Back to Journals » OncoTargets and Therapy » Volume 11
D-limonene exhibits antitumor activity by inducing autophagy and apoptosis in lung cancer
Authors Yu X, Lin H, Wang Y, Lv W, Zhang S, Qian Y, Deng X, Feng N, Yu H , Qian B
Received 1 November 2017
Accepted for publication 6 February 2018
Published 4 April 2018 Volume 2018:11 Pages 1833—1847
DOI https://doi.org/10.2147/OTT.S155716
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ingrid Espinoza
Xiao Yu,1,* Hongyan Lin,1,* Yu Wang,1 Wenwen Lv,1 Shuo Zhang,1 Ying Qian,1 Xiaobei Deng,1 Nannan Feng,1 Herbert Yu,2 Biyun Qian1
1Hongqiao International Institute of Medicine, Shanghai Tongren Hospital & Faculty of Public Health, Shanghai Jiao Tong University School of Medicine, Shanghai, China; 2Cancer Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI, USA
*These authors contributed equally to this work
Purpose: D-limonene is a plant extract with widespread application, and it has been recently reported to have antiproliferative and proapoptotic effects on cancer cells. However, the mechanisms by which D-limonene achieves these effects, especially in lung cancer, are not entirely clear. Therefore, the goal of this study was to examine the effects of D-limonene on lung cancer and explore its mechanisms of action.
Methods: We examined the therapeutic effects of D-limonene on lung cancer cells and in a xenograft animal model by characterizing its effects on the pathways of apoptosis and autophagy. Cell proliferation was measured using the Cell Counting Kit-8, and apoptosis was determined by flow cytometric analysis. Levels of LC3 puncta, an autophagy marker, were analyzed by laser scanning confocal microscopy. Autophagy and apoptosis-related gene expression were assessed by real-time quantitative polymerase chain reaction and Western blot.
Results: D-limonene inhibited the growth of lung cancer cells and suppressed the growth of transplanted tumors in nude mice. Expression of apoptosis and autophagy-related genes were increased in tumors after treatment with D-limonene. Furthermore, the use of chloroquine, an autophagy inhibitor, and knockdown of the atg5 gene, suppressed the apoptosis induced by D-limonene.
Conclusion: D-limonene may have a therapeutic effect on lung cancer as it can induce apoptosis of lung cancer cells by promoting autophagy.
Keywords: D-limonene, lung cancer, apoptosis, autophagy
Introduction
Lung cancer is the most common cancer in the world. According to the World Health Organization, there are approximately 1.8 million new cases and 1.6 million deaths worldwide from lung cancer each year.1 Surgical resection, postoperative radiation, and chemotherapy are the standard treatments for lung cancer; however, the efficacies of these treatments are poor, and only 30%–40% of patients with non–small cell lung cancer are responsive to chemotherapies, with a limited improvement in survival time.2–4 Thus, developing more effective drugs or agents for the prevention and treatment of lung cancer is a research priority.
Studies have shown that some plant or vegetable extracts, such as isothiocyanates5 and baicalin,6 may possess chemopreventive or anticancer properties. A natural product found in essential oils of citrus fruits, D-limonene (1-methyl-4-isopropyl-cyclohexene; structure shown in Figure S1) has been reported to have chemopreventive and anticancer activities. Multiple in vitro studies have reported that D-limonene can induce tumor cell apoptosis and has cytotoxic effects on malignant tumor cells.7–9 In addition, animal experiments show that D-limonene has therapeutic effects on breast,10 liver,11 kidney,12 and skin cancers.13 Moreover, a Phase I clinical trial reported that D-limonene could be well-tolerated by cancer patients.14 Despite the mounting evidence, mechanisms underpinning the antitumor property of D-limonene remain unclear. Several possible actions have been proposed, including cell-cycle arrest,15 induction of apoptosis,16 and suppression of the PI3K/Akt/mTOR17 and NF-κB pathways.18 However, it is unknown whether D-limonene has these effects on lung cancer and, if yes, what mechanisms mediate these effects.
Autophagy is a eukaryotic cellular degradation pathway in which bulk intracellular components are sequestered and targeted for lysosomal degradation to maintain cell homeostasis.19 In recent years, accumulating evidence has suggested that dysregulated autophagy is linked to cancer.20 Furthermore, extensive studies have suggested that induction of autophagy suppresses tumorigenesis via various pathways, including inducing apoptosis of cancer cells.21–23 Because D-limonene has been reported to induce apoptosis in many types of tumor cells and increase autophagy in neuroblastoma cells,15,17,24 it is important to explore the role of D-limonene in autophagy and apoptosis in lung cancer.
The goal of this study was, therefore, to examine the effects of D-limonene on lung cancer and explore its mechanisms of action. To accomplish these goals, we established a xenograft model in nude mice that were treated with D-limonene. Moreover, we investigated the effects of D-limonene on apoptosis and the autophagy signaling pathway in animal models and lung cancer cell lines.
Materials and methods
Materials
D-limonene was purchased from Sigma-Aldrich (St Louis, MO, USA). Antibody against β-actin (A1978) was purchased from Sigma-Aldrich, and antibodies against BAX (32503), cleaved PARP (32064), LC3B (ab48394), and Atg5 (ab108327) were purchased from Abcam (Cambridge, UK). Small-interfering RNA (siRNA) against atg5 and the mRFP-GFP-LC3 plasmid were obtained from GenePharma (Shanghai, China). The BD FITC Annexin V Apoptosis Detection Kit I was obtained from BD Biosciences (San Jose, CA, USA). The Cell Counting Kit-8 was purchased from Dojindo Laboratories (Kumamoto, Japan). Matrigel® was purchased from Corning (Coring, NY, USA). BALB/c mice were obtained from Shanghai SLAC Laboratory Animal Co. (Shanghai, China). Rhodamine 123 was purchased from Beyotime (Shanghai, China).
Cell lines and cell culture
The A549 and H1299 cell lines were obtained from the American Type Culture Collection (Manassas, VA). All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 25 U/mL penicillin, and 25 μg/mL streptomycin at 37°C in a 5% CO2 atmosphere.
D-limonene treatment
D-limonene was diluted with dimethyl sulfoxide at a ratio of 1:19 and then dissolved in the cell culture medium. Final concentrations of 0.5 and 0.75 mM D-limonene were used for cell treatment. The solution was freshly prepared before each experiment.
In vitro cell growth assays
Cells were plated in 96-well plates at a density of 2,000 cells per well. After overnight growth, the medium was removed and 100 μL fresh medium with different doses of D-limonene were added. After incubation, the medium was removed and new fresh medium with the Cell Counting Kit-8 reagent was added to the wells. After 2 hours of incubation, fluorescence was measured using a spectrophotometer (Thermo Fisher, Waltham, MA, USA).
Colony-formation assay
Five hundred cells were seeded in 10-cm dishes. After overnight growth, the culture medium was removed and fresh media with different treatment agents were added. After 10 days, the culture media were removed, and 2 mL of 4% paraformaldehyde was added to each dish to fix the cells. After washing twice with PBS, Crystal Violet was added and the cells were incubated for 20 minutes; thereafter, the cells were washed with water and photographed.
Cell apoptosis assay
Cells (1 × 105) were seeded into 24-well plates, followed by overnight incubation and 24-hour treatment. Cells were then digested with 0.25% trypsin and collected. The BD FITC Annexin V Apoptosis Detection Kit I was used to determine cell apoptosis, according to the manufacturer’s instructions.
Detection of mitochondrial membrane potential
Cells (1 × 105) were seeded into 24-well plates, followed by overnight incubation and 24-hour treatment. Cells were then digested with 0.25% trypsin and collected. After washing twice with PBS, the cells were incubated with 2 μM Rhodamine 123 for 15 minutes and subjected to flow cytometry.
Experiments on xenograft animals
Female BALB/c mice (5 weeks old) were kept in a specific-pathogen-free grade environment. All animal experiments were approved by the Animal Care and Use Committee of Shanghai Jiaotong University School of Medicine (Shanghai, China). All applicable guidelines of the Animal Care and Use Committee of Shanghai Jiaotong University School of Medicine for the care and use of animals were followed. A549 cells were collected and resuspended in PBS at a concentration of 1 × 108 cells/mL and mixed with Matrigel® at the ratio of 1:1. The mixture (0.1 mL) was subcutaneously injected into the hind flank regions of the mice. After 2 weeks, the mice were randomly divided into three groups, with eight mice in each group. Mice in different groups were treated with D-limonene diluted in milk (Mengniu, China) by gavage at a concentration of 400 or 600 mg/kg once a day. The control group was treated with milk only. Tumor size was measured once every 4 days using a Vernier caliper across its two perpendicular diameters, and tumor volume was calculated using the following formula: V = 1/2*a*b2; where V is the tumor volume, a is the largest diameter, and b is the smallest diameter. After 4 weeks of treatment, all mice were sacrificed and their tumors were collected and weighed. Histological evaluation of the tumor samples was undertaken. RNA and protein were extracted from tumors.
Histopathological analyses
Tumor tissues from animals were fixed in 4% paraformaldehyde (BOSTER, Wuhan, China) for 48 hours at room temperature. The fixed tissues were then dehydrated in a graded series of alcohol, cleaned in xylene, and embedded in paraffin. A rotary microtome was used to section paraffin blocks into 4-μm thicknesses. The sections were deparaffinized and stained with hematoxylin and eosin (H&E). A light microscope (Olympus, Tokyo, Japan) was used to examine the stained sections.
Plasmid and siRNA transfections
Cells (3 × 105) were seeded into six-well plates. The cells were transfected with plasmid and siRNA after overnight incubation at 37°C. Plasmid and siRNA transfections were undertaken according to the manufacturer’s protocol for the Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). The sequences of siRNA are listed in Table S1.
LC3 puncta analyses
Cells were transfected with an monomeric Red Fluorescent Protein-Green Fluorescent Protein-LC3 (mRFP-GFP-LC3) plasmid, and the images were captured by a fluorescence confocal microscope (Carl Zeiss, Dresden, Germany), using 63× oil objectives. GFP+/RFP+ LC3 puncta were yellow, and GFP-/RFP+ LC3 puncta were red. The average numbers of LC3 puncta per cell were determined on the basis of the data collected from >10 cells. The experiments were repeated three times.
RNA extraction and quantitative polymerase chain reaction
Total RNA was extracted from cells and tissues using the TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. RNA (2 μg) was reverse-transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Apoptosis- and autophagy-related gene expressions were measured by qPCR using the Roche LightCycler® 480 Software and Power SYBR® Green PCR Master Mix (Applied Biosystems) under the following conditions: denaturation at 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. Gene expression levels were normalized to β-actin and calculated according the formula 2−ΔΔCT. The primer sequences are listed in Table S2.
Western blotting analysis
Cells were harvested after experiments. To collect cell lysates, a cell lysis buffer with a protease inhibitor was added to dissociate the cells; thereafter, the supernatant was collected after centrifugation at 12,000× g for 10 minutes at 4°C. The protein samples were added to loading buffer and boiled at 100°C for 5 minutes. Depending on the size of the protein, different percentages of polyacrylamide gels were used for electrophoresis, followed by transferring the resolving proteins in the gel onto the polyvinylidene difluoride membranes. The transferred membrane was incubated in low-fat milk (5%) in PBS at room temperature for 1 hour to block nonspecific protein binding. The membranes were incubated overnight with primary antibodies at 4°C, and then were washed with PBS containing 0.1% Tween (PBST) three times (10 minutes per wash cycle). After that, a secondary antibody was added, and the mixture was incubated for 1 hour at room temperature. The membranes were washed again three times with PBST. ImageJ sofware (National Institutes of Health, Bethesda, MD, USA) was used to analyze the images obtained.
Immunohistochemical assay
Xenograft tumor tissue was fixed with 4% paraformaldehyde and cut into 4 μm slices. The slides were dried for 30 minutes at 65°C and soaked in xylene for 15 minutes, followed by treatment in sequence with anhydrous ethanol, 95% ethanol, 85% ethanol, and 75% ethanol for 5 minutes. After washing three times for 3 minutes with PBS (0.02 mol/L; pH = 7.4), the slides were incubated with 3% hydrogen peroxide for 10 minutes. Then, the slides were incubated with the antibody against cleaved PARP overnight at 4°C. After washing, the slides were incubated with goat anti-rabbit antibody in PBS for 30 minutes at room temperature, and stained with 3,3-diaminobenzidine tetrahydrochloride (DAB). Three views (200× magnification) were randomly chosen of each group. The positively stained area was used to analyze protein expression.
Statistical analysis
All data were analyzed using SPSS, version 19.0 (SPSS, Chicago, IL, USA), and the results are expressed as mean ± standard error. Two-tailed Student’s t-test and one-way analysis of variance were used for data analysis. P < 0.05 was considered indicative of statistical significance.
Results
Inhibition of tumor cell proliferation by D-limonene
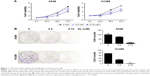
Cell growth in A549 and H1299 cells was inhibited in a dose- and time-dependent manner after treatment with D-limonene when compared to the control cells (Figure 1A). Figure 1B shows that 0.5 and 0.75 mM D-limonene suppressed the colony formation of A549 and H1299 cells in a dose-dependent manner. Moreover, D-limonene inhibits the proliferation and clone formation of the other three lung cancer cell lines H1975, H520, and PC9 (Figure S2). Taken together, these data indicate that D-limonene inhibits the proliferation of lung cancer cells.
Antitumor effect of D-limonene in xenograft animals
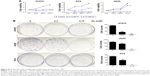
After 4 weeks of D-limonene treatment, tumor sizes decreased in two treatment groups (both, P < 0.05) when compared to the control group (Figure 2A). A dose–response relationship was observed on comparing tumor size among the three groups: 1,251.99, 867.44, and 577.08 mm3 in the control, low-dose, and high-dose groups, respectively (Figure 2A). Pictures of the removed tumors verified these measurements (Figure 2B). Furthermore, tumor weight was significantly different among the three groups: 0.77, 0.48, and 0.31 g in the control, low-dose, and high-dose groups, respectively (Figure 2C). The extent of tumor growth inhibition in the treatment groups was calculated according to the tumor weight. The rate of inhibition in the low-dose group was 37.08% and reached as high as 60.03% in the high-dose group (Figure 2D). H&E staining showed that tumors in the treatment groups possessed less moderately differentiated cancer cells (Figure 2E). Together, these results indicate that D-limonene inhibits the growth of lung cancer in vivo.
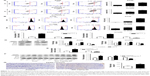
Promotion of apoptosis by D-limonene in lung cancer
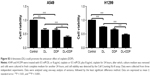
The cell apoptosis assay showed that D-limonene induced cell apoptosis in A549 and H1299 cells (Figure 3A). The mitochondrial membrane potential declined in lung cancer cells treated with D-limonene (Figure 3B). Figure 3C and D show that the expression of Bax mRNA and protein were elevated in D-limonene–treated A549 and H1299 cells, and there was increased protein expression of cleaved PARP. However, the expressions of Bcl2 mRNA and protein decreased after D-limonene treatment. To verify the results of cell experiments, we analyzed the levels of apoptosis-related genes in xenograft tumors and found that the changes were consistent with the in vitro results (Figure 3E–G). These results show that D-limonene affects the processes of apoptosis.
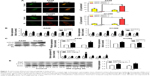
Promotion of autophagy by D-limonene in lung cancer
Redressing the autophagic activity is an emerging concept in apoptosis; we investigated the status of autophagy in non–small cell lung cancer cells. Figure 4A shows that the puncta in D-limonene–treated cells increased and tended to be GFP-/RFP+ (red). Furthermore, we quantified the expression of autophagy-associated genes. Figure 4B shows that lc3b, beclin1, atg3, atg5, atg7, atg12, and atg14 expression were all increased after treatment with D-limonene. Moreover, the expression of the autophagy-related proteins – LC3-II and Atg5 – were affected by D-limonene (Figure 4C). The same trend of changes was detected in xenograft tumors (Figure 4D and E). Together, these results show that D-limonene affects the processes of autophagy.
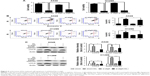
Role of autophagy in mediating the effects of D-limonene on cell growth and apoptosis
Chloroquine and si-atg5 alone had no significant effect on cell proliferation; however, the growth-inhibitory effect of D-limonene was significantly reduced after chloroquine or si-atg5 treatment of cells (Figures 5A and 6A), suggesting that autophagy may play an important role in D-limonene–induced inhibition of cancer cell growth. Compared to the controls, cells treated with chloroquine or si-atg5 showed no change in the percentage of apoptosis. However, apoptosis after D-limonene treatment combined with chloroquine or si-atg5 was significantly lower than that of cells treated with D-limonene alone (Figures 5B and 6B), indicating that autophagy played a role in D-limonene–induced apoptosis. Western blotting analyses were then conducted to show the expression of cleaved PARP and LC3-II. Because chloroquine blocked the binding of autophagosomes and lysosomes, LC3-II expression was increased as autophagy was inhibited. Whereas Atg5 participates in the formation of autophagosomes, treatment with si-atg5 resulted in a reduced level of LC3-II. However, in both of the autophagy-inhibited treatments, cleaved PARP levels decreased when cells were treated with D-limonene combined with chloroquine or si-atg5, as compared to those treated with D-limonene alone (Figures 5C and 6C). Taken together, these results indicate that autophagy positively regulates D-limonene–induced apoptosis.
Discussion
Our experiments showed that D-limonene could inhibit lung cancer growth, both in vivo and in vitro. In addition, we explored the possible mechanism involved by showing that D-limonene could induce apoptosis through promotion of autophagy.
Previous reports from in vivo and in vitro studies stated that D-limonene was involved in the induction of apoptosis.15,17,25,26 Ji et al found increased expression of Bax in human leukemia cells after limonene treatment.16 Moreover, our study showed an increase in Bax and cleaved PARP during the treatment, suggesting that the mitochondria-mediated intrinsic death pathway may play an important role in D-limonene–induced death of lung cancer cells, which is consistent with its role in the treatment of other cancers.
In cancer cells, autophagy results in both cell survival and cell death, and the role of autophagy in cancer therapy has drawn increasing attention.27 Many commonly used anticancer agents such as arsenic trioxide,28 tamoxifen,29 and imatinib30 have been reported to modulate autophagy in tumor cells. Currently, a promising strategy in the search for anticancer drugs is to target the autophagic pathway. For example, rapamycin, one of the best-studied autophagy inducers, has been used for the clinical treatment of multiple cancers, including lung cancer.31 Better anticancer effects were found when classic anticancer drugs such as cisplatin were combined with rapamycin.32–35 D-limonene has been reported to induce autophagy in neuroblastoma cells,24 which is consistent with our findings in lung cancer cells. In addition, we treated cells with a combination of D-limonene and cisplatin and found that lung cancer cells showed a slower level of proliferation than when treated with D-limonene and cisplatin alone (Figure S3). We, therefore, speculate that D-limonene may have the potential to be used in combination with classic anticancer drugs in treating lung cancer.
The interplay of autophagy and apoptosis in cancer cells is complicated. Autophagy may have different effects on apoptosis, depending on the surrounding microenvironment, cancer stage, and therapeutic intervention.36 Autophagy may be conducive to maintaining cell homeostasis, so that cancer cells do not undergo apoptosis.37 For example, autophagy induced by the ruthenium (II) imidazole complex may inhibit apoptosis in lung cancer cells.38 However, many studies have reported that autophagy can promote apoptosis. In the breast cancer cell line MCF-7, autophagy preceded apoptosis when the cells were treated with oridonin.39 Liu et al reported that autophagy-mediated apoptosis was induced by parthenolide.40 Our study showed that chloroquine – an autophagy inhibitor – inhibited the fusion of autophagosomes and lysosomes, and decreased cell apoptosis. The same trend was also observed with the blockage of autophagy by si-atg5. Atg5 is involved in the formation of autophagosomes. Overall, our results strongly support the possibility that D-limonene may increase autophagy, which further promotes apoptosis. In our experiments, we found significant increases in Atg5 after D-limonene treatment. Because the N-terminal cleavage product of Atg5 binds to Bcl-X1, leading to apoptosis,41,42 we speculated that the upregulation of Atg5 may be partially involved in D-limonene–induced apoptosis. However, the precise mechanisms underlying the role of autophagy in D-limonene–induced apoptosis need to be more fully investigated in future studies.
Conclusion
In summary, we evaluated the anticancer effect of D-limonene on lung cancer in vitro and in vivo. Our experiments showed that D-limonene could suppress lung cancer growth and induce apoptosis via a mechanism involving autophagy.
Acknowledgments
The authors would like to thank Rongna Ma and Sumeng Gao for their technical help and Xue Li for her writing assistance. This work was supported by the National Natural Science Foundation of China (grant no 81573231), Three-year Action Plan on Public Health, Phase IV, Shanghai, China (grant no 15GWZK0801), and the Scientific Project of Shanghai Health and Planning Commission (grant no 20154Y0197).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Field JK, Oudkerk M, Pedersen JH, Duffy SW. Prospects for population screening and diagnosis of lung cancer. Lancet. 2013;382(9893):732–741. | ||
Huber RM, Reck M, Thomas M. Current status of and future strategies for multimodality treatment of unresectable stage III nonsmall cell lung cancer. Eur Respir J. 2013;42(4):1119–1133. | ||
Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–731. | ||
Abbaoui B, Riedl KM, Ralston RA, et al. Inhibition of bladder cancer by broccoli isothiocyanates sulforaphane and erucin: characterization, metabolism, and interconversion. Mol Nutr Food Res. 2012;56(11):1675–1687. | ||
Yu Y, Pei M, Li L. Baicalin induces apoptosis in hepatic cancer cells in vitro and suppresses tumor growth in vivo. Int J Clin Exp Med. 2015;8(6):8958–8967. eCollection 2015. | ||
Afshordel S, Kern B, Clasohm J, et al. Lovastatin and perillyl alcohol inhibit glioma cell invasion, migration, and proliferation – impact of Ras-/Rho-prenylation. Pharmacol Res. 2015;91:69–77. | ||
Bai J, Zheng Y, Wang G, Liu P. Protective Effect of D-Limonene against oxidative stress-induced cell damage in human lens epithelial cells via the p38 pathway. Oxid Med Cell Longev. 2016;2016:5962832. | ||
Ma Y, Bian J, Zhang F. Inhibition of perillyl alcohol on cell invasion and migration depends on the Notch signaling pathway in hepatoma cells. Mol Cell Biochem. 2016;411(1–2):307–315. | ||
Chen TC, Cho HY, Wang W, et al. A novel temozolomide-perillyl alcohol conjugate exhibits superior activity against breast cancer cells in vitro and intracranial triple-negative tumor growth in vivo. Mol Cancer Ther. 2014;13(5):1181–1193. | ||
Victor Antony Santiago J, Jayachitra J, Shenbagam M, Nalini N. Dietary d-limonene alleviates insulin resistance and oxidative stress-induced liver injury in high-fat diet and L-NAME-treated rats. Eur J Nutr. 2012;51(1):57–68. | ||
Dietrich DR, Swenberg JA. The presence of alpha 2u-globulin is necessary for d-limonene promotion of male rat kidney tumors. Cancer Res. 1991;51(13):3512–3521. | ||
Chaudhary SC, Siddiqui MS, Athar M, Alam MS. D-Limonene modulates inflammation, oxidative stress and Ras-ERK pathway to inhibit murine skin tumorigenesis. Hum Exp Toxicol. 2012;31(8):798–811. | ||
Vigushin DM, Poon GK, Boddy A, et al. Phase I and pharmacokinetic study of D-limonene in patients with advanced cancer. Cancer Research Campaign Phase I/II Clinical Trials Committee. Cancer Chemother Pharmacol. 1998;42(2):111–117. | ||
Satomi Y, Ohara K, Yazaki K, Ito M, Honda G, Nishino H. Production of the monoterpene limonene and modulation of apoptosis-related proteins in embryonic-mouse NIH 3T3 fibroblast cells by introduction of the limonene synthase gene isolated from Japanese catnip (Schizonepeta tenuifolia). Biotechnol Appl Biochem. 2009;52(Pt 3):185–190. | ||
Ji J, Zhang L, Wu YY, Zhu XY, Lv SQ, Sun XZ. Induction of apoptosis by d-limonene is mediated by a caspase-dependent mitochondrial death pathway in human leukemia cells. Leuk Lymphoma. 2006;47(12):2617–2624. | ||
Jia SS, Xi GP, Zhang M, et al. Induction of apoptosis by D-limonene is mediated by inactivation of Akt in LS174T human colon cancer cells. Oncol Rep. 2013;29(1):349–354. | ||
Chi G, Wei M, Xie X, Soromou LW, Liu F, Zhao S. Suppression of MAPK and NF-kB pathways by limonene contributes to attenuation of lipopolysaccharide-induced inflammatory responses in acute lung injury. Inflammation. 2013;36(2):501–511. | ||
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. | ||
Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. | ||
Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59(2):505–517. | ||
Liu XD, Yao J, Tripathi DN, et al. Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene. 2015;34(19):2450–2460. | ||
Lozy F, Cai-McRae X, Teplova I, et al. ERBB2 overexpression suppresses stress-induced autophagy and renders ERBB2-induced mammary tumorigenesis independent of monoallelic Becn1 loss. Autophagy. 2014;10(4):662–676. | ||
Russo R, Cassiano MG, Ciociaro A, et al. Role of D-Limonene in autophagy induced by bergamot essential oil in SH-SY5Y neuroblastoma cells. PLoS One. 2014;9(11):e113682. | ||
Song X, Xie L, Wang X, et al. Temozolomide-perillyl alcohol conjugate induced reactive oxygen species accumulation contributes to its cytotoxicity against non-small cell lung cancer. Sci Rep. 2016;6:22762. | ||
Zhang XZ, Wang L, Liu DW, Tang GY, Zhang HY. Synergistic inhibitory effect of berberine and d-limonene on human gastric carcinoma cell line MGC803. J Med Food. 2014;17(9):955–962. | ||
Isakson P, Bjørås M, Bøe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116(13):2324–2331. | ||
Jiao G, Ren T, Guo W, Ren C, Yang K. Arsenic trioxide inhibits growth of human chondrosarcoma cells through G2/M arrest and apoptosis as well as autophagy. Tumour Biol. 2015;36(5):3969–3977. | ||
Li Y, Jiang B, Zhu H, et al. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour Biol. 2017;39(6):1010428317705790. | ||
Bilir A, Erguven M, Oktem G, et al. Potentiation of cytotoxicity by combination of imatinib and chlorimipramine in glioma. Int J Oncol. 2008;32(4):829–839. | ||
Chi KH, Ko HL, Yang KL, Lee CY, Chi MS, Kao SJ. Addition of rapamycin and hydroxychloroquine to metronomic chemotherapy as a second line treatment results in high salvage rates for refractory metastatic solid tumors: a pilot safety and effectiveness analysis in a small patient cohort. Oncotarget. 2015;6(18):16735–16745. | ||
Wong SW, Tiong KH, Kong WY, et al. Rapamycin synergizes cisplatin sensitivity in basal-like breast cancer cells through up-regulation of p73. Breast Cancer Res Treat. 2011;128(2):301–313. | ||
Han L, Wu JL, Yang LX. Effect of combination of rapamycin and cisplatin on human cervical carcinoma Hela cells. Contemp Oncol (Pozn). 2012;16(6):512–515. | ||
Lei WB, Jia T, Su ZZ, Wen WP, Zhu XL. [Effect of chemotherapy with cisplatin and rapamycin against Hep-2 cells in vitro]. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28(10):1838–1841. Chinese [with English abstract]. | ||
Liu J, Zhang L, Zhang X, Xing X. Rapamycin enhanced the antitumor efficacy of oxaliplatin in cisplatin-resistant ovarian cancer cells A2780cis both in vitro and in vivo. J Chemother. 2015;27(6):358–364. | ||
Liu G, Pei F, Yang F, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18(2). pii: E367. | ||
Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. | ||
Chen L, Li G, Peng F, et al. The induction of autophagy against mitochondria-mediated apoptosis in lung cancer cells by a ruthenium (II) imidazole complex. Oncotarget. 2016;7(49):80716–80734. | ||
Cui Q, Tashiro S, Onodera S, Minami M, Ikejima T. Autophagy preceded apoptosis in oridonin-treated human breast cancer MCF-7 cells. Biol Pharm Bull. 2007;30(5):859–864. | ||
Liu W, Wang X, Sun J, Yang Y, Li W, Song J. Parthenolide suppresses pancreatic cell growth by autophagy-mediated apoptosis. Onco Targets Ther. 2017;10:453–461. | ||
Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132. | ||
Aporta A, Catalán E, Galán-Malo P, et al. Granulysin induces apoptotic cell death and cleavage of the autophagy regulator Atg5 in human hematological tumors. Biochem Pharmacol. 2014;87(3):410–423. |
Supplementary materials
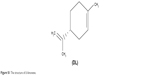  | Figure S1 The structure of D-limonene. |
  | Table S1 Sequences of siRNA |
  | Table S2 Sequences of primers |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.