Back to Journals » Cancer Management and Research » Volume 10
Decreased expression of TRIM21 indicates unfavorable outcome and promotes cell growth in breast cancer
Authors Zhou WB, Zhang YY, Zhong CN, Hu JT, Hu H , Zhou DX, Cao MQ
Received 27 May 2018
Accepted for publication 25 June 2018
Published 20 September 2018 Volume 2018:10 Pages 3687—3696
DOI https://doi.org/10.2147/CMAR.S175470
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Wenbin Zhou,1 Yayuan Zhang,1 Caineng Zhong,1 Jintao Hu,2 Hong Hu,1 Dongxian Zhou,1 MeiQun Cao3
1Department of Breast Surgery, Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, Shenzhen, Guangdong Province, China; 2Department of Pathology, Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, Shenzhen, Guangdong Province, China; 3Shenzhen Institute of Geriatrics, Shenzhen Second People’s Hospital, The First Affiliated Hospital of Shenzhen University, Shenzhen, Guangdong Province, China
Background: Tripartite motif-containing protein 21 (TRIM21), an E3 ubiquitin ligase, has been implicated in autoimmune diseases. Dysregulation of TRIM21 contributes to the progression of human malignancies, but its role and clinical significance in breast cancer remain unclear.
Methods: The expression of TRIM21 was examined by quantitative real-time PCR, Western blot, and immunohistochemistry. The role of TRIM21 in the progression of breast cancer was determined using in vitro and in vivo models. The upstream regulation of TRIM21 was investigated by luciferase reporter assay.
Results: Here, we showed that TRIM21 expression in breast cancer tissues was decreased at both the mRNA and protein levels in comparison to that in nontumorous tissues. TRIM21 expression was closely associated with tumor size, estrogen receptor, human epidermal growth factor receptor 2, and clinical stage. Low TRIM21 expression was correlated with poor overall and disease-free survival in two independent cohorts containing 1,219 patients with breast cancer. A multivariate Cox regression model suggested TRIM21 as an independent factor for overall survival. In vitro data revealed that TRIM21 expression was suppressed by miR-494-3p directly targeting the 3′ untranslated region of TRIM21. Overexpression of TRIM21 impeded cell proliferation and tumor growth in breast cancer, whereas TRIM21 depletion enhanced these capacities.
Conclusion: Collectively, our findings indicate that TRIM21 serves as a potential prognostic biomarker and functions as a tumor suppressor in breast cancer.
Keywords: TRIM21, prognosis, cell proliferation, breast cancer
Introduction
Although several promising molecular classification and therapeutic strategies have been applied in clinical settings, the outcome of patients with breast cancer rarely improves.1 The morbidity and mortality of breast cancer rank among the highest in females worldwide.2 This is because the mechanisms of breast cancer tumor initiation and progression of are still unclear. The identification of biomarkers with potent proliferative capability and prognostic value has recently attracted increasing interest.
Ubiquitylation and proteasome-dependent degradation, modulated by ubiquitin ligases, are responsible for the control of cellular protein expression.3 The tripartite motif-containing (TRIM) superfamily consists of more than 80 members that are multidomain ubiquitin E3 ligases. They share evolutionary conserved domain structures including an N-terminal RING domain with E3 ubiquitin ligase activity, a B-box domain, a coiled-coil domain, and a C-terminal PRYSPRY domain.4 TRIM21, also called Ro52 or RNF81, was identified as an antibody-binding protein involved in the autoimmune diseases systemic lupus erythematosus and Sjögren’s syndrome and exerts critical functions in innate immunity to neutralize viral infection.5 It is ubiquitously expressed by both hematopoietic and nonhematopoietic cells. The known function of TRIM21 is binding to internalized antibody-coated viruses, targeting them for degradation.6 For example, BECLIN1, Bcl-2, and p62 have been identified as the binding proteins of TRIM21 during cellular processes, such as autophagy and cell proliferation.6 Mounting evidence has shown that TRIM21 is involved in the progression of human cancers.7 Decreased expression of TRIM21 has been found in hepatocellular carcinoma and diffuse large B-cell lymphoma.8,9 TRIM21 enhances the ubiquitination of Oct-1 to participate in the maintenance and self-renewal of cancer stem cells.10 TRIM21 confers drug resistance to cisplatin to colon cancer cells via the suppression of Par-4.11 In breast cancer, TRIM21 was found to be capable of downregulating the expression of Sal-like 4 (SALL4) and Lifeguard.12,13 However, the clinical significance of TRIM21 and its role in cell proliferation in breast cancer remain elusive.
Using tissue microarray (TMA)-based immunohistochemistry (IHC) and in vitro experiments, we evaluated the expression of TRIM21 and its clinical value in two independent cohorts of 1,219 patients with breast cancer. We also investigated the upstream regulation of TRIM21 and its effect on cell growth. Our data suggest that TRIM21, regulated by miR-494-3p, serves as a potential prognostic biomarker and functions as a tumor suppressor in breast cancer.
Materials and methods
Patients
A cohort consisting of 169 female patients with primary breast invasive ductal carcinoma was recruited from May 2005 to December 2010 at Shenzhen People’s Hospital cohort. Written informed consent was obtained from each patient. None of the patients received adjuvant therapies before surgical resection. The paraffin-embedded tissues, along with the clinical information, were collected for the construction of TMA. The patients were 21–75 years old. The median follow-up was 74.5 months. Another 21 cases with matched nontumorous and tumorous tissues were obtained. The use of human samples was approved by the Research Ethics Committee of Second Clinical Medical College of Jinan University.
Cell lines and transfection
Breast cancer cell lines (MCF7, MDA-MB-231, and SKBR3) purchased from the Cell Resource Center, Chinese Academy of Science Committee (Shanghai, China) were cultured with DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% heat-inactivated FBS (HyClone, Logan, UT, USA). Following the instructions for Lipofectamine 2000 (Thermo Fisher Scientific), cells were transfected with TRIM21-overexpressing vector or shRNA for 24 hours (sc-40917-SH, Santa Cruz Biotechnology Inc., Dallas, TX, USA) and then selected in G418-contained medium for another 2 weeks to construct stable cell lines.
Quantitative real-time PCR (qRT-PCR)
Complementary DNA was synthesized from the total RNA using the PrimeScript RT reagent Kit (Takara, Kyoto, Japan). qRT-PCR was performed with SYBR Premix ExTaq (Takara). The expression of the endogenous gene β-actin served as a control for the relative expression of TRIM21. The −ΔCt method was used to calculate the expression. Conditions for RT-PCR were set as the following: 95°C for 10 minutes, 40 cycles of 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension of 10 minutes at 72°C. The primers used in this study were as the following. TRIM21, forward: 5′-TCAGCAGCACGCTTGACAAT-3′ and reverse: 5′-GGCCACACTCGATGCTCAC-3′. β-actin, forward: 5′-TGGCACCCAGCACAATGAA-3′ and reverse: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
MTT
Stable cells were cultured in 96-well plates for 5 days. A total of 20 µL of MTT (5 mg/mL) was added into the wells for 3 hours. The formazan crystals were dissolved in DMSO (150 µL/well). The absorbance at 490 nm of each sample was measured. The cell growth rate was calculated.
Colony formation
Five hundred cells were incubated in six-well plates with DMEM medium plus 500 mg/L G418. After 2 weeks, the colonies were fixed with 4% paraformaldehyde and stained with 0.05% crystal violet solution for 10 minutes, washed with PBS, and counted.
Transient transfection and luciferase assays
SKBR3 cells were transfected with empty vector, psiCHECK2-TRIM21-3′ untranslated region (UTR)-wt or psiCHECK2- TRIM21-3′UTR-mutant using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The luciferase assays were conducted with the Luciferase Assay System (Promega Corporation, Fitchburg, WI, USA).
Western blot
Proteins extracted from fresh tissues or cells with various treatments were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and then incubated with a primary specific antibody for TRIM21 (1:1,000, #92043, Cell Signaling Technology) and GAPDH (1:1,000, sc-293335, Santa Cruz Biotechnology) in 5% nonfat milk, followed by a horse radish peroxidase-conjugated anti-rabbit/mouse second antibody. Enhanced chemiluminescence detection reagent (Amersham Life Science, Piscataway, NJ, USA) was used to show the results.
IHC and evaluation
Formalin-fixed and paraffin-embedded sample sections were dewaxed in xylene and graded alcohols, hydrated, and washed in PBS. After pretreatment in a microwave oven, endogenous peroxidase was inhibited by 3% hydrogen peroxide in methanol for 20 minutes, followed by avidin–biotin blocking using a biotin-blocking kit (DAKO, Darmstadt, Germany). Slides were then incubated with TRIM21 antibody overnight in a moist chamber at 4°C, washed in PBS, and incubated with biotinylated goat anti-rabbit/mouse antibodies. Slides were developed with DAB and counterstained with hematoxylin. Semiquantitative IHC detection was used to determine the TRIM21 protein levels. Using the H-score method, the expression levels were scored as proportion of immune-positive staining area (0%, 0; 1%–25%, 1; 26%–50%, 2; 51%–75%, 3; 76%–100%, 4) multiplied by the intensity of staining (0, negative; 1, weak; 2, moderate; 3, intense). The scores were independently rendered by two pathologists. The median IHC score was chosen as the cutoff value for defining high and low expression.
IHC was also used to assess the expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (Her2), and Ki67 in breast cancer. Positivity for ER, PR, and Ki67 was recorded if the percentage of positive cells was more than 10% of the cancer cells. Positivity for Her2 was determined by an IHC score 3+. For cases with an IHC score 2+, FISH was used for the determination of Her2-positive cases.
Animal model
Four-week-old male BALB/c-nude mice were randomly separated into two groups. A total of 5×106 stable cells were subcutaneously injected into the right flanks of the mice. Tumors formed by stable cells were measured with calipers and calculated using the following formula: volume (mm3=[width2 (mm2×length (mm)]/2. At day 27, the tumors were dissected and weighed. All animal studies were approved by the Research Ethics Committee of Second Clinical Medical College of Jinan University. Animal experiments complied with the ARRIVE guidelines and were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical analysis
Continuous variables were analyzed using Student’s t-test (two-tailed) and presented as the mean with SEM. Kaplan–Meier analysis (the log-rank test) was used for survival analysis and univariate analysis. The Cox proportional hazards regression model was used to evaluate the independent prognostic value of TRIM21 in breast cancer. A P-value <0.05 was considered statistically significant.
Results
TRIM21 expression is decreased in breast cancer
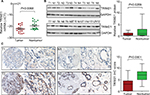
To determine the expression of TRIM21 in breast cancer, 21 pairs of specimens were collected. The qRT-PCR results showed that TRIM21 mRNA in breast cancer was significantly downregulated compared with the corresponding adjacent nontumorous samples in 61.9% (13/21) of cases (Figure 1A). Consistently, the protein expression of TRIM21 was, on average, 6.18-fold higher in the tumor tissues than that in the nontumorous tissues (Figure 1B).
To further evaluate the expression of TRIM21 in breast cancer, a cohort containing 169 archived paraffin-embedded samples, along with their clinical and pathologic features, was collected to construct a TMA. TMA-based IHC demonstrated that TRIM21 was mainly identified in the cytoplasm of tumor and nontumor cells (Figure 1C). According to IHC scores, the expression of TRIM21 was significantly decreased in breast cancer tissues compared to that in nontumorous tissues (Figure 1C). Negative staining of TRIM21 in tumor tissue was found in 29.9% (50/169) of cases, whereas it was found in 9.6% (16/169) of nontumor cases. Downregulation of TRIM21 in breast cancer compared with that in the corresponding adjacent breast tissues was present in 76.9% (130/169) of the cases.
Low expression of TRIM21 is correlated with poor outcomes in breast cancer
To determine the clinical implication of TRIM21 in breast cancer, the cases were divided into two groups (high TRIM21 expression and low TRIM21 expression) according to the median IHC score (3.0). Statistical analyses indicated that high expression of TRIM21 was significantly associated with small tumor size (P=0.016), positive ER (P=0.003), negative Her2 (P=0.005), and early clinical stage (P=0.001) (Table 1). The median overall survival time was 36.27 months for patients with low TRIM21 expression and 48.0 for those with high TRIM21 expression. Kaplan–Meier analyses revealed that patients with low TRIM21 expression were likely to have shorter survival times accompanied by a shorter period of tumor relapse (Figure 2A, B). The prognostic value of TRIM21 in breast cancer was confirmed in The Cancer Genome Atlas cohort containing 1,050 cases, showing that patients with low TRIM21 mRNA expression had a higher propensity for disease progression and cancer-related death (Figure 2C, D). Furthermore, multiple Cox regression analysis suggested TRIM21 as an independent factor for unfavorable overall survival (hazard ratio =0.372, 95% CI: 0.193–0.715, P=0.003) (Table 2).
TRIM21 expression is downregulated by miR-494-3p in breast cancer
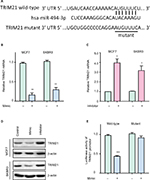
To investigate the upstream regulation of TRIM21, two bioinformatic algorithms (miRanda and TargetScan) were used to search for the potential regulator of TRIM21 in breast cancer. A putative binding site for miR-494-3p was found at 244–250 base pairs of the 3′UTR of TRIM21 (Figure 3A). In MCF7 and SKBR3 cells, TRIM21 mRNA was reduced by overexpression of miR-494-3p (Figure 3B) but induced by the miR-494-3p inhibitor (Figure 3C). The protein expression of TRIM21 was consistently altered by miR-494-3p (Figure 3D). To determine whether miR-494-3p modulated TRIM21 by directly targeting the 3′UTR of TRIM21, a dual-luciferase reporter assay was performed. The results showed that relative luciferase activity was significantly reduced in cells with miR-494-3p mimics and wild-type TRIM21 3′UTR. This reduction was not found in the cells with mutant TRIM21 3′UTR (Figure 3E). Collectively, these results demonstrated that miR-494-3 p directly targeted the 3′UTR of TRIM21 to suppress its expression in breast cancer.
TRIM21 suppresses cell growth in breast cancer
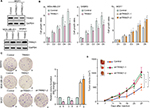
To explore the biologic function of TRIM21 in breast cancer, stable cells were established according to the basal expression of TRIM21 (Figure S1). The overexpression of TRIM21 in SKBR3 and MDA-MB-231 cells and the silence of TRIM21 in MCF7 were validated by Western blot (Figure 4A). MTT assays showed that the growth rates were accelerated in cells with TRIM21 overexpression but slowed down in cells with TRIM21 silencing (Figure 4B). The effect of TRIM21 on cell proliferation was confirmed by colony formation assays. More foci were formed by depletion of TRIM21, whereas exogenous TRIM21 markedly inhibited cell growth (Figure 4C). Nude mouse model data demonstrated that TRIM21 was capable of suppressing breast cancer growth in vivo. Xenografts formed by cells with TRIM21 silencing grew much faster than those in the control group (Figure 4D). These findings indicated that TRIM21 might function as a tumor suppressor in breast cancer.
Discussion
Dysregulation of ubiquitin ligase has been demonstrated to contribute to the progression of human cancers.14 Increasing attention has been paid to the potent implications of the function of E3 ubiquitin ligase in the clinical management of breast cancer.15 In this study, we showed that TRIM21 was downregulated in breast cancer and associated with poor outcomes. The decrease of TRIM21, partly attributed to the miR-494-3p-mediated post-transcriptional suppression, facilitated cell growth in breast cancer.
The prognostic value of TRIM proteins in human cancers has been documented. TRIM24 was correlated with poor survival and functioned as an oncogene in prostate and breast cancer.16,17 The oncogenic nature of TRIM25 was identified in prostate and breast cancer. High expression of TRIM25 was linked to unfavorable prognosis.18,19 TRIM29 was upregulated and associated with tumor progression and served as a robust pathogenic driver in bladder cancer.20 However, reduced expression of TRIM29 was found to correlate with poorer survival in breast cancer.21 Aberrant expression of TRIM65 has been shown to connect with poor overall and disease-free survival in hepatocellular carcinoma.22 The literature reports that decreased expression of TRIM21 predicts shorter survival times in patients with hepatocellular carcinoma and diffuse large B-cell lymphoma8,9 but better prognosis in patients with pancreatic cancer.11 TRIM21 has also been identified as a diagnostic biomarker for breast cancer.23 In our study, TRIM21 was noticeably downregulated in breast cancer. Patients with low TRIM21 expression were likely to have shorter survival times and experience a shorter interval of tumor progression in two independent cohorts consisting of 1,219 patients. This might be due to the correlation of reduced expression of TRIM21 and larger tumor size, advanced clinical stage, and the presence of Her2 amplification. In addition, after adjusting for the prognostic factors established in the univariate analysis, a significant correlation between TRIM21 expression and overall survival was confirmed. Collectively, these data indicate that TRIM proteins are of promising prognostic value in human cancers.
Uncontrolled cell proliferation mediated by oncogene overexpression or depletion of tumor suppressors is responsible for malignant progression. Our data demonstrated that ectopic expression of TRIM21 markedly inhibited, whereas knockdown of TRIM21 significantly facilitated, breast cancer growth in vitro and in vivo. While further studies are required to disclose the mechanism through which TRIM21 promotes cell proliferation in breast cancer, currently published data may offer some possibilities. First, TRIM21 was capable of ubiquitylating and degrading oncoproteins involved in the maintenance of cell stemness, such as SALL4 and OCT1.10,12 Second, TRIM21 was able to affect the progression of the cell cycle, autophagy, and apoptosis via modulating expression of p53, SQSTM1/p62, and Par-4.11,24,25 Third, TRIM21 is involved in the regulation of glycolysis and DNA repair by downregulating DDX41 and phosphofructokinase 1 platelet isoform.26,27 Taken together, TRIM21 functions as an important regulator of several cellular processes that enhance the cell growth.
In summary, our data showed that TRIM21 was downregulated by miR-494-3p in breast cancer. The decrease of TRIM21 was closely associated with clinical features and poor survival and promoted cell proliferation in vitro and tumor growth in vivo. Our findings suggest that TRIM21 serves as a potential prognostic and therapeutic biomarker and functions as a tumor suppressor in breast cancer.
Acknowledgment
This study was supported by Medical Scientific Research Foundation of Guangdong Province China (A2016490).
Disclosure
The authors report no conflicts of interest in this work.
References
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Qian G, Hu X, Li G, et al. Smurf1 restricts the antiviral function mediated by USP25 through promoting its ubiquitination and degradation. Biochem Biophys Res Commun. 2018;498(3):537–543. | ||
Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–860. | ||
Fukuda-Kamitani T, Kamitani T. Ubiquitination of Ro52 autoantigen. Biochem Biophys Res Commun. 2002;295(4):774–778. | ||
Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. | ||
Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. | ||
Brauner S, Zhou W, Backlin C, et al. Reduced expression of TRIM21/Ro52 predicts poor prognosis in diffuse large B-cell lymphoma patients with and without rheumatic disease. J Intern Med. 2015;278(3):323–332. | ||
Ding Q, He D, He K, et al. Downregulation of TRIM21 contributes to hepatocellular carcinoma carcinogenesis and indicates poor prognosis of cancers. Tumour Biol. 2015;36(11):8761–8772. | ||
Du L, Li YJ, Fakih M, et al. Role of SUMO activating enzyme in cancer stem cell maintenance and self-renewal. Nat Commun. 2016;7:12326. | ||
Nguyen JQ, Irby RB. TRIM21 is a novel regulator of Par-4 in colon and pancreatic cancer cells. Cancer Biol Ther. 2017;18(1):16–25. | ||
Itou J, Li W, Ito S, et al. Sal-like 4 protein levels in breast cancer cells are post-translationally down-regulated by tripartite motif-containing 21. J Biol Chem. 2018;293(17):6556–6564. | ||
Müller J, Maurer V, Reimers K, Vogt PM, Bucan V. TRIM21, a negative modulator of LFG in breast carcinoma MDA-MB-231 cells in vitro. Int J Oncol. 2015;47(5):1634–1646. | ||
Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18(2):69–88. | ||
Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19(3):130–140. | ||
Groner AC, Cato L, de Tribolet-Hardy J, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29(6):846–858. | ||
Tsai WW, Wang Z, Yiu TT, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–932. | ||
Takayama KI, Suzuki T, Tanaka T, et al. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene. 2018;37(16):2165–2180. | ||
Walsh LA, Alvarez MJ, Sabio EY, et al. An integrated systems biology approach identifies TRIM25 as a key determinant of breast cancer metastasis. Cell Rep. 2017;20(7):1623–1640. | ||
Tan ST, Liu SY, Wu B. TRIM29 overexpression promotes proliferation and survival of bladder cancer cells through NF-κB signaling. Cancer Res Treat. 2016;48(4):1302–1312. | ||
Ai L, Kim WJ, Alpay M, et al. TRIM29 suppresses TWIST1 and invasive breast cancer behavior. Cancer Res. 2014;74(17):4875–4887. | ||
Yang YF, Zhang MF, Tian QH, Zhang CZ. TRIM65 triggers β-catenin signaling via ubiquitylation of Axin1 to promote hepatocellular carcinoma. J Cell Sci. 2017;130(18):3108–3115. | ||
Wang J, Figueroa JD, Wallstrom G, et al. Plasma autoantibodies associated with basal-like breast cancers. Cancer Epidemiol Biomarkers Prev. 2015;24(9):1332–1340. | ||
Reddy BA, van der Knaap JA, Bot AG, et al. Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol Cell. 2014;53(3):458–470. | ||
Pan JA, Sun Y, Jiang YP, et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell. 2016;61(5):720–733. | ||
Lee JH, Liu R, Li J, et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun. 2017;8(1):949. | ||
Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14(2):172–178. |
Supplementary material
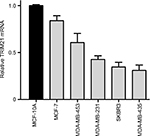  | Figure S1 TRIM21 mRNA expression in breast cancer cell lines. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.