1 and 1 blaKPC-2 and blaVIM-1 co-carriers (Table 1) were collected. One hundred twenty-six pre-verified CRE clinical strains, mainly consisting of Klebsiella pneumoniae, Escherichia coli and Enterobacter cloacae isolated during 2015–2020, were further collected from the first affiliated Hospital of Chongqing Medical University (Table 2). Carba NP test23 was performed on all the isolates to determine if there were any bacteria that appeared to produce carbapenemases by phenotypic methods but were negative by genotypic methods, or vice versa.
 |
Table 1 “Standard Strains” Verified by Whole-Genome Sequencing* |
 |
Table 2 Pre-Verified Clinical CRE Strains Tested in This Study |
Preparation of Bacterial DNA
Bacterial DNA was extracted by TIANamp Bacteria DNA kit (TIANGEN BIOTECH (BEIJING) CO., LTD; Cat#DP302-02; Lot#S8230) according to the manufacturer’s instructions.
Primer Design
The reference sequences of the five major carbapenemase gene families (blaNDM, blaKPC, blaVIM, blaIMP, and blaOXA-48-like) were downloaded from GeneBank website (https://www.ncbi.nlm.nih.gov/genbank/), and LAMP primers were designed by PrimerV5 (http://primerexplorer.jp/lampv5e/index.html). With the pre-defined primer designing principles: no mutation sites to be involved at the 3ʹ-end of the forward primer, optimal TM values, terminal stability of the primers, and no secondary structures between the primers, the selected primers could theoretically be used to detect much more reported variants of each carbapenemase family gene group as compared to those of the previous studies. So far, according to the Beta-Lactamase DataBase (BLDB) (http://www.bldb.eu/Enzymes.php), blaNDM has 31 variants, blaKPC has 79 variants, blaIMP has 89 variants and blaVIM has 73 variants. The sequences of the mutants of blaOXA-48-like were compared with the sequence of blaOXA-48. The primer sets of this study could detect all the reported variants of blaNDM and blaKPC, the blaIMP group including blaIMP-1-blaIMP-14, and blaIMP-22, blaIMP-32, blaIMP-33, blaIMP-48, and blaIMP-68, the blaVIM group including all the reported variants except for blaVIM-7, blaVIM-10, blaVIM-51 and blaVIM-65, and the blaOXA-48-like group including blaOXA-48, blaOXA-181, blaOXA-232, blaOXA-204, blaOXA-162, and blaOXA-244. The LAMP primers were listed in Table 3 and synthesized by Sangon Biotech (Shanghai, China) Co., Ltd.
 |
Table 3 Nucleotide Sequences of the LAMP Primers for Detection of blaNDM, blaKPC, blaVIM, blaIMP and blaOXA-48-Like Gene Families |
Colorimetric LAMP Assay
LAMP reaction was performed in a total volume of 25 µL, comprising of 2.5 µL 10×LAMP Primer Mix [2 µM each of outer primer (F3, B3), 16 µM each of inner primer (FIP, BIP), 8 µM each of loop primer (LF, LB)], 12.5 µL WarmStart Colorimetric LAMP 2X Master Mix (NEW ENGLAND BioLabs® Inc), 9 µL dH2O, 1 µL target DNA template. The amplification was carried out at 65°C for 30 minutes. Aseptic distilled water was used as the negative control. Color change from bright pink to yellow indicates a positive reaction.
Comparison of the Specificity and Sensitivity Between the Colorimetric LAMP and PCR
To estimate the specificity of the colorimetric LAMP assay for the detection of the five major carbapenemase gene families, we used the standard strain containing only one specific carbapenemase gene. Strains without the specific genes being detected were also selected to verify the specificity of the LAMP primers. Experimental sensitivity of the LAMP assay was ascertained by a series of DNA template extracted from 101 to 108 CFU/reaction from an initial concentration of 109 CFU/mL bacteria. The lowest value of positive result is the limit of detection. As a standard for assessing the colorimetric LAMP system, PCR was used to detect the potential presence of the five carbapenemase gene families. The evaluation experiments of sensitivity and specificity were repeated thrice to ensure reproducibility.
Ethical Considerations
For this study, samples were collected at the microbiology laboratory of the hospital, with no contact with the patient. This study was retrospective with no patient identification performed during data collection. Therefore, the Chongqing Medical University Institutional Review Board and the Biomedical Ethics Committee determined that this research was exempt from approval.
Results
Specificity of the Colorimetric LAMP
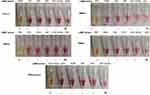
The spanning sites where the LAMP primers are located in each carbapenemase gene family are listed as in Figure 1. As shown in Figure 2, we only observed color change from bright pink to yellow when the specific LAMP primers for the target gene were added, thus demonstrating 100% specificity of the colorimetric LAMP system in detecting the five carbapenemase gene families.
Much Higher Sensitivity of the Colorimetric LAMP Compared with Traditional PCR
As depicted in Figure 3, while more than 103 CFU/reaction was required for PCR amplification of all the five carbapenemase gene families, positive LAMP results could be achieved with only 101 CFU/reaction for blaKPC (Figure 3B), blaIMP (Figure 3D), and blaOXA-48-like (Figure 3E), and 102 CFU/reaction for blaNDM (Figure 3A) and blaVIM (Figure 3C).
Higher Proficiency of the Colorimetric LAMP in Detecting Carbapenemase Gene Families in Clinical Isolates
The five carbapenemase gene families in a total of 126 pre-verified clinical CRE isolates were detected by both the colorimetric LAMP assay and the traditional PCR assay. Both of them identified 65 carbapenemase-carriers, including 23 strains with blaNDM, 37 with blaKPC, 2 with blaOXA-48-like, 2 with blaIMP and 1 with blaKPC and blaVIM, indicating 100% concordance in the detection proficiency between the two methods. Of note, with its simplicity and rapidity, the colorimetric LAMP assay demonstrated much higher proficiency in detecting carbapenemase genotypes in clinical isolates when compared with traditional PCR and subsequent sequencing.
Discussion
With the wide spread of carbapenemase genes, CPE is becoming an increasingly serious world health concern. Infections caused by CPE will not only prolong the length of hospital stays but also increase the medical expenses of patients whose immune systems are compromised. Rapid and accurate identification of drug resistance genotypes can provide a basis for rational clinical use of antibiotics.
In the past decade, bacterial culture must precede the routine diagnostic methods used to detect the carbapenem-resistant organisms. MHT,7 EDTA inhibition test,8 MALDI-TOF MS,9 CNPt10,11 and CIM12,13 were commonly employed to detect the carbapenemase activity. However, some of these phenotypic tests usually require expensive instruments and special cultural conditions, and furthermore, the time required to complete some of these tests is usually 16–20 hours.21 Molecular diagnostic methods remain the gold standard for the identification of carbapenemase genes. However, quantitative measurements of target genes are not always straightforward in PCR analyses, and the presence of the amplified products has to be confirmed by both gel electrophoresis and subsequent sequencing. Therefore, the LAMP method that requires only water bath or heating block to obtain a constant temperature is an alternative molecular tool for identifying carbapenemase genes in the primary-level laboratories.
As a simple and rapid identification method, LAMP has been widely used in the diagnosis of carbapenemase genes. However, as the key in DNA amplification using the LAMP method, appropriate primer design is essential. Over 89 IMP, 73 VIM, 79 KPC, 31 NDM, and 26 OXA-48-like carbapenemases have been described as of the date of March 01, 2021 (http://www.bldb.eu/F-BLDB.php). Many of the variations within the carbapenemase gene families matter when it complicates the design of comprehensive PCR detection methods, necessitating repeated “tweaking” as new variants are added to the detection repertoire. Through Beta-Lactamase DataBase (http://www.bldb.eu/F-BLDB.php) search and review, we comprehensively concluded all the information on the mutation sites and the primers spanning these mutation sites were discarded during the optimal primer screening, so as to cover as many as possible the gene variants by the designed primers. Previous studies showed that the number of genotypes that could be detected by the reported LAMP system was limited, from the initial blaNDM-1 by Liu et al,22 to the detection of blaOXA-23, blaVIM-2, and blaIMP-1 by Kim et al,21 and the detection of blaNDM-1 through blaNDM-9, blaKPC-2 through blaKPC-15, blaVIM-2, and blaIMP-4 by Chen et al.20 Later, LAMP-HNB was used to detect blaNDM-1 through blaNDM-9, blaNDM-11 through blaNDM-16, blaOXA-48, blaVIM-1 through blaVIM-46 (except for blaVIM-7, blaIMP-14) and blaKPC-2 through blaKPC-24 (except for blaKPC-20 and blaKPC-23) groups by Srisrattakarn et al.19 The LAMP system of the present study was demonstrated to be capable of specifically detecting all the reported subtypes of the five major carbapenemase genes as described in this work, which was consistent with the results of PCR and subsequent sequencing. After demonstration of the specificity of the respective LAMP primers for blaNDM, blaKPC, blaIMP, blaVIM, and blaOXA-48-like, we further tested the sensitivity of the LAMP system, and the results showed higher sensitivity for LAMP when compared with that of the conventional PCR method, which was consistent with the results of previous studies.19,20 In this study, using the WarmStart master LAMP mixed with the chromogenic reagent, primers and templates were added to the sealed reaction tank and amplified at 65°C for 30 minutes, which greatly reduced the possible pollution caused by repeated lid opening operations. And compared with other detection methods, such as turbidity, gel electrophoresis, real-time turbidimeter, SYBR Green or calcein, using this reagent to visually observe the color change to read the results is safer, more simple and rapid. Previous studies used betaine, magnesium sulfate and other substances, but other aspects such as the operational factors during reagent preparation may increase the unreliability of the results. Therefore, our results further supported the use of more convenient chromogenic mixed reagents for LAMP detection.
The colorimetric LAMP system has been demonstrated to be a simple and fast carbapenemase identification method that can be carried out in conventional laboratories without special instruments, and the results provided can lead to the rational use of antibiotics in clinic. Nevertheless, one major limitation of the present LAMP method is that amplification for each carbapenemase gene has to be run individually (ie 1 reaction/gene) as no multiplex LAMP reaction is possible at present. Then, 5 reactions are necessary to define what carbapenemase gene(s) is/are present in one isolate vs 1 reaction in a multiplex amplification, leading to a complication when a high number of isolates have to be tested.
Acknowledgments
We thank all the staffs from the Microbiology Laboratory of the first affiliated Hospital of Chongqing Medical University for providing technical support.
Disclosure
All authors reported no conflicts of interest in this work.
References
1. Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36(3):182–186. doi:10.1053/j.semdp.2019.04.011
2. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi:10.1179/2047773215Y.0000000030
3. van Loon K, Voor in ‘t holt AF, Vos MC. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2017;62(1):e01730–17. doi:10.1128/AAC.01730-17
4. Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15–21. doi:10.1177/2049936115621709
5. Lutgring JD, Limbago BM. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol. 2016;54(3):529–534. doi:10.1128/JCM.02771-15
6. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. doi:10.1016/j.molmed.2012.03.003
7. Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitouta JDD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50(12):3877–3880. doi:10.1128/JCM.02117-12
8. Bartolini A, Frasson I, Cavallaro A, Richter SN, Palù G. Comparison of phenotypic methods for the detection of carbapenem non-susceptible Enterobacteriaceae. Gut Pathog. 2014;6:13. doi:10.1186/1757-4749-6-13
9. Yu J, Liu J, Li Y, et al. Rapid detection of carbapenemase activity of Enterobacteriaceae isolated from positive blood cultures by MALDI-TOF MS. Ann Clin Microbiol Antimicrob. 2018;17(1):22. doi:10.1186/s12941-018-0274-9
10. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503–1507. doi:10.3201/eid1809.120355
11. Segawa T, Matsui M, Suzuki M, et al. Utilizing the Carba NP test as an indicator of expression level of carbapenemase genes in Enterobacteriaceae. J Microbiol Methods. 2017;133:35–39. doi:10.1016/j.mimet.2016.12.015
12. Van Der Zwaluw K, De Haan A, Pluister GN, Bootsma HJ, De Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the carba NP test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS One. 2015;10(3):e0123690. doi:10.1371/journal.pone.0123690
13. Kuchibiro T, Komatsu M, Yamasaki K, et al. Evaluation of the modified carbapenem inactivation method for the detection of carbapenemase-producing Enterobacteriaceae. J Infect Chemother. 2018;24(4):262–266. doi:10.1016/j.jiac.2017.11.010
14. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi:10.1093/nar/28.12.e63
15. Endo S, Komori T, Ricci G, et al. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol Lett. 2004;234(1):93–97. doi:10.1016/j.femsle.2004.03.015
16. Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett. 2005;253(1):155–161. doi:10.1016/j.femsle.2005.09.032
17. Imai M, Ninomiya A, Minekawa H, et al. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24(44–46):6679–6682. doi:10.1016/j.vaccine.2006.05.046
18. Kong QM, Lu SH, Tong QB, et al. Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasit Vectors. 2012;5(1):1–7. doi:10.1186/1756-3305-5-2
19. Srisrattakarn A, Lulitanond A, Wilailuckana C, et al. Rapid and simple identification of carbapenemase genes, bla NDM, bla OXA-48, bla VIM, bla IMP-14 and bla KPC groups, in Gram-negative bacilli by in-house loop-mediated isothermal amplification with hydroxynaphthol blue dye. World J Microbiol Biotechnol. 2017;33(7):130. doi:10.1007/s11274-017-2295-5
20. Cheng C, Zheng F, Rui Y. Rapid detection of blaNDM, blaKPC, blaIMP, and blaVIM carbapenemase genes in bacteria by loop-mediated isothermal amplification. Microb Drug Resist. 2014;20(6):533–538. doi:10.1089/mdr.2014.0040
21. Kim HJ, Kim HS, Lee JM, Yoon SS, Yong D. Rapid detection of Pseudomonas aeruginosa and Acinetobacter baumannii harboring blaVIM-2, blaIMP-1 and blaOXA-23 genes by using loop-mediated isothermal amplification methods. Ann Lab Med. 2016;36(1):15–22. doi:10.3343/alm.2016.36.1.15
22. Liu W, Zou D, Li Y, et al. Sensitive and rapid detection of the new delhi metallo-beta- lactamase gene by loop-mediated isothermal amplification. J Clin Microbiol. 2012;50(5):1580–1585. doi:10.1128/JCM.06647-11
23. Patel JB. M100-S25 performance standards for antimicrobial susceptibility testing; Twenty-Fifth Informational Supplement. Clin Lab Stand Inst. 2015.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.