Back to Journals » International Journal of Nanomedicine » Volume 15
Design and Fabrication of Dual Redox Responsive Nanoparticles with Diselenide Linkage Combined Photodynamically to Effectively Enhance Gene Expression
Authors Fang Y, Lin X, Jin X, Yang D, Gao S, Shi K , Yang M
Received 19 June 2020
Accepted for publication 24 August 2020
Published 1 October 2020 Volume 2020:15 Pages 7297—7314
DOI https://doi.org/10.2147/IJN.S266514
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Yan Fang,1 Xiaojie Lin,2 Xuechao Jin,2 Dongjuan Yang,2 Shan Gao,2 Kai Shi,2 Mingshi Yang1,3
1Wuya College of Innovation, Shenyang Pharmaceutical University, Shenyang 110016, People’s Republic of China; 2Department of Pharmaceutics, School of Pharmaceutical Science, Shenyang Pharmaceutical University, Shenyang 117004, People’s Republic of China; 3Department of Pharmacy, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen O DK-2100, Denmark
Correspondence: Kai Shi; Mingshi Yang Email [email protected]; [email protected]
Background: PEI is currently the most used non-viral gene carrier and the transfection efficiency is closely related to the molecular weight; however, the prominent problem is that the cytotoxicity increased with the molecular weight.
Methods: A novel redox responsive biodegradable diselenide cross-linked polymer (dPSP) was designed to enhance gene expression. ICG-pEGFP-TRAIL/dPSP nanoparticles with high drug loading are prepared, which have redox sensitivity and plasmid protection. The transfection efficiency of dPSP nanoparticle was evaluated in vitro.
Results: The plasmid was compressed by 100% at the N/P ratio of 16, and the particle size was less than 100 nm. When explored onto high concentrations of GSH/H2O2, dPSP4 degraded into small molecular weight cationic substances with low cytotoxicity rapidly. Singlet oxygen (1O2) was produced when indocyanine green (ICG) was irradiated by near-infrared laser irradiation (NIR) to promote oxidative degradation of dPSP4 nanoparticles. Under the stimulation of NIR 808 and redox agent, the particle size and PDI of ICG-pDNA/dPSP nanoparticle increased significantly.
Conclusion: Compared with gene therapy alone, co-transportation of dPSP4 nanoparticle with ICG and pEGFP-TRAIL had better antitumor effect. Diselenide-crosslinked polyspermine had a promising prospect on gene delivery and preparation of multifunctional anti-tumor carrier.
Keywords: diselenide, polyspermine, biodegradable, gene delivery, single-line oxygen
Introduction
PEI was successfully used by Behr for gene therapy in 1995 firstly and now has been widely used in gene delivery.1 As a gold standard for gene delivery vector, the high proton sponge effect and transfection efficiency of PEI (25 KDa) have been confirmed,2,3 however, the material toxicity is also a major drawback.4 Strong gene compression, effective intracellular transportation, and strong endo/lysosomal escaping are the three characters of an ideal gene carrier. High-level concentration of GSH, reactive oxygen (ROS) and ATP is accumulated in the microenvironment of tumor cells, which lead to low pH, besides, multiple enzymes are also distributed there.5,6 Compared with normal cells, in addition to producing high concentrations of glutathione, cancer cells produce high concentrations of ROS due to DNA changes, chronic inflammation and cell proliferation,7,8 which now has attracted great attention.9,10 High concentrations of GSH and ROS are tumor microenvironment activatable agents, so GSH or ROS-triggered vectors have been constructed.11,12 Based on this, disulfides13–15 and diselenides16–18 cross-linked polymers responsive to redox signals in tumor cells are prepared, which can be used for controlled release and targeted delivery, thereby improving efficacy. ROS-sensitive groups such as thioethers,19,20 phenylboronic acid-based compounds,21–23 tellurium-containing compounds,24,25 thioketals,26 and selenium-containing compounds27–29 have been studied.
Spermine (SP), like histidine, has a proton buffering effect, which can cause endosomal rupture and promote gene transfection.30,31 Spermine, a small polyamine molecule with fully agglutinated genes in sperm, is an endogenous substance of human body, safe and non-toxic, but has a weak ability to agglutinate genes. In order to improve transfection efficiency and reduce toxicity, cationic polymers bridged by redox sensitive bonds were designed and synthesized.32,33 The small molecular weight PEI bridged by disulfide bonds to deliver RNA or DNA, which reduces the cytotoxicity of large molecular weight PEI and improves transfection efficiency.34,35 Disulfides-containing polymers are rapidly degraded under the stimulation of tumor cells and are stable in the extracellular environment.34,36,37 Selenium, a necessary nutrient, has great potential value in biomaterials due to its high sensitivity for GSH and H2O2.28,38–40 The similarity between selenium and sulfur is manifested in atomic size, electronegativity and accessible oxidation state.41 The bond lengths of the disulfide and diselenide are 206 and 232 pm, respectively.29,42,43 The bond energy of S-C (272 kJ/mol) and S-S (240 kJ/mol) are stronger than that of Se-C (244 kJ/mol) and Se-Se (172 kJ/mol), which results in selenium compounds more easily oxidized and sensitive than sulfur compounds.28,29,40,44,45 Since diselenide is more sensitive to GSH and ROS, diselenyl-containing polymers may have dual redox responses.9,29 Utilizing the redox reactivity of dienyl ether, the dienyl ether structure is introduced into the polymer chain to achieve drug encapsulation and controlled release.28,46–48 Based on this, the researchers are committed to designing and synthesizing diselenides crosslinked polymers as gene victors to improve transfection efficiency.29,42,46,49–51 Photodynamics is that the photosensitizer will produce single-line oxygen to directly kill tumor cells under the infrared light.52 ICG has been approved by the US Food and Drug Administration, has significant near-infrared (NIR) optical properties, and is safe and non-toxic.53,54
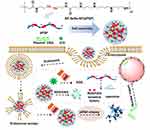
In this paper, a high molecular weight polymer dPSPs was prepared by cross-linking endogenous spermine through ROS/GSH-sensitive cross-linking agent (-Se-Se-). Polymers (dPSPs and PSP) were synthesized from Michael addition reactions of spermine to a reducible selenocystamine bisacrylamide or irreversible N, N-methylenebisacrylamide linker. Compared with small molecules of spermine, dPSPs are characterized by complete DNA compression, low toxicity and excellent proton buffering capacity. The dPSP nanoparticles formed with DNA protect nucleic acids from degradation until the high concentration of GSH/ROS successfully triggers the release of DNA in the intracellular cytoplasmic compartment. The structure of the polymer was characterized by nuclear magnetic resonance (13CNMR and 1H NMR), infrared spectroscopy (FT-IR) and gel permeation chromatography (GPC). The feasibility of dPSPs as a gene carrier was evaluated by comparing the relative polymerization degree, proton sponge effect, ROS/GSH sensitivity, cytotoxicity, and ability to compress DNA of each polymer. The formation of nanoparticles was analyzed by dynamic light scattering and gel electrophoresis. The active oxygen produced by ICG under-infrared radiation can not only kill tumors55 but also promote the degradation of dPSP4. Therefore, ICG and pDNA co-transport nanoparticles were prepared to investigate the ability of dPSP as a gene delivery vehicle to deliver DNA (Figure 1). The redox sensitivity of ICG-DNA/dPSP nanoparticles was further evaluated by comparing the particle size, PDI, ICG and DNA release ability of ICG-DNA/dPSP nanoparticles under near-infrared light and GSH/H2O2. The ability of singlet oxygen generation and anti-tumor of ICG-pEGFP-TRAIL/dPSP4 nanoparticle were studied in vitro. Co-localized uptake of ICG and YOYO-1-DNA was used to evaluate the ability of co-transportation in vitro. After NIR 808 irradiation, the transfection results of ICG-pEGFP-TRAIL (TRAIL, TNF related apoptosis-inducing ligand) loaded nanoparticles in 4T1 cells were evaluated by flow cytometry (FCM) and confocal microscopy (CLMS). All the research results prove that the ICG and DNA co-transport nanoparticle combined with infrared light irradiation can significantly improve the anti-tumor effect and gene expression.
Materials and Methods
Materials and Cells
2,2ʹ-diselenobisethanaminen (97%), N, N-methylenebisacrylamide (97%) and spermine (98%) were purchased from ENERGY (Shanghai, China); pCMV-Luc, pEGFP-Tnfsf10 (39–291), 4T1 cells and NIH/3T3 cells were obtained from the Feng Hui Bios (Changsha, China); glutathione, Lyso-TrackTM Red, 4ʹ,6-Diamidino-2-Phenylindole (DAPI), Thiazolyl Blue Tetrazolium Bromide (MTT), fetal bovine serum (FBS) (Gibco) and RPMI 1640 (Gibco) were purchased from Mei Lun Bios (Dalian, China). All the chemicals used in the experiment were from commercial sources and can be used without purification.
Synthesis of Redox Diselenide-Containing Polyethyleneimine (dPSP)
Firstly, 2, 2ʹ-diselenylbisphenyldiamine (0.05 mol, 10.6 g) was dissolved in a three-necked flask with 60 mL of water. Secondly, after the solution was cooled to 0–5°C in an ice-water bath, NaOH aqueous solution (0.4 mol, 20 mL) and acryloyl chloride dichloromethane solution (0.2 mol, 10 mL) were added simultaneously. Finally, after stirring in an ice water bath for 1.5 h, the temperature was raised to room temperature and the reaction was carried out overnight. The reaction solution was extracted with dichloromethane and then removed under reduced pressure. The product was washed repeatedly with water, and the crude product of selenocystamine bisacrylamide (SCBA) was collected. The crude product was further purified and crystallized.
Pure product was vacuum-dried and analysed by NMR (Bruker AVANCE-600) and FT-IR (Bruker, Switzerland). The yield of selenocystamine bisacrylamide was approximately 70%.
1H-NMR (SCBA, 400MHz, DMSO): δ: 6.21, 6.07, 5.58 (dd, 6H, J=1.6, 6.8, 11.2 Hz, -CH2=CH2), δ: 3.42 (t, 4H, -CH2-NH-), δ: 2.82 (m, 4H, -CH2-Se-). 13C-NMR (SCBA, 600MHz, DMSO): δ: 165.88(-CO-), δ: 131.59, 126.59 (-C=CH2), δ: 38.37, (-CH2-NH-), δ: 37.50, (-CH2-Se-). SCBA (KBr): 3252.4 cm−1 (-NH-, stretching), 3066.7 cm−1 (-CH=, stretching), 2954.3, 2872.9 cm−1 (-CH-, stretching), 1667.6 cm−1(-CO-, stretching), 1554.3 cm−1(-C-N-, stretching).
Briefly, 0.8 g of spermine was dissolved in 30 mL of a 10% methanol aqueous solution and added to a three-necked flask equipped with a condenser. Under the protection of nitrogen, selenocystamine bisacrylamide was slowly added dropwise to the reaction solution and reacted at 50°C. The dPSPs with different molecular weights were obtained by adjusting the molar ratio of reactants and reaction time. Before the reaction solution gelled, excess amine was added to terminate the reaction. After the reaction was terminated, the reaction solution was diluted with 20 mL of water and adjusted to pH 4 with 10 M HCl, and then dialyzed (MWCO: 3500) to remove unreacted spermine. Finally, the purified dPSP was obtained by lyophilizing the dialysate and stored at −20°C. The product was subsequently analyzed by NMR and FT-IR. The yields of dPSP1, dPSP2, dPSP3 and dPSP4 were about 70%, 60%, 55% and 45%, respectively.
1H-NMR (Spermine, 400MHz, D2O): δ: 2.62 (t, 4H, -CH2-NH2), δ: 2.55(m, 8H, -CH2-NH-), δ: 1.59 (quint, 4H, -CH2-CH2-NH-), δ: 1.47 (quint, 4H, -CH2-CH2-CH2-NH-). 13C-NMR (Spermine, 600MHz, D2O): δ: 41.62 (-CH2-NH2), δ: 49.13, 51.41 (-CH2-NH-), δ: 34.50, 29.34 (-CH2-CH2-CH2-NH-). Spermine (KBr): 3348.0 cm−1 (-NH-, stretching), 2927.9 cm−1, 2856.9 cm−1(-CH2-, stretching), 1592.0 cm−1 (-C-N-, stretching). 1H-NMR(dPSP, 400MHz, D2O):δ: 3.48 (m, 4H, -CH2-NH-CO-), δ: 3.07 (t, 4H, -CH2-Se-), δ: 2.92–2.82 (m, 16H, -CH2-NH-), δ: 1.75–1.58 (m, 4H, -NH-CH2-CH2-CH2-CH2-NH-). 13C-NMR(dPSP, 600MHz, D2O): δ: 164.53 (-CO-), δ: 44.77–47.33 (8C, -CH2-NH-), δ: 37.62 (-CH2-Se-), δ: 36.52–37.57 (2C, -CH2-NH-), δ: 23.37–27.13 (6C, -CH2-CH2-). dPSP (KBr): 3422.9 cm−1 (-NH-, stretching), 2926.6, 2853.2 cm−1 (-CH2-, stretching), 1630.5 cm−1 (-CO-, stretching), 1562.9 cm−1 (-C-N-, stretching).
Polymer Characterizations
Proton Buffering Capacity
The proton buffering capacity of the polymer can be determined by acid-base titration.56,57 Briefly, 2 mg of dPSPs was dissolved in 5 mL of 150 mM NaCl (0.4 mg/mL). The pH of polymer solution was first titrated to the initial 10.0 with NaOH and then to 3.0 with 0.1 M HCl. After stirring thoroughly, leave at room temperature for 3min. The change of pH value with the volume of HCl solution added can be measured by a pH meter. Spermine and 150 mM NaCl solution were used as positive control and negative control, respectively. The percentage of protonated amine groups from pH 7.4 to 5.1 is the buffer capacity and is calculated using the following formula:33,58
Buffering capacity (%)=(ΔVHCl×0.1M)/N×100%
Here ΔVHCl is the volume of HCl solution (0.1 M) required to lower the pH from 7.4 to 5.1, and N is the total moles of protonable amine in dPSPs and spermine.
GPC Analysis
According to the previously reported method, the molecular weight and polydispersity of dPSPs (Mw/Wn) were determined by GPC.59,60 Sodium acetate buffer (0.3 M NaAc, pH 4.4) was used as the mobile phase, polyethylene glycol (PEG) was used as the standard, and the flow rate was 1 mL/min to determine the molecular weight of the polymer. The biodegradability of polymer was also determined by GPC analysis.
In vitro Cytotoxicity Assays of dPSPs
NIH/3T3 cells and 4T1 cells were seeded in 96-well plates at a density of 5 × 104 cells/well to analyze the cytotoxicity of the dPSPs at four polymerization degrees. After cells were incubated with polymer for 24 hours, 10 µL of MTT (concentration of 5 mg/mL) were added to each well of the 96-well plate and the incubated at 37°C for another 4 h to form formazan dye. Then, 200 μL of DMSO was used to dissolve formazan in each well and shaken at room temperature for 15 minutes. Cells that only received fresh medium were the negative control group, and PEI-containing medium were the positive control group. The percentage of cell viability of dPSPs-treated cells relative to untreated cells is the final result (in 100% viability). The following formula represents the relative cell viability:
Cell Viability (%)=A490 (treated)/A490 (untreated)×100%
Here A490 (treated) is the absorbance of each well and A490 (untreated) is the absorbance of the negative control well.
Nanoparticles Characterizations
Preparation of dPSP Nanoparticles
Positively charged dPSPs can compress DNA to form regular-sized nanoparticles. Briefly, the nanoparticles were prepared by combining aqueous solutions of polycations (dPSP1, dPSP2, dPSP3 and dPSP4) with equal volumes of DNA (100 μg/mL) at various N/P (dPSP nitrogen/DNA phosphate) ratios. Incubate for 30 min before the analysis, and the nanoparticles were all freshly prepared.
ICG-pDNA/dPSP nanoparticle was prepared as described above, as shown in Figure 1. ICG-pDNA/dPSP nanoparticles were prepared in four steps by mixing ICG and DNA solution, vortexing with an equal volume of polymer, incubating at room temperature for 30 minutes, and centrifuging to remove free ICG.
Electrophoretic Mobility Shift Assay
The DNA condensation capability was examined by gel electrophoresis. One percent agarose gel electrophoresis was prepared to analyze the mobility of DNA/dPSPs nanoparticles with different N/P ratios. The dPSPs nanoparticles containing 1 μg DNA were prepared, mixed with the loading buffer at 5:1, run at 85 V for 45 min, and observed with Mini Gel (SAGECREATION, China).
Particle Size and Zeta Potential Measurements
The dPSP4 nanoparticle or PSP nanoparticle loaded with 5 μg DNA was prepared as above. The effect of N/P ratio on the particle size and zeta potential of the nanoparticle was investigated by dynamic light scattering (DLS) (Zetasizer Nano ZS90 (Malvern Instruments, U.K.)). Under redox conditions, the changes in particle size and PDI of dPSP or PSP nanoparticles were measured to evaluate the biodegradability of nanoparticles. ICG-DNA/dPSP4 nanoparticle was incubated with PBS containing GSH/H2O2 at 37°C or under infrared light for different times to determine the Z-averaged diameter and PDI.
Transmission Electron Microscopy (TEM)
The morphological characteristics of the nanoparticles formed by dPSPs of four molecular weights were observed and imaged using TEM with JEM-1200EX (Tokyo, Japan). The nanoparticle solution was dropped onto a 200-mesh carbon-coated copper grid and simmered for 8 min at room temperature.
DNA Release Ability Assay
ICG-pDNA/dPSP4 nanoparticles were collected in small EP tubes and incubated with PBS containing different redox agents or under certain laser at 37°C to study DNA release. After taking out samples, each sample was irradiated with specific laser (0.5W/cm2, 1.5W/cm2 and 2.5W/cm2) for 2 minutes to study the effect of laser irradiation on the release rate. As for monitoring the redox-triggered drug release, six kinds of PBS containing a certain amount of GSH or H2O2 were added into the drug-loaded nanoparticles solutions. The ability of nanoparticles to release DNA was evaluated using a dye-exclusion technique with PicoGreenTM.61 According to the manufacturer’s instructions, 100 μL of the release solution was mixed with the diluted PicoGreen dsDNA reagent (LIFE IL BIO) at intervals. After the mixed solution was incubated in dark for 8 minutes, the fluorescence intensity was measured by a multi-mode microplate reader TECAN 200 at a standard fluorescence wavelength (λex = 480 nm, λem = 520 nm). The relative fluorescence was calculated as the ratio of the fluorescence of the release medium to the fluorescence induced by the free DNA. The fluorescence of PicoGreenTM incubated with blank PBS was used as a blank control. All the release experiments data were measured at least six times for drawing figures.
ICG Release Ability Assay
ICG-loaded nanoparticles were placed in a dialysis bag (MW: 12,000 Da) and incubated in PBS containing redox agents to quantitatively determine the release profile of ICG. The 500 μL release solution was taken out at fixed points, and the same volume of fresh release medium was replenished. The concentration of ICG in the release medium was determined by UV absorption spectrophotometer, and the cumulative release percentage was calculated.
Quantitative Determination of 1O2
The singlet oxygen fluorescence probe (SOSG) was used to characterize the 1O2 produced by ICG and ICG-DNA/dPSP4 under infrared light irradiation. ICG-DNA/dPSP4 or free ICG was mixed with the degassed SOSG solution and irradiated with NIR 808 nm at power of 1.5 W/cm2 for 10 min to produce 1O2. Every 1 min, the fluorescence intensity of SOSG at 530 nm under excitation at 490 nm was recorded.
Cytotoxicity of ICG-pEGFP-TRAIL/dPSP4 Nanoparticle
Two nanoparticles containing 2.5 μg/mL pEGFP-TRAIL or ICG-pEGFP-TRAIL (ICG 0.3 μg/mL) were prepared. 4T1 cells were incubated with 100 μL of fresh medium containing free ICG or nanoparticle for 4 hours. After replacing the drug-containing medium with complete medium, some cells were irradiated with a laser at power of 1.5 W/cm2 for 3 min, and then incubated for another 44 hours. MTT assay was used to determine the cytotoxicity, and the effects of laser irradiation and non-irradiation on cell survival rate were compared.
Cell Uptake and Intracellular Colocalization
The cellular uptake of YOYO-1-DNA can be observed with FCM and CLSM (YOYO-1 λex = 491 nm, λem = 509 nm). 4T1 cells were seeded into 24-well plates at a density of 1 × 105 cells peer well and incubated at 37°C for 24 h in a 5% CO2 atmosphere. A part of the cells was replaced with fresh medium containing ICG-DNA/dPSP4 or ICG-DNA/PSP nanoparticles and continuously cultured for 1 hour or 4 hours (ICG λex=780 nm, λem=840 nm). The other part was placed in a fresh medium containing YOYO-1-DNA-loaded nanoparticles for different time periods (1 hour and 4 hours) and then labeled with LysoTrackerTM Red for 40 minutes. After removing the medium, the cells were carefully rinsed three times with PBS and then fixed with 4% paraformaldehyde solution. The cells were washed twice with PBS, stained with DAPI for 8 minutes, washed again and then checked by CLSM for fluorescence intensity.
4T1 cells were seeded into 6-well plates at a density of 1×106 cells/well and incubated for 24 hours at 37°C, then naked YOYO-1-DNA or DNA-loaded nanoparticles were added and incubated for 1 or 4 hours. The cells were triple washed repeatedly with ice PBS, then digested with trypsin without phenolphthalein and collected into EP tubes, centrifuged at 800 rpm for 3 minutes, resuspended with 0.5 mL PBS containing 1% serum, and finally checked by FCM using a FAC scanner.
Transfection Studies
Luciferase Assay
The pCMV-Luc reporter plasmid was used to evaluate the transfection ability of dPSP4 nanoparticles in 4T1 cells in vitro. Logarithmic 4T1 cells were seeded in a 6-well plate at 4.0×105 cells/well, and the transfection efficiency of nanoparticles with a N/P ratio of 1 to 32 (DNA concentration of 1 µg/mL) was investigated. When the cells grew to 80% confluence, rinsed with 1×PBS, added medium containing DNA-loaded nanoparticles or free DNA, and incubated for 4 hours. Subsequently, the culture was continued with serum-containing fresh medium for 44 hours, the cells were washed repeatedly with 1×PBS, and finally treated with cell lysate. Luciferase quantification was performed using the luciferase reporter gene detection substrate kit-firefly. The total protein in the lysate was measured by the BCA protein kit (Meilun), and the luciferase gene expression was quantitatively analyzed using a luciferase reporter assay substrate kit-firefly (Beyotime).
Confocal Microscopy
In order to evaluate the effect of infrared light on the transfection efficiency of dPSP4 and PSP nanoparticles, 4T1 cells were seeded at a density of 1×104 cells/well into a 24-well plate and cultured for 24 h at 37°C. The ICG-pEGFP-TRAIL/dPSP4 and ICG-pEGFP-TRAIL/PSP nanoparticles with an N/P ratio of 16 were prepared. After 4T1 cells grew to about 80% confluence, the original medium was replaced with FBS-free medium containing nanoparticles (0.2 μg pEGFP-TRAIL/well and 0.1μg ICG/well). After incubating for 4 hours, the medium of all cells was replaced with fresh medium and some cells received NIR808 irradiation at 1.5W/cm2 for 3 minutes, and then incubated at 37°C for another 24 h. The intensity of green fluorescence in each group was observed by CLSM.
Flow Cytometry
4T1 cells were seeded in a 6-well plate at a density of 1 × 106 cells/well and incubated at 37°C until 80% the cells fused. Firstly, cells were cultured in medium containing naked pEGFP-TRAIL or ICG-pEGFP-TRAIL-loaded nanoparticles for 4 hours, then washed with PBS for 3 times and replaced with fresh medium. The cells were irradiated with NIR 808 irradiation at power density of 1.5 w/cm2 and cultured for 24 hours. The expression of green fluorescent protein and the number of positive cells was checked by FCM.
Hemolytic Study of pDNA/dPSP4 Nanoparticle
Briefly, PEI 25 KDa and dPSP4 were, respectively, diluted into different centrifuge tubes at the same ratio, and an equal volume of 1.2% red blood cell suspension was added for incubation at 37°C. After 4 h, the erythrocyte suspension was centrifuged, the absorbance at 545 nm was measured, and the hemolysis ratio was calculated. PBS was used as negative control.
Statistical Analysis
The data were statistically analyzed by ANOVA and expressed as mean standard deviation (SD).
Results and Discussion
Characterization of Diselenide-Containing Polyspermine
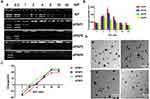
The dPSPs were synthesized by SP and SCBA through Michael addition reaction (Figure 2). The viscosity of the reaction solution was observed, the reaction was terminated with more than 10% of amine monomer before gelation. The acrylamide functional group is completely consumed, and the characteristic peak between δ = 5.6 and 6.2 (-CH = CH2) no longer exists (data not shown) (Figure 3A); there is no δ = 131.6 and 126.6 [- CH = CH2] (Figure 3B), which proves that the acrylamide functional group of SCBA is completely consumed; δ = 164.63 [-C = O] amide bond carbonyl peak indicates the presence of SCBA in the polymer. From the infrared spectroscopy data (Figure 3C), it can be seen that -CONH retractable vibration occurs, and -C = CH2 tensile vibration disappears, thus verifying the formation of polymer dPSP.
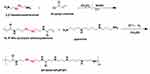 |
Figure 2 Synthesis of diselenide-containing poly(spermine)s (dPSPs). |
The molecular weight of dPSP was determined by GPC. As the molar ratio of spermine to SCBA and the time of the crosslinking reaction increased, the Mw and Mn of the product and the polydispersity increased accordingly. It was found that all copolymers have different molecular weights varying from 4500 to 21,800 Da (Table 1). It is worth noting that these dPSPs have considerable polydispersity (Mw/Mn = 1.36–2.33). The Mw and PDI of dPSP4 are significantly larger than those of the other three polymers, indicated that the degree of polymerization is uneven. The synthesis of dPSP is a classic Michelson addition reaction, and the degree of polymerization of the reaction is related to the molar ratio of the reactants and the reaction time. Theoretically, the molar ratio of the crosslinker to the monomer spermine is 1:1; in actual experiments, the reaction will form a gel after 12 hours. The degree of polymerization of the polymer synthesized by the addition reaction is related to the crosslinking agent and the crosslinking time. The greater the amount of crosslinking agent, the greater the molecular weight of the polymer formed. On the premise that the crosslinking agent remains unchanged, as the reaction time increases, the degree of polymerization of the polymer also increases. When the molar ratio is 1:4, the reaction is carried out for 12 hours to obtain a low molecular weight polymer with small molecular weight and good water solubility. After the reaction is continued for 48 hours, the viscosity of the reaction solution increases and a large molecular weight product is obtained. When the molar ratio is 1:2, after 48 hours of reaction, the reaction solution is very viscous and gelatinized, and polymers of different molecular weights can be obtained by controlling the reaction time.
 |
Table 1 Physical Characteristics of Diselenide Cross-Linked Polyspermines (dPSPs) |
Although the larger the molecular weight, the stronger the ability to compress the plasmid. But there are some reasons why we choose dPSP4 as the best. Firstly, as the time of crosslinking is prolonged, dPSP will gelatinize and is not suitable for use as a cationic carrier. Secondly, the gelled dPSP (higher molecular weight) is insoluble and cannot compress DNA. Finally, it is reported that PEI 25 KDa is the standard for traditional cationic gene carriers, and the molecular weight of dPSP4 is equivalent to that of PEI 25 KDa. Because molecular weight is related to transfection efficiency, dPSP4 of the four dPSPs might have higher gene transfection efficiency.
According to the “proton sponge” hypothesis, an essential property of cationic polymers is buffering capacity, which enhances gene expression by promoting the endosomal escaping of the nanocomposite.60,62 The buffering capacity of spermine and dPSPs were measured in 150 mM NaCl solution by acid–base titration. The dPSPs with 22–38% amine groups protonation showed better buffering capacity in the endosomal–lysosomal pH range (7.4–5.1). As shown in Figure 3D, spermine showed a relatively flat slope in the titration curve, and its buffering is 30% (Table 1). The buffering capacity of dPSP4 is 38%, higher than others (dPSP2 22%, dPSP3 25% and dPSP1 28%). These results indicated that in addition to the molecular weight, the primary, secondary and tertiary amine groups on polymer also play a key role on the buffering capacity. The molecular weights of dPSP1, dPSP2 and dPSP3 increase with the increase of diselenide bonds, the density of diselenide bonds increases, and the density of amino groups decreases, so their buffering capacity is lower than spermine. When the degree of polymerization is not too large, the density of amino groups is the dominant factor, so the proton buffering capacity of low molecular weight polymers will be lower than that of the monomer spermine. The reason why the buffering capacity of SP is superior to dPSP1, dPSP2 and dPSP3 is that the amino density of dPSPs decreases after the introduction of diselenide.
The impact of the increased molecular weight of dPSP4 is much greater than the increase of the diselenide bond, so its buffering capacity is greater than that of spermine.
To study the biodegradability of the polymer, after incubating for 1 hour with 1×PBS containing 0.5 mM GSH or 10 μM H2O2, the polymer was analyzed by GPC. Briefly, 20 mg dPSP was incubated with 2 mL of PBS containing GSH/H2O2, and then 200 μL of solution was taken out every other 10 minutes for GPC analysis, PEG was selected as a standard. For the redox-response study, GPC analysis was performed and the degradation curves of dPSPs treated with GSH/H2O2 are shown in Figure 3E and F. The degradation ability of novel cationic polymer is essential for both cytotoxicity and gene expression efficiency. Due to its unique character, the Se-Se bond can be oxidized into selenic acid (-SeOOH) by an oxidizing agent (such as H2O2) or reduced into selenol (-SeH) by a reducing agent (such as GSH).50,63 High molecular weight dPSPs degraded rapidly into spermine monomer and oligomer after GSH/H2O2 incubation. Changes on the Mw of dPSPs in the simulating the intracellular environment testify the feasibility of cationic polymer delivery genes. It can be rapidly degraded under 0.5 mM GSH or 10 μM H2O2, which is applicative for tumor cell, for that the concentration of GSH and H2O2 in tumor cells can reach the mmol level.7,64
MTT method was used to detect the effect of cationic polymer on cell viability. As shown in Figure 3G and H, the cytotoxicity of the four dPSPs was very low, which was safe enough for gene delivery. The survival rate of PEI was less than 50% at the concentration of 30 μg/mL. When the concentration was 200 μg/mL, the relative viability of largest molecular weight dPSP4 to 3T3 cells was 75%, and that of 4T1 cells was 80%. Compared with the similar molar weight of PEI, dPSP is less cytotoxic. As a traditional cationic polymer, PEI has a higher total charge number and charge density than dPSP. PEI cannot be degraded into small non-toxic molecules in tumor cells, so it has considerable cytotoxicity. According to reports, the charge density and molecular weight of cationic polymers had a greater effect on cell survival than the total charge.65 The difference of cytotoxicity caused by four biodegradable polymers is acceptable, mainly due to the effect of redox environment on the molecular weight of dPSPs. Due to the low molecular weight and low charge of dPSP degradation products, its cytotoxicity was very low. The high cytotoxicity of traditional cationic materials seriously impaired the efficiency of gene delivery, but dPSPs could be degraded into small molecular weight non-toxic spermine in the tumor microenvironment, which might greatly improve the efficiency of gene delivery.
Characterization of Nanoparticles
The ability of cationic polymers to compress DNA into nanoparticles is the primary prerequisite for successful gene delivery.33 As shown in Figure 4A, agarose gel electrophoresis was used to detect the ability of polycations to compress DNA. Agarose gel results revealed that dPSP1, dPSP2, dPSP3 and dPSP4 exhibited excellent DNA compression ability. When the N/P ratio was greater than 2, DNA migration was completely blocked. The ability of dPSP to agglutinate DNA increased with the increasing of molecular weight (dPSP4≥dPSP3> dPSP2> dPSP1). These results suggested that dPSPs has outstanding ability to condense DNA, which is consistent with previous research results that small-molecule cationic compounds cannot effectively compress genes, but cross-linking into high molecular weight polymers can effectively compress genes.35
Comprehensive characterization of dPSPs suggested that dPSP4 is the most promising DNA delivery vector. Firstly, dPSP4 has the best proton sponge, which can achieve the escape of endosomes to ensure effective gene transfection. Secondly, dPSP4 has the strongest ability to compress plasmids, which is the most essential part of gene delivery.
For gene loaded nano-delivery system, particle size and zeta potential are the main factors affecting gene distribution and transfection.3 When the increase of N/P ratio, the size of the nanoparticle increases firstly and then decreases (Figure 4B), and the zeta potential transforms from negative to positive (Figure 4C). When the N/P ratio is 1, since the negative charge of DNA is greater than that of the cationic material, the nanoparticle is negatively charged and z-averaged size is about 178 nm. As the N/P ratio increases, the potential begins to decrease and the particle size increases, because nanoparticles aggregate and grow at a potential of almost zero, with a maximum particle size greater than 300 nm. The effective endocytosis of nanoparticles requires a particle size less than 200 nm. When the N/P ratio is greater than 8, four nanoparticles can meet the requirements. It is found that when the N/P ratio is 16, the zeta potential of dPSP/pDNA nanoparticle is 18.9–25.9 mV, indicating that the N/P ratio with a significant influence on the zeta potential. TEM images showed that dPSPs nanoparticles were smooth and spherical, with the increasing of molecular weight, the particle size gradually decreased at the N/P ratio of 16 (Figure 4D). The particle size of dPSP4/pDNA was about 80 nm when N/P was 16, and the potential was greater than 15 mV, which satisfied the requirements of gene delivery for the vector. Based on the results mentioned above, dPSP4 was selected for subsequent research. The PSP was selected as the control for that both its molecular weight and the synthetic method were similar to dPSP4.
Evaluation of Biodegradability of Nanoparticles
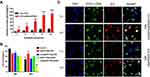
Nanoparticles were incubated in PBS containing various concentrations of GSH/H2O2 at 37°C to detect the redox sensitivity. Nanoparticles were prepared according to the previous method, and nanoparticles with N/P ratio of 16 were subjected to the redox-sensitivity experiments. ICG-DNA/dPSP nanoparticles were incubated for 1 h under NIR 808 irradiation or GSH/H2O2, and then the two nanoparticles were incubated for 24 h. The changes of the particle size and PDI with time were monitored by DLS. Before reaching the targeted site for endocytosis, the stability of the nanoparticles is an important guarantee for gene expression.56 As shown in Figure 5A and B, after incubated in PBS, dPSP4 nanoparticles showed quite good stability. But under the irradiation of NIR 808, the particle size and PDI of nanoparticle increased rapidly. The possible reason was that ICG-DNA/dPSP4 generates single-line oxygen through infrared light, which could promote the dissociation of diselenides and the aggregation of nanoparticles. After irradiation with different power NIR for 3 min, the change of nanoparticles was different. The particle size and PDI of nanoparticles changed fastest when irradiated with 2.5W/cm2 power, but there was no strong effect of high concentration of H2O2 directly. Between low GSH/H2O2 and high GSH/H2O2 environments, both the particle size and PDI of dPSP4 nanogels suggested significant differences. Time-depended particle sizes and PDI were also investigated. Under low-level concentration of H2O2/GSH, the particle size and PDI of the dPSP4 nanoparticles kept constant, which was consistent with dPSP4 nanoparticles under PBS conditions. After treated with high GSH/H2O2, ICG-DNA/dPSP4 nanoparticles quickly aggregated into a particle close to size 900 mm at 60 min, while ICG-DNA/dPSP4 nanoparticles only grew from about 100 nm to 600 nm after incubation with moderate GSH/H2O2. The ICG-DNA/dPSP4 nanoparticles still showed excellent stability at low GSH/H2O2 (normal tissue level). The averaged-size and PDI of dPSP4 nanoparticles increased about 3 and 5 times at high GSH levels after 24 h, respectively, while showed little change at low GSH levels (Figure 5C and D). After irradiated by infrared light, the particle size and PDI of PSP nanoparticles were consistent with incubated in PBS after 24 hours. The ICG-DNA/dPSP4 nanoparticles showed obvious redox-sensitivity compared with ICG-DNA/PSP nanoparticles. Therefore, dPSP4 nanoparticles might have outstanding endo/lysosomal escape ability. The results showed that the ICG-loaded dPSP4 nanoparticles exhibited the same biodegradability as the GSH/H2O2 stimulation under the irradiation of NIR808.
The concentration of ROS/GSH in tumor cells is substantially higher than extracellular levels. It was known that ICG-DNA/dPSP4 nanoparticles can produce single-line oxygen under the irradiation of infrared light, and the structure of dPSP4 was easily destroyed at high GSH/H2O2 levels. It was initially predicted that the drug release behavior of nanoparticles corresponded to changes in polymers (Figure 1). Figure 5E shows that at higher GSH levels, about 100% of the DNA of dPSP4 nanoparticles was released at 24 h, which was five times the value at low GSH levels, demonstrating the dPSP4 nanoparticles very sensitive to GSH. The DNA release behavior of dPSP4 nanoparticles at high H2O2 levels was similar to that from dPSP4 nanoparticles at high GSH surroundings. As shown in Figure 5F, at high H2O2 concentration, approximately 85 wt% of DNA in dPSP4 nanoparticles was released, which was 3 times that of low H2O2 levels, indicating that the dPSP4 nanoparticles were also effective and sensitive to ROS. However, almost 20 wt% or 25 wt% of DNA was released from PSP nanoparticles at high H2O2/GSH levels in 24 h. The DNA release of PSP polymer at low H2O2/GSH levels was similar to that at high H2O2/GSH levels (in Figure 5I and J). As shown in Figure 5G, after explored onto infrared light irradiation, dPSP4 nanoparticle quickly releases DNA, with the increasing of infrared light power, the release behavior was also accelerated. Under infrared light, the release rate of PSP nanoparticles was still very slow, less than 35% at 24 h, but slightly faster than PBS (Figure 5K). The release result of ICG is shown in Figure 5H and L. The release rate of PSP nanoparticles was less than 30% under H2O2/GSH, while that of dPSP nanoparticles was 80%, which was much higher than PSP nanoparticles. The results showed that ICG-DNA/dPSP4 nanoparticle was more sensitive to infrared light and H2O2/GSH, while PSP was insensitive. In the simulated tumor microenvironment, dPSP4 nanoparticles can release DNA and ICG quickly, which plays an essential role for efficient gene delivery.
Generation of Singlet Oxygen in Nanoparticles
The results of the singlet oxygen generation ability of ICG in dPSP4 nanoparticles are shown in Figure 6A. Free ICG continuously produced singlet oxygen, with the extension of NIR illumination time, the fluorescence intensity of SOSG gradually increased. The SOSG fluorescence intensity of ICG-DNA/dPSP4 nanoparticle was 7.9 times that of free ICG. The possible reason was that ICG was unstable in aqueous solution and was easy to be quenched, while nanoparticle had a protective effect on ICG, and ICG would not be quenched before light irradiation. Furthermore, even if the ICG aqueous solution was saturated with oxygen, the dPSP4 nanoparticle might carry a large amount of oxygen. Singlet oxygen is the lowest excited state of molecular oxygen and is the main cytotoxic ROS produced. When the photosensitizer is irradiated with laser light, it transfers energy to molecular oxygen to produce singlet oxygen, causing oxidative stress, which can cause apoptosis or necrosis of target tissue cells.
Cytotoxicity of ICG-pEGFP-TRAIL/dPSP4 Nanoparticle
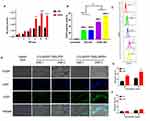
The results of degradation experiments and release experiments showed that dPSP4 was a redox-sensitive polymer. ICG was added as a photosensitizer to the carrier to prepare ICG-pEGFP-TRAIL nanoparticles to increase the level of ROS and trigger drug release. It was known that under laser irradiation, ICG could produce high concentration of singlet oxygen to kill tumor cells. Singlet oxygen (1O2) is a general term for the paramagnetic state of molecular oxygen. It belongs to active oxygen and is the excited state of ordinary oxygen (3O2). Singlet oxygen has strong active oxygen radicals and cytotoxic effects. The cell membrane and mitochondria are most sensitive to it. It can interact with a variety of biological macromolecules in the cell and cause damage to the cell membrane system by binding with molecules. As shown in Figure 6B, free ICG had almost no toxicity without infrared light irradiation, but ICG generated cytotoxic singlet oxygen under infrared light irradiation, which reduced the cell survival rate. Without infrared light irradiation, there was no difference in cell survival rate between ICG-pEGFP-TRAIL/PSP and pEGFP-TRAIL/PSP group. However, under infrared light irradiation, ICG-pEGFP-TRAIL/PSP was higher cytotoxicity than pEGFP-TRAIL/PSP group. The cell survival rate of the pEGFP-TRAIL/dPSP4 group was lower than that of the PSP group. The ICG-pEGFP-TRAIL/dPSP4 group had the lowest cell survival rate under infrared light irradiation. This was because the ROS produced by ICG can not only promote the apoptosis of tumor cells but also promote the degradation of dPSP4 to increase the expression of TRAIL protein.
Cellular Uptake
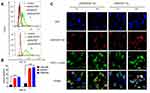
The cell uptake of ICG-pDNA nanoparticles was investigated by CLSM and FCM. As shown in Figure 6C, the fluorescence intensity of the two groups of nanoparticles increased significantly over time, and the red and green fluorescence coincided, indicating that the nanoparticles can co-transport ICG and DNA. The uptake results of ICG-pDNA/dPSP4 nanoparticles showed that PSP and dPSP4 could be easily taken up by 4T1 cells, and the absorption efficiency between PSP and dPSP4 was slightly different. As shown in Figure 7A, the proportion of positive cells in the PSP and dPSP4 nanoparticle groups was significantly higher than that in the naked pDNA group, indicating effective cell uptake after 1 h or 4 h of incubation, which was consistent with CLSM data. After incubating the cells with nanoparticles for 4 hours, the proportion of positive cells in the experimental group (PSP and dPSP4) was 87.65% and 91.25%, respectively, while the proportion of positive cells in naked pDNA was 15.05% (Figure 7B). The uptake of dPSP4 and PSP was time dependent, and the uptake at 4 hours was significantly stronger than that at 1 hour, while the uptake of naked DNA did not change significantly.
The CLSM and FCM results indicated that PSP and dPSP4 nanoparticles-mediated DNA delivery promotes cell uptake through positive charge and achieves endosome escape through the “proton sponge effect”. Cell uptake results indicated that polyspermine (with or without diselenides) can be used as a carrier for ICG and DNA co-transportation, which can quickly and efficiently deliver drugs to cells.
Intracellular Localization of YOYO-1-pDNA Loaded Nanoparticles
To study the subcellular localization of YOYO-1-pDNA/dPSP4 nanoparticles, intracellular co-localization experiments were performed using LysoTrackerTM Red and DAPI. As shown in Figure 7C, the intracellular distribution of YOYO-1-pDNA further illustrated that the delivery process of the two nanoparticles after cell uptake was different. After 4 hours of incubation of 4T1 cells with the nanoparticles, high levels of intracellular ROS/GSH increased lysosome escape of the dPSP4 nanoparticle group, which was not observed in the PSP control. Under the stimulation of high GSH/ROS in tumor cells, the diselenides in dPSP4 structure were broken, and the affinity with pDNA was reduced, thereby improving the lysosomal escape of gene. Higher GSH/ROS levels in tumor cells would ensure that sufficient pDNA is released into the cytoplasm in the dPSP4 group, but not in the PSP group. Cell uptake results indicated that the release of pDNA/dPSP4 nanoparticles in the cells was time dependent. Although both PSP and dPSP4 could guarantee sufficient cellular uptake, the lysosomal escape phenomenon of the two groups was significantly different. The dPSP4 containing redox-sensitive diselenides was quickly degraded into non-toxic small molecules in tumor cells and releases plasmids. PSP was non-degradable, highly toxic and could not quickly and completely release plasmids. Therefore, dPSP4 might have a higher gene transfection efficiency compared to PSP.
Transfection Efficiency of dPSP4 Nanoparticles in vitro
To evaluate the transfection efficiency of dPSP4 nanoparticles, the expression of two reporter genes (pEGFP-TRAIL and pCMV-Luc) were detected in 4T1 cells. As shown in Figure 8A, there was a significant difference in luciferase gene expression between nanoparticles with and without biodegradable diselenides. The expression of luciferase increased with the increase of N/P ratio. The reason was that the greater the N/P ratio, the nanoparticles prepared were of greater positive charge, smaller particle size and more condensed plasmid. This was consistent with the results of the in vitro properties investigation of nanoparticles above. The expression of luciferase gene in 4T1 cells showed that dPSP4 had relatively efficient luciferase expression, and its expression efficiency was significantly improved compared to PSP. The transfection efficiency reached its maximum when the N/P ratio was 16, and then gradually decreased.
According to the above experimental results, the optimal N/P ratio of 16 was selected for subsequent experiments. As shown in Figure 8D, the expression of green fluorescent protein in the dPSP4 group was significantly enhanced after infrared light irradiation, which was significantly stronger than the group without infrared light irradiation. However, the intensity of green fluorescence was almost the same in the PSP group with or without infrared light.
The reason why the green fluorescence intensity of the dPSP4 group was significantly stronger than that of the PSP group was that dPSP4 containing diselenide was easily destroyed under redox stimulation. The high concentration of ROS/GSH in tumor cells and the singlet oxygen produced by ICG under infrared light irradiation accelerated the decomposition of diselenides.
The percentage of EGFP positive cells and the average fluorescence value was measured by flow cytometry. As shown in Figure 8B and C, the transfection result of dPSP4 was obviously stronger than that of PSP. When irradiated by infrared light, the transfection efficiency of dPSP4 was 70.9%, while that of PSP was only 29.0%. In the absence of infrared light, the transfection efficiency of dPSP was 55.3%, while that of PSP was only 26.4%. For dPSP4 nanoparticles, the number of positive cells was about 2.45 times that of PSP (NIR808 irradiation) and 2.09 times that of PSP (no infrared light), which was consistent with the CLSM results. These results indicated that dPSP4 was an excellent non-viral gene vector, which was stable under normal physiological environment, while was completely degraded in tumor cells environment. Under the irradiation of infrared light, singlet oxygen generated by dPSP nanoparticles combined with photosensitizer accelerated gene release and improved transfection efficiency.
Hemolytic Analysis of pDNA/dPSP4 Nanoparticle
The blood compatibility of the carrier is an indicator of the safety of the gene carrier, which is particularly important for subsequent in vivo experiments. As shown in Figure 8E and F, the hemolysis rate of PEI and dPSP increased with the increase of sample concentration and incubation time. The reason of the result is that as time increases, the degree of reaction with serum proteins increases, causing more serious hemolysis effects. With the increase of sample concentration, the possibility of reaction between sample and red blood cells was increased, and the hemolysist is also improved. After 24 hours of incubation, the hemolysis rate of 100 μg/mL of PEI was 3.89 times that of dPSP. The charge density of cleavable dPSP is lower than that of PEI, and the electrostatic reaction between positive and negative phospholipids is weaker than that of PEI, thereby improving the safety and efficiency of the gene transfection.
Conclusions
In summary, we successfully prepared a diselenides-bridged cationic polymers as a gene vector with controlled release in tumor cells. The diselenides were easily broken, which endowed the nanoparticles stronger redox sensitivity. In addition, dPSPs showed good biocompatibility and non-toxicity to both normal cells and tumor cells. Nanoparticles loaded with photosensitizer can not only promote tumor apoptosis but also improve gene expression. Compared with ICG, ICG-pEGFP-TRAIL/dPSP4 nanoparticles produce more singlet oxygen, which can kill tumor cells directly and promote the dissociation of diselenides. The ICG-pDNA/dPSP4 nanoparticle had stronger redox sensitivity and faster release profile of pDNA and ICG when compared with the PSP nanoparticle under simulated tumor cell condition. The cytotoxicity, uptake and transfection of dPSP4 nanoparticles were evaluated in vitro, which showed sufficient cell uptake rates, successful lysosomal escape, high luciferase and pEGFP-TRAIL expression. The transfection results of ICG-pEGFP-TRAIL/dPSP4 nanoparticle revealed that the co-transportation nanoparticle delivery system combined with laser irradiation can significantly improve gene expression. In summary, the preparation of redox stimulus-responsive dPSP4 nanoparticles highlights the promising prospect of dPSP polymers in design of safe, stable, and intelligent gene vectors for anti-tumor.
Funding
The authors wish to thank the National Natural Science Foundation of China (No. 31671020, 81971729) for financial support. The authors declare no competing financial interest.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–7301. doi:10.2307/2368231
2. Liu W, Xue Y, Na P, He W, Zhuo R, Huang S. Dendrimer modified magnetic iron oxide nanoparticle/DNA/PEI ternary magnetoplexes: a novel strategy for magnetofection. J Mater Chem. 2011;21(35):13306–13315. doi:10.1039/c1jm11460c
3. Wong SP, Argyros O, Howe SJ, Harbottle RP. Systemic gene transfer of polyethylenimine (PEI)–plasmid DNA complexes to neonatal mice. J Control Release. 2011;150(3):298–306. doi:10.1016/j.jconrel.2010.12.010
4. Pandey AP, Sawant KK. Polyethylenimine: a versatile, multifunctional non-viral vector for nucleic acid delivery. Mat Sci Eng C Mater. 2016;68(1):904–918. doi:10.1016/j.msec.2016.07.066
5. Chen B, Dai W, He B, et al. Current multistage drug delivery systems based on the tumor microenvironment. Theranostics. 2017;7(3):538–558. doi:10.7150/thno.16684
6. Grek CL, Tew KD. Redox metabolism and malignancy. Curr Opin Pharmacol. 2010;10(4):362–368. doi:10.1016/j.coph.2010.05.003
7. Chen H, He W, Guo Z. An H2O2-responsive nanocarrier for dual-release of platinum anticancer drugs and O2: controlled release and enhanced cytotoxicity against cisplatin resistant cancer cells. Chem Commun. 2014;50(68):9714–9717. doi:10.1039/c4cc03385j
8. Wen AM, Steinmetz NF. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem Soc Rev. 2016;45(15):4074–4126. doi:10.1039/C5CS00287G
9. Deepagan VG, Kwon S, You DG, et al. In situ diselenide-crosslinked polymeric micelles for ROS-mediated anticancer drug delivery. Biomaterials. 2016;103:56–66. doi:10.1016/j.biomaterials.2016.06.044
10. Qu C, Li J, Zhou Y, et al. Targeted delivery of doxorubicin via CD147-mediated ROS/pH dual-sensitive nanomicelles for the efficient therapy of hepatocellular carcinoma. APPS J. 2018;20(2):20–34. doi:10.1208/s12248-018-0195-8
11. Chen H, Tian J, He W, Guo Z. H2O2-activatable and O2-evolving nanoparticles for highly efficient and selective photodynamic therapy against hypoxic tumor cells. J Am Chem Soc. 2015;137(4):1539–1547. doi:10.1021/ja511420n
12. Dai L, Cai R, Li M, et al. Dual-targeted cascade-responsive prodrug micelle system for tumor therapy in vivo. Chem Mater. 2017;29(16):6976–6992. doi:10.1021/acs.chemmater.7b02513
13. Peng H, Huang X, Melle A, Karperien M, Pich A. Redox-responsive degradable prodrug nanogels for intracellular drug delivery by crosslinking of amine-functionalized poly(N-vinylpyrrolidone) copolymers. J Colloid Interf Sci. 2019;540:612–622. doi:10.1016/j.jcis.2019.01.049
14. Pandey B, Patil NG, Bhosle GS, Ambade AV, Gupta SS. Amphiphilic glycopolypeptide star copolymer-based cross-linked nanocarriers for targeted and dual stimuli-responsive drug delivery. Bioconjuga Chem. 2019;30(3):633–646. doi:10.1021/acs.bioconjchem.8b00831
15. Zhang Y, Wu K, Sun H, Zhang J, Yuan J, Zhong Z. Hyaluronic acid-shelled disulfide-crosslinked nano-polymersomes for ultrahigh-efficiency reactive encapsulation and CD44-targeted delivery of mertansine toxin. ACS Appl Mater Inter. 2018;10(2):1597–1604. doi:10.1021/acsami.7b17718
16. Sun T, Zhu C, Xu J. Multiple stimuli-responsive selenium-functionalized biodegradable starch-based hydrogels. Soft Matter. 2018;14(6):921–926. doi:10.1029/C7SM02137B
17. Hailemeskel BZ, Hsu WH, Addisu KD, et al. Diselenide linkage containing triblock copolymer nanoparticles based on Bi(methoxyl poly(ethylene glycol))-poly(ε-carprolactone): selective intracellular drug delivery in cancer cells. Mat Sci Eng C Mater. 2019;103:1–16. doi:10.1016/j.msec.2019.109803
18. Hailemeskel BZ, Addisu KD, Prasannan A, Mekuria SK, Kao CY, Tsai HC. Synthesis and characterization of diselenide linked poly(ethylene glycol) nanogel as multi-responsive drug carrier. Appl Surf Sci. 2018;449(15):15–22. doi:10.1016/j.apsusc.2017.12.058
19. Wang J, Sun X, Mao W, et al. Tumor redox heterogeneity-responsive prodrug nanocapsules for cancer chemotherapy. Adv Mater. 2013;25(27):3670–3676. doi:10.1002/adma.201300929
20. Allen BL, Johnson JD, Walker JP. Encapsulation and enzyme-mediated release of molecular cargo in polysulfide nanoparticles. ACS Nano. 2011;5(6):5263–5272. doi:10.1021/nn201477y
21. Wang M, Sun S, Neufeld CI, Perez-Ramirez B, Xu Q. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew Chem Int Ed. 2014;53(49):13444–13448. doi:10.1002/anie.201407234
22. Major Jourden JL, Cohen SM. Hydrogen peroxide activated matrix metalloproteinase inhibitors: a prodrug approach. Angew Chem Int Ed. 2010;49(38):6795–6797. doi:10.1002/ange.201003819
23. Liu X, Xiang J, Zhu D, Jiang L, Shen Y. Fusogenic reactive oxygen species triggered charge-reversal vector for effective gene delivery. Adv Mater. 2016;28(9):1743–1752. doi:10.1002/adma.201504288
24. Fang R, Xu H, Cao W, Yang L, Zhang X. Reactive oxygen species (ROS)-responsive tellurium-containing hyperbranched polymer. Polym Chem. 2015;6(15):2817–2821. doi:10.1039/C5PY00050E
25. Cao W, Gu Y, Li T, Xu H. Ultra-sensitive ROS-responsive tellurium-containing polymers. Chem Commun. 2015;51(32):7069–7071. doi:10.1039/C5CC01779C
26. Shim MS, Xia Y. A reactive oxygen species (ROS)-responsive polymer for safe, efficient, and targeted gene delivery in cancer cells. Angew Chem Int Ed. 2013;52(27):6926–6929. doi:10.1002/ange.201209633
27. Ma N, Li Y, Ren H, Xu H, Li Z, Zhang X. Selenium-containing block copolymers and their oxidation-responsive aggregates. Polym Chem. 2010;1(10):1521–1708. doi:10.1039/C0PY00144A
28. Xu H, Cao W, Zhang X. Selenium-containing polymers: promising biomaterials for controlled release and enzyme mimics. Acc Chem Res. 2013;46(7):1647–1658. doi:10.1021/ar4000339
29. Ma N, Li Y, Xu H, Wang Z, Zhang X. Dual redox responsive assemblies formed from diselenide block copolymers. J Am Chem Soc. 2010;132(2):442–443. doi:10.1021/ja908124g
30. Won YW, Yoon SM, Lee KM, Kim YH. Poly(oligo-D-arginine) with internal disulfide linkages as a cytoplasm-sensitive carrier for siRNA delivery. Mol Ther. 2011;19(2):372–380. doi:10.1038/mt.2010.242
31. Woon YW, Kim KM, An SS, Lee M, Ha Y, Kim YH. Suicide gene therapy using reducible poly (oligo-d-arginine) for the treatment of spinal cord tumors. Biomaterials. 2011;32(36):9766–9775. doi:10.1016/j.biomaterials.2011.08.089
32. Yin Q, Shen J, Chen L, Zhang Z, Gu W, Li Y. Overcoming multidrug resistance by co-delivery of Mdr-1 and survivin-targeting RNA with reduction-responsible cationic poly(β-amino esters). Biomaterials. 2012;33(27):6495–6506. doi:10.1016/j.biomaterials.2012.05.039
33. Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30(29):5804–5814. doi:10.1016/j.biomaterials2009.06050
34. Jeong JH, Christensen LV, Yockman JW, et al. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi:10.1016/j.biomaterials.2006.12.019
35. Christensen LV, Chang CW, Kim WJ, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene. Bioconjuga Chem. 2006;17(5):1233–1240. doi:10.1021/bc0602026
36. Lee YS, Kim SW. Bioreducible polymers for therapeutic gene delivery. J Control Release. 2014;190:424–439. doi:10.1016/j.jconrel.2014.04.012
37. Gao S, Wang X, Tai Z, et al. Reducible chimeric polypeptide consisting of octa-d-arginine and tetra-l-histidine peptides as an efficient gene delivery vector. Int J Nanomedicine. 2015;10:4669–4690. doi:10.2147/IJN.S83507
38. Zhou W, Wang L, Li F, et al. Selenium-containing polymer@metal-organic frameworks nanocomposites as an efficient multiresponsive drug delivery system. Adv Funct Mater. 2017;27(6):1–8. doi:10.1002/adfm.201605465
39. Li W, Zhang P, Zheng K, Hu Q, Wang Y. Redox-triggered intracellular depegylation based on diselenide-linked polycations for DNA delivery. J Mater Chem B. 2013;1(46):6418–6426. doi:10.1039/c3tb21241f
40. Cheng G, He Y, Xie L, et al. Development of a reduction-sensitive diselenide-conjugated oligoethylenimine nanoparticulate system as a gene carrier. Int J Nanomedicine. 2012;7:3991–4006. doi:10.2147/IJN.S32961
41. Ma G, Liu J, He J, Zhang M, Ni P. Dual-responsive polyphosphoester-doxorubicin prodrug containing a diselenide bond: synthesis, characterization, and drug delivery. ACS Biomater Sci Eng. 2018;4(7):2443–2452. doi:10.1021/acsbiomaterials.8b00429
42. Liu J, Pang Y, Chen J, et al. Hyperbranched polydiselenide as a self assembling broad spectrum anticancer agent. Biomaterials. 2012;33(31):7765–7774. doi:10.1016/j.biomaterials.2012.07.003
43. Blom R, Haaland A. A modification of the schomaker—stevenson rule for prediction of single bond distances. J Mol Struct. 1985;128(1–3):21–27. doi:10.1016/0022-2860(85)85036-5
44. Xia J, Ji S, Xu H. Diselenide covalent chemistry at the interface: stabilizing asymmetric diselenide-containing polymer via micelle formation. Polym Chem. 2016;7(44):6708–6713. doi:10.1039/C6PY01610C
45. Zhang Y, Xu Y, Wei C, Zhang Y, Lang M. Diselenide-containing poly(ε-caprolactone)-based thermo-responsive hydrogels with oxidation and reduction-triggered degradation. Mater Today Chem. 2017;4:172–179. doi:10.1016/j.mtchem.2017.03.004
46. Cheng X, Jin Y, Sun T, Qi R, Fan B, Li H. Oxidation- and thermo-responsive poly(N-isopropylacrylamide-co-2-hydroxyethyl acrylate) hydrogels cross-linked via diselenides for controlled drug delivery. RSC Adv. 2015;5(6):4162–4170. doi:10.1039/C4RA13500H
47. Gao Y, Dong C. Triple redox/temperature responsive diselenide-containing homopolypeptide micelles and supramolecular hydrogels thereof. J Polym Sci. 2018;56(10):1067–1077. doi:10.1002/pola.28984
48. Feng C, Zhu D, Chen L, et al. Targeted delivery of chlorin e6 via redox sensitive diselenide-containing micelles for improved photodynamic therapy in cluster of differentiation 44-overexpressing breast cancer. Front Pharmacol. 2019;10:1–9. doi:10.3389/fphar.2019.00369
49. Zhai S, Hu X, Hu Y, Wu B, Xing D. Visible light-induced crosslinking and physiological stabilization of diselenide-rich nanoparticles for redox-responsive drug release and combination chemotherapy. Biomaterials. 2017;121:41–54. doi:10.1016/j.biomaterials.2017.01.002
50. Sun C, Ji S, Li F, Xu H. Diselenide-containing hyperbranched polymer with light-induced cytotoxicity. ACS Appl Mater Inter. 2017;9(15):12924–12929. doi:10.1021/acsami.7b02367
51. Saravanakumar G, Kim J, Kim WJ. Reactive-oxygen-species-responsive drug delivery systems: promises and challenges. Adv Sci. 2017;4(1):1–19. doi:10.1002/advs.201600124
52. Turan ISYD, Turksoy A, Gunaydin G, Akkaya EU. A bifunctional photosensitizer for enhanced fractional photodynamic therapy: singlet oxygen generation in the presence and absence of light. Angew Chem Int Ed. 2016;55(8):2875–2878. doi:10.1002/ange.201511345
53. Li W, Zhang H, Guo X, et al. Gold nanospheres-stabilized indocyanine green as a synchronous photodynamic–photothermal therapy platform that inhibits tumor growth and metastasis. ACS Appl Mater Interfaces. 2017;9(4):3354–3367. doi:10.1021/acsami.6b13351
54. Zheng M, Yue C, Ma Y, et al. Single-step assembly of DOX/ICG loaded lipid–polymer nanoparticles for highly effective chemo-photothermal combination therapy. ACS Nano. 2013;7(3):2056–2067. doi:10.1021/nn400334y
55. Hannah A, Luke G, Wilson K, Homan K, Emelianov S. Indocyanine green-loaded photoacoustic nanodroplets: dual contrast nanoconstructs for enhanced photoacoustic and ultrasound imaging. ACS Nano. 2014;8(1):250–259. doi:10.1021/nn403527r
56. Benns JM, Mahato RI, Kim SW. Optimization of factors influencing the transfection efficiency of folate–PEG–folate-graft-polyethylenimine. J Control Release. 2002;79(1–3):255–269. doi:10.1016/s0168-3659(01)00513-2
57. Piest M, Lin C, Mateos-Timoneda MA, et al. Novel poly(amido amine)s with bioreducible disulfide linkages in their diamino-units: structure effects and in vitro gene transfer properties. J Control Release. 2008;130(1):38–45. doi:10.1016/j.jconrel.2008.05.023
58. Lin C, Zhong Z, Lok MC, et al. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjuga Chem. 2007;18(1):138–145. doi:10.1021/bc062001
59. Wu D, Xian JL, Wu Y, Lay CL, Liu Y. ‘Living’ controlled in situ gelling systems: thiol-disulfide exchange method toward tailor-made biodegradable hydrogels. J Am Chem Soc. 2010;132(43):15140–15143. doi:10.1021/ja106639c
60. Sun Y, Zeng X, Meng Q, Zhang X, Cheng S, Zhuo R. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29(32):4356–4365. doi:10.1016/j.biomaterials.2008.07.045
61. Ping Y, Wu D, Kumar JN, Cheng W, Lay CL, Liu Y. Redox-responsive hyperbranched poly(amido amine)s with tertiary amino cores for gene delivery. Biomacromolecules. 2013;14(6):2083–2094. doi:10.1021/bm400460r
62. Lin C, Engbersen JFJ. Effect of chemical functionalities in poly(amido amine)s for non-viral gene transfection. J Control Release. 2008;132(3):267–272. doi:10.1016/j.jconrel.2008.06.022
63. Wang L, Cao W, Yi Y, Xu H. Dual redox responsive coassemblies of diselenide-containing block copolymers and polymer lipids. Langmuir. 2014;30(19):5628–5636. doi:10.1021/la501054z
64. DP CJ J, Samiec PS, Sternberg P, Mody VC, Reed RL, Brown LAS. Glutathione measurement in human plasma: evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin Chim Acta. 1998;275(2):175–184. doi:10.1016/S0009-8981(98)00089-8
65. Fischer D, Dautzenberg H, Kunath K, Kissel T. Poly(diallyldimethylammonium chlorides) and their N-methyl-N-vinylacetamide copolymer-based DNA-polyplexes: role of molecular weight and charge density in complex formation, stability, and in vitro activity. Int J Pharm. 2004;280(1–2):253–269. doi:10.1016/j.ijpharm.2004.05.018
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.