Back to Journals » OncoTargets and Therapy » Volume 9
Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients
Authors Iliev R, Stanik M , Fedorko M, Poprach A, Vychytilova-Faltejskova P, Slaba K, Svoboda M, Fabian P, Pacik D, Dolezel J, Slaby O
Received 27 June 2015
Accepted for publication 19 September 2015
Published 8 January 2016 Volume 2016:9 Pages 217—222
DOI https://doi.org/10.2147/OTT.S91295
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Robert Iliev,1,2 Michal Stanik,3 Michal Fedorko,4 Alexandr Poprach,1 Petra Vychytilova-Faltejskova,1,2 Katerina Slaba,2 Marek Svoboda,1 Pavel Fabian,5 Dalibor Pacik,1 Jan Dolezel,2 Ondrej Slaby3,4
1Department of Comprehensive Cancer Care, Masaryk Memorial Cancer Institute, 2Central European Institute of Technology, Masaryk University, 3Department of Urologic Oncology, Masaryk Memorial Cancer Institute, 4Department of Urology, University Hospital Brno, Masaryk University Brno, 5Department of Diagnostic and Experimental Pathology, Masaryk Memorial Cancer Institute, Brno, Czech Republic
Abstract: Piwi-interacting RNAs (piRNAs) are a newly discovered class of small non-coding RNAs involved in silencing of transposable elements and in sequence-specific chromatin modifications. PIWI proteins (PIWIL) belong to the family of Argonaute genes/proteins, bind piRNAs and their functioning have been described mainly in germ-line cells and more recently also in stem cells and cancer cells. There are four PIWI proteins PIWIL1, PIWIL2, PIWIL3, and PIWIL4 discovered in human till now. Recent studies suggested that deregulated the expression of Piwi proteins and selected piRNAs is common to many types of cancers. We found significantly lower expression of PIWIL1 (P<0.0001) and piR-823 (P=0.0001) in tumor tissue in comparison to paired renal parenchyma. Further, we observed progressive decrease in PIWIL1 (P=0.0228), PIWIL2 (P=0.0015), and PIWIL4 (P=0.0028) expression levels together with increasing clinical stage. PIWIL2 (P=0.0073) and PIWIL4 (P=0.0001) expression progressively decreased also with increasing Fuhrman grade. Most importantly, low-expression levels of PIWIL1 (P=0.009), PIWIL2 (P<0.0001), and PIWIL4 (P=0.0065) were significantly associated with worse overall survival in renal cell carcinoma (RCC) patients. Our results suggest the involvement of PIWIL genes and piR-823 in RCC pathogenesis, and indicate PIWIL1, PIWIL2, and PIWIL4 as potential prognostic biomarkers in patients with RCC.
Keywords: kidney cancer, PIWIL, piRNA
Introduction
Renal cell carcinoma (RCC) represents approximately 75% of all kidney tumors and 3% of all cancers in the adult population.1 Piwi-interacting RNAs (piRNAs) are a newly discovered class of small non-coding RNAs with a length of 26–31 nucleotides. piRNAs were first identified in the germ cells of various animal species.2–4 In the human genome, there were more than 24,000 distinct piRNA sequences identified till now,5 and their deregulated expression was observed in breast,6 bladder7 and gastric cancer,8 and multiple myeloma.9 piRNAs bind to a specific subfamily of Argonaute proteins called PIWI proteins. Argonaute proteins are a highly conserved group of proteins divided into two main classes: AGO and PIWI. In humans, four PIWI genes were identified. PIWI proteins in humans include PIWIL1/HIWI, PIWIL2/HILI, PIWIL3, and PIWIL4/HIWI2.10 They bind piRNAs and amplify them in the so-called ping-pong cycle, which is endoribonuclease Dicer independent. Their physiological role is transposon repression through base-pairing and direct degradation.11 They are also involved in DNA methylation but the exact mechanism of action remains unknown.12 These days, there is increasing evidence to show that PIWIL proteins are linked to the hallmarks of cancer defined by Weinberg and Hanahan, such as deregulated cell proliferation, altered apoptosis, genomic instability, invasion, and metastasis.13 Recent studies also suggested that the deregulated expression of PIWIL proteins is common in tumor tissue, and PIWIL expression levels are correlated to clinicopathological features and survival in patients with cancer and PIWIL proteins. Therefore, they present promising cancer biomarkers.13 In our study, we determined the expression levels of PIWIL genes in clear cell RCC tumors and paired renal parenchyma, and evaluated their association with clinicopathological features in patients with RCC. Further, we investigated whether the piR-823 expression, observed to be downregulated in gastric cancer,8 is similarly deregulated in RCC tumors, and if there is a correlation between piR-823 and PIWIL gene expression due to their involvement in piRNA biogenesis.
Materials and methods
Subjects
The patients with RCC who underwent radical nephrectomy at the Masaryk Memorial Cancer Institute (Brno, Czech Republic) and University Hospital Brno (Brno, Czech Republic) were included. Tissue samples were collected after signing an informed consent and stored at the Bank of Biological Material (Masaryk Memorial Cancer Institute). The samples were obtained from 57 patients (36 men and 21 women) with clear cell RCC with median age 64 (range 35–80) years at the time of diagnosis. In 38 cases, tissue samples of adjacent non-tumor renal parenchyma was available for the expression analysis. Clinical and pathological characteristics including clinical stage and Fuhrman grade are summarized in Table 1. The study has been approved by the local Ethical Committee of Masaryk Memorial Cancer Institute.
RNA extraction and qPCR
RNA isolation was performed using mirVana miRNA Isolation Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA yield and purity were measured using a spectrophotometer NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer’s recommendations. Quantitative polymerase chain reaction (qPCR) was performed in a total volume of 20 μL using specific probes for PIWIL1 (Hs01041737_m1), PIWIL2 (Hs00216263_m1), PIWIL3 (Hs00908825_m1), PIWIL4 (Hs00381509_m1; Life Technologies), and peptidylprolyl isomerase A (PPIA, 99999904_m1 Hs) as an endogenous control. cDNA synthesis of piR-823 was carried out using TaqMan MicroRNA Reverse Transcription Kit (Life Technologies). The primers and probe were designed and synthesized (Life Technologies) accordingly to the piR-823 sequence 5′-AGCGUUGGUGGUAUAGUGGUGAGCAUAGCUGC-3′. The expression level of piR-823 was computed relatively to endogenous control RNU44. qPCR was performed on an ABI 7500 (Life Technologies). Relative expression was calculated using a ΔCt method.
Statistic analysis
To calculate the significance of differences in PIWIL and piR-823 expression levels in cases of different clinical stages or Fuhrman grades, we used non-parametric Kruskal–Wallis test. To calculate the differences in expression between paired tumor tissues and adjacent non-tumor renal parenchyma, a nonparametric Wilcoxon matched pair test was used. For the sensitive discrimination of RCC patients with short and long overall survival cut-off values were identified for PIWIL genes and piR-823 by receiver operating curve analysis. Kaplan–Meier survival curves and long-rank test were used for survival analysis. Calculations were performed in GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
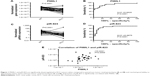
We determined the expression levels of PIWIL genes and piR-823 in tumor tissue of 57 patients with RCC and 38 samples of matched non-tumor renal parenchyma. First, we compared the expression levels of PIWIL genes and piR-823 in paired samples of RCC tumor tissue and non-tumor renal parenchyma. We found significant downregulation in the expression levels of PIWIL1 (P<0.0001) and piR-823 (P=0.0001; Figure 1A and B). Receiver operating curve analysis revealed that the tissue levels of both PIWIL1 and piR-823 could serve as appropriate biomarkers for differentiating tumor tissue from non-tumor renal parenchyma with the area under the curve of 0.8620 (95% confidence interval: 0.7690–0.9543, P<0.0001) reaching the 86% sensitivity and 75% specificity for PIWIL1, and 0.9975 (95% confidence interval: 0.7908–0.9842, P<0.0001) reaching the 89% sensitivity and 78% specificity for piR-823, respectively (Figure 1C and D). Expression levels of piR-823 were significantly correlated with PIWIL1 expression levels (r=0.4242, P=0.0065; Figure 1E) supporting its involvement in piR-823 biogenesis. We have observed no significant differences in the expression levels of PIWIL2, PIWIL3, and PIWIL4 between tumors and paired renal parenchyma (Table 1). Further, we evaluated correlations of PIWIL genes and piR-823 with clinicopathological features and survival in patients with RCC (summarized in Table 1). We observed progressive decrease in PIWIL1 (P=0.0007, Figure 2A), PIWIL2 (P=0.0174, Figure 2B) and PIWIL4 (P=0.00403, Figure 2C) expression levels together with increasing clinical stage. PIWIL1 (P=0.0213, Figure 2D), PIWIL2 (P=0.0033, Figure 2E) and PIWIL4 (P<0.0001, Figure 2F) expression progressively decreased also with increasing Fuhrman grade. Most importantly, low-expression levels of PIWIL1 (P=0.009, Figure 3A), PIWIL2 (P<0.0001, Figure 3B) and PIWIL4 (P=0.0065, Figure 3C) were significantly associated with worse overall survival in patients with RCC. PIWIL3 and piR-823 were not correlated with any evaluated clinicopathological feature of RCC.
  | Figure 3 Expression of PIWIL1, PIWIL2, and PIWIL4 gene is associated with overall survival of renal cell carcinoma patients (A–C). |
Discussion
In our study, we quantified the expression levels of PIWIL1, PIWIL2, PIWIL3, and PIWIL4 genes and piR-823 in paired samples of RCC tumor tissue and non-tumor renal parenchyma. Significant differences were observed only in the case of PIWIL1 and piR-823, whose levels were significantly decreased in tumor tissue. Accordingly, decreased levels of piR-823 were also observed in gastric cancer.8 The strong correlation between PIWIL1 and piR-823 expression levels observed in our study suggest involvement of PIWIL1 in piR-823 biogenesis. In agreement with our results, Al-Janabi et al described the trend of PIWIL1 expression to be decreased in RCC tumors (P=0.089).14 In addition, they observed a significantly higher level of PIWIL4 in tumors (P<0.001), which was not shown in tumor tissues in our cohort of patients with RCC. Analogically, the expression of PIWIL genes is downregulated in seminoma and non-seminoma testicular tumors, probably due to the existence of promoter CpG island hypermethylation-associated silencing.15 Contrary to this observation and our results, upregulation of PIWIL1 (HIWI) in tumor tissue was shown in glioma,16 cervical cancer,17 breast cancer,18 and non-small cell lung cancer, while PIWIL2 and PIWIL4 genes were downregulated.19
Further, we observed expression levels of PIWIL1, PIWIL2, and PIWIL4 to be progressively decreased with increasing clinical stage, and Fuhrman grade and lower levels of those genes to be significantly associated with the worse overall survival in patients with RCC. In agreement with our observations, Taubert et al described decreased levels of PIWIL2 protein to be significantly associated with poor prognosis in cohort of 202 bladder cancer patients (P=0.005).20 A higher level of PIWIL4 mRNA was found to be associated with longer overall survival also in non-small cell lung cancer patients.19 In patients with soft tissue sarcoma, Greither et al identified significant association between the low-expression levels of PIWIL2, PIWIL3, and PIWIL4 mRNAs and shorter tumor-specific survival.21 Contrary to these observations and also our results, the increased expression levels of PIWIL1 were correlated with tumor grade in gliomas22 and associated with worse survival in colorectal cancer,23 hepatocellular carcinoma,24 and gastric cancer patients.25 In gastric cancer it was also reported that PIWIL2 expression levels are associated with worse overall survival.25
To conclude, we observed deregulation of PIWIL1 and piR-823 in tumor tissue of patients with RCC and significant association of PIWIL1, PIWIL2, and PIWIL4 expression with the survival of patients with RCC. It seems that PIWIL mRNA levels are commonly deregulated in tumor tissue and correlated with clincopathological features of tumors, however, the tendency of changes varies according to the tissue of origin. In urological tumors (RCC and bladder cancer), the expression of PIWIL genes is more likely to be downregulated and the loss of their expression is associated with a more aggressive phenotype of tumors and worse survival.
Acknowledgments
The project was supported by MZCR NT/13547-4/2012, AZV NV15-34678A, CEITEC Central European Institute of Technology (CZ.1.05/1.1.00/02.0068), the Project MZCR RVO (MOU, 00209805), and The Ministry of Education, Youth and Sports for the Project BBMRI CZ (LM2010004).
Disclosure
The authors report no conflicts of interest in this work.
References
Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. World Health Organization Classification of Tumours. Lyon: IARC Press; 2004:7. | ||
Horwich MD, Li C, Matranga C, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. | ||
Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. | ||
Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–345. | ||
Zhang P, Si X, Skogerbø G, et al. piRBase: a web resource assisting piRNA functional study. Database (Oxford). 2014;2014:bau110. | ||
Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. | ||
Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356:561–567. | ||
Cheng J, Deng H, Xiao B, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. | ||
Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. | ||
Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323–330. | ||
Gunawardane LS, Saito K, Nishida KM, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. | ||
Watanabe T, Tomizawa S, Mitsuya K, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. | ||
Tan Y, Liu L, Liao M, et al. Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin (Shanghai). 2015;47(5):315–324. | ||
Al-Janabi O, Wach S, Nolte E, et al. Piwi-like 1 and 4 gene transcript levels are associated with clinicopathological parameters in renal cell carcinomas. Biochim Biophys Acta. 2014;1842(5):686–690. | ||
Ferreira HJ, Heyn H, Garcia del Muro X, et al. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics. 2014;9(1):113–118. | ||
Wang X, Tong X, Gao H, et al. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45(6):2385–2392. | ||
Liu WK, Jiang XY, Zhang ZX. Expression of PSCA, PIWIL1 and TBX2 and its correlation with HPV16 infection in formalin-fixed, paraffin-embedded cervical squamous cell carcinoma specimens. Arch Virol. 2010;155(5):657–663. | ||
Wang DW, Wang ZH, Wang LL, Song Y, Zhang GZ. Overexpression of hiwi promotes growth of human breast cancer cells. Asian Pac J Cancer Prev. 2014;15(18):7553–7558. | ||
Navarro A, Tejero R, Viñolas N, et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget. 2015;6:31544–31556. | ||
Taubert H, Wach S, Jung R, et al. Piwil 2 expression is correlated with disease-specific and progression-free survival of chemotherapy-treated bladder cancer patients. Mol Med. 2015;21:371–380. | ||
Greither T, Koser F, Kappler M, et al. Expression of human Piwi-like genes is associated with prognosis for soft tissue sarcoma patients. BMC Cancer. 2012;12:272. | ||
Sun G, Wang Y, Sun L, et al. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–188. | ||
Zeng Y, Qu LK, Meng L, et al. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chin Med J (Engl). 2011;124(14):2144–2149. | ||
Zhao YM, Zhou JM, Wang LR, et al. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer. 2012;118(10):2708–2717. | ||
Wang Y, Liu Y, Shen X, et al. The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol. 2012;5(4):315–325. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.