Back to Journals » Journal of Inflammation Research » Volume 13
Cytokine Response in SARS-CoV-2 Infection in the Elderly
Authors Sherwani S , Khan MWA
Received 12 August 2020
Accepted for publication 29 September 2020
Published 20 October 2020 Volume 2020:13 Pages 737—747
DOI https://doi.org/10.2147/JIR.S276091
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Subuhi Sherwani,1 Mohd Wajid Ali Khan2
1Department of Biology, College of Sciences, University of Ha’il, Ha’il 2440, Saudi Arabia; 2Department of Chemistry, College of Sciences, University of Ha’il, Ha’il 2440, Saudi Arabia
Correspondence: Subuhi Sherwani
Department of Biology, College of Sciences, University of Ha’il, Ha’il 2440, Saudi Arabia
Tel +966-590172770
Email [email protected]
Abstract: The last few months of 2019 witnessed the emergence, rise and rapid spread of a novel coronavirus known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), causing an acute respiratory disease called coronavirus disease 2019 or Covid-19. Severe pathological manifestations of the disease in the infected population with comorbidities are linked to acute respiratory distress syndrome (ARDS), associated with an exaggerated synthesis and expression of cytokines, leading to a systemic inflammatory response also known as a cytokine storm (CS). Elderly patients (> 60 years of age) showed more deaths in Covid-19 infection. Age-related immune imbalance increases patient susceptibility to CS. In acute Covid-19 infection, it is difficult to minimize or control the overproduction of cytokines; hence, limited medical treatments are effective. This review aims to provide an overview of the current knowledge of involvement of cytokines in SARS-CoV-2 infection, susceptibility factors for the accompanying cytokine storm in severe Covid-19 cases and possible treatment strategies.
Keywords: SARS-CoV-2, Covid-19, coronavirus, cytokine storm, inflammation, elderly
Introduction
The emergence of the highly contagious Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in the latter half of 2019, followed by its rapid spread worldwide and its ability to cause severe infections in humans, often with fatal consequences,1 led the World Health Organization (WHO) to declare it as a global pandemic on 11 March 2020.2 By 25 September, the WHO report confirmed more than 32 million cases of infections and over 979,212 confirmed deaths worldwide.3
In the past few decades, three new Coronaviruses have posed a global threat to public health. Severe acute respiratory syndrome coronavirus (SARS-CoV) identified in 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) emergent in 2012, and the newly identified SARS-CoV-2 in the latter half of 2019, all share similarities of animal reservoirs, unpredictable emergence, rapid spread and serious infectious outcomes in humans, collectively leading to high rates of mortality in the human population.4
Out of all Covid-19 infected patients, significantly large numbers (80%) showed mild symptoms and only 20% of these infected individuals exhibited severe complications associated with comorbidities requiring hospitalization. Moreover, out of these 20%, 5% patients experienced critical symptoms associated with excess cytokine production such as hypoxia and ARDS, which lead to multiple organ failure and eventually death.5
Several factors such as easy mode of transmission of SARS-CoV-2, infectivity in the incubation period, differing clinical course ranging from asymptomatic forms to severe pathological manifestations of the disease in the infected population associated with comorbidities, lack of clear treatment options and delay in vaccine production, all point to an unprecedented health-care crisis.
One of the main reasons of death in SARS-CoV-2-infected patients is abnormal immune response, which include uncontrolled secretion of cytokines also referred to as CS. With the numbers of infected individuals needing hospitalization still high in most countries around the world, a greater understanding of the virus, its pathogenesis and host cytokine response is required.
Receptors for Coronaviruses
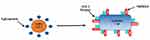
The host innate immune system detects viral infections through pattern recognition receptors (PRRs). Host PRRs detect viral components, pathogen-associated molecular patterns, which include genomic DNA, single-stranded (ss) RNA, double-stranded (ds) RNA, RNA with 5′-triphosphate ends and viral proteins. Currently, three classes of PRRs are known to be involved in the recognition of virus-specific components in innate immune cells, namely toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) and NOD-like receptors (NLRs). Among these receptor types, TLRs and RLRs are important for the production of type I interferons (IFNs) and various cytokines, whereas NLRs are known to regulate interleukin (IL)-1β maturation through activation of caspase-1.6,7 A study found that the SARS-CoV-2 isolated from human airway epithelial cells was closer to bat-SL-CoVZC45 and bat-SL-CoVZXC21 at the whole-genome level.8 Additionally, the external subdomain of the Covid-19 receptor-binding domain (RBD) bore greater similarity to severe SARS-CoV.9 Angiotensin-converting enzyme 2 (ACE2), a metallopeptidase, is a recognized host cell receptor of SARS-CoV-2 and plays a crucial role in virus and cellular membrane fusion via the spike protein, facilitating infection.10 This is also the receptor for SARS-CoV and HCoV-NL63.11,12 Zhou et al proved that the SARS-CoV-2 does not use other coronavirus receptors such as aminopeptidase N, and dipeptidyl peptidase-4.13 Another study found that the RBD domain of the SARS-CoV-2 and S-protein support strong interaction with human ACE2 molecules.14 High levels of ACE2 expression have been identified in type II alveolar cells of lung,15,16 upper and stratified epithelial cells of esophagus, absorptive enterocytes from ileum and colon,16 cholangiocytes,17 myocardial cells, kidney proximal tubule cells, and bladder urothelial cells.15 Since studies suggest that ACE2 plays a pivotal role in cellular entry; hence, SARS-CoV-2 may preferentially target ACE2-expressing cells leading to infection Figure 1. These findings indicate that organs with high ACE2-expressing cells may potentially be at a higher risk for Covid-19 infection.
Additionally, a transmembrane serine protease 2 (TMPRSS2) is another key protein involved in the SARS-CoV-2 cell entry process Figure 1. It is the main host cell serine protease, which cleaves the S protein of human CoVs on the cell membrane. This enables virus to release fusion peptide for membrane fusion. Both receptors are co-expressed in lung epithelial cells, the upper epithelial and gland cells of oesophagus and absorptive enterocytes from ileum and colon; suggesting an alternate mode of infection.18 The expression of ACE2 increases in individuals who have type I and II diabetes, as treatment options include ACE inhibitors and angiotensin-receptor blockers (ARBs). Additionally, hypertension, also treated with ACE inhibitors and ARBs, results in increased expression of ACE2.19 The increase in ACE2 receptors from hypertension medications, and diabetes medications is the basis of the initial hypotheses that suggest ACE2 upregulation may explain the correlation between these conditions. However, there is certainly not a consensus on these conclusions, with other evidence contradicting these claims to show that ACE2 protects patients from severe acute lung failure.20 The only known is that there is a correlation between severity of Covid-19 cases and patients with hypertension, diabetes, and obesity.
Cytokines in Viral Infections
Cytokines play an important role in establishing an antiviral state as the nonspecific first line of defense in viral infections. The network of cytokines in inflammatory diseases is a very complex system. These immune-modulatory peptides, produced by a broad range of cells such as B-lymphocytes, T lymphocytes, macrophages, mast cells, stromal cells and endothelial cells, etc., to regulate different cellular processes and interactions, such as cellular development, repair, hematopoiesis, inflammation, etc., through the transduction of signals mediated by binding to cellular receptors.21 In addition, cytokines have pleiotropic activity, i.e. they often have multiple functions, acting on different target cells to affect the function of other cytokines in an additive, synergistic or antagonistic manner.22 Therefore, cytokines are critical communication mediators for the immune system and are essential for host defense against pathogens. Intracellular signaling cascades are activated by the detection of viral components by immune cells with the resultant secretion of type I IFNs, proinflammatory cytokines and chemokines, as well as an increased expression of co-stimulatory molecules such as CD40, CD80 and CD86.6 Type I IFNs induce cell-intrinsic antimicrobial states in both virus infected and neighbouring cells to resolve viral infection, by activating intracellular signaling pathways via a type I IFN receptor.23 They also regulate gene expression of a set of genes, such as protein kinase R and 2′-5′-oligoadenylate synthase, which are involved in eliminating viral components from infected cells, inducing apoptosis of infected cells and conferring resistance against viral infection in uninfected cells.6 Type I IFNs are produced by immune cells such as dendritic cells (DCs), macrophages, NK cells as well as other cells like fibroblasts.6 Proinflammatory cytokines such as IL-1β, IL-6, and tumor necrosis factor (TNF)-α are predominantly secreted by monocytes and activated macrophages as well as by nonimmune cells, such as fibroblasts and endothelial cells. They are critical for pathological pain as well as eliminating virus infection by up-regulation of inflammatory reactions and recruiting innate and acquired immune cells.24 Aberrant induction and/or imbalance of various proinflammatory cytokines may induce severe systemic inflammatory response syndrome,25 resulting in systemic inflammation, organ dysfunction and organ failure. Co-stimulatory molecules are essential for natural killer (NK) cell activation. As NK cells are involved in the recognition of altered cells, they can directly destroy infected cells as well as promote recruitment and response of the other components of the immune system by the release of cytokines and chemokines.26 Similarly, co-stimulatory molecules are also involved in the activation of T cells, leading to acquired immune reactions. Failure to mount effective cytokine responses against viral agents can sometimes lead to systematic infection, while excessive cytokine production may lead to pathogenicity, severe symptoms or even fatality such as seen in SARS, MERS and SARS-CoV-2 infected patients. A fine balance of cytokine production and inhibition is required in countering viral infections, the tilting of which may result in severe pathogenesis.
Cytokines in Coronavirus
SARS-CoV infects airway and alveolar epithelial cells. Infection of hematopoietic cells such as DCs, monocyte-macrophages, and other peripheral blood mononuclear cell-derived cells by the virus is abortive. Infection of DCs by SARS-CoV induces low-level expression of antiviral cytokines IFN-αβ, moderate up-regulation of pro-inflammatory cytokines such as TNF and IL-6, and a significant up-regulation of inflammatory chemokines CCL3, CCL5, CCL2, and CXCL10.27,28 Similarly, SARS-CoV-infected macrophages and airway epithelial cells (AECs) show delayed and increased levels of IFN and other pro-inflammatory cytokines.28,29 The delayed but exaggerated production of these cytokines and chemokines induces a dysregulation of innate immune response. Elevated levels of pro-inflammatory cytokines (IFN-γ, IL-1, IL-6, IL-12, and TGFβ) and chemokines (CCL2, CXCL10, CXCL9, and IL-8) were found in the sera of SARS patients with severe disease compared to individuals with uncomplicated SARS.30,31 Whereas SARS patients with severe disease had very low levels of the anti-inflammatory cytokine, IL-10,31 individuals with lethal SARS showed increased levels of pro-inflammatory cytokines and chemokines such as IFN (IFN-α and IFN-γ) and IFN-stimulated genes (ISGs) (CXCL10 and CCL-2) compared to healthy controls or individuals with mild-moderate disease.32 These results gave early suggestions for a possible role for IFNs and ISGs in the immunopathogenesis of SARS in humans. Thus, dysregulated and/or exaggerated cytokine and chemokine responses by SARS-CoV-infected AECs, DCs, and macrophages play an important role in SARS pathogenesis.
Similar to SARS, MERS-CoV infection of human airway epithelial cells induces significant and delayed IFN and proinflammatory cytokine (IL-1β, IL-6, and IL-8) responses.33 MERS-CoV replicates in naïve and activated human monocyte-macrophages.34 This contrasts with SARS-CoV, which abortively infects monocyte-macrophages, DCs and T cells. MERS-CoV infection of human leukemic cell line (THP-1) cells, a monocyte cell line, and human peripheral blood monocyte-derived macrophages and DCs exhibited delayed but increased levels of pro-inflammatory cytokines and chemokines (CCL-2, CCL-3, CCL-5, IL-2, and IL-8).35 However, induction of IFN-α/β by monocyte, macrophages and DCs was not significant except for plasmacytoid dendritic cells, which produced large amounts of IFNs upon MERS-CoV infection.36 Studies showed elevated levels of serum pro-inflammatory cytokines (IL-6 and IFN-α) and chemokines (IL-8, CXCL-10, and CCL5) in individuals with severe MERS compared to those with mild to moderate MERS infection.37 Elevated serum cytokine levels in patients infected with MERS correlated with lung pathology due to high neutrophil and monocyte numbers in lungs and in the peripheral blood.37,38 In conclusion, SARS-CoV and MERS-CoV infect human airway epithelial cells, inducing proinflammatory cytokines.
Cytokines in SARS-CoV-2
SARS-CoV-2 shares phylogenetic similarities with two other betacoronaviruses, i.e. SARS-CoV and MERS-CoV as well as some clinical features of beta-coronavirus infections.39 SARS-CoV-2 and host cell interactions lead to the strong production of immune mediators. The secretion of large quantities of chemokines and cytokines such as IL-1, IL-6, IL-8, IL-21, TNF-b and MCP-1, in turn recruit lymphocytes and leukocytes to the site of infection. Infected individuals with SARS-CoV-2 can broadly be classified as asymptomatic or confirmed cases experiencing mild respiratory symptoms (cold-like symptoms to mild pneumonia), severe respiratory illness (serious pneumonia and shortness of breath) or fatal cases with multi-organ and systemic manifestations (sepsis, septic shock, and multiple organ dysfunction syndromes, hypoxia and blood clotting) often resulting in death. A study of the inflammatory cytokines and C-Reactive Protein (CRP) profiles of serum samples from a hospital in Wuhan, found higher values of cytokines such as IFN-γ, TNF-α, IL-3, IL-4, IL-6, IL-10 and CRP in patients with Covid-19 compared to healthy controls, suggesting an activation of immune response to SARS-CoV-2 infection.40 Also, only IL-6, IL-10 and CRP showed increased expression along with disease severity. Another study from China showed that about 20% of patients developed severe disease. Out of these, individuals with serious underlying health conditions were at a higher risk of death. A fatality of 4% was reported in a minority of patients presented with respiratory failure, septic shock and multi-organ dysfunction.41 All these studies ascertained increased level or proinflammatory cytokines are linked to SARS-CoV-2 related deaths.
Cytokine Storm in Covid-19
Historically coronaviruses exhibited relatively mild, self-limiting common cold-like phenotypes in humans. Four beta coronaviruses are, i.e. HCoV 229E, NL63, OC43, and HKU1, globally endemic and account for 10–30% of upper respiratory tract infections in adults. However, recently two highly pathogenic viruses, SARS-CoV and MERS-CoV, have caused global epidemics with resultant lung pathology and high rates of morbidity and mortality linked to excessive and aberrant non-effective host immune responses associated with a cytokine storm.42 Cytokine storm or cytokine storm syndrome (CSS), is an acute and systemic expression of a vigorous immune system, resulting in the excessive synthesis and release of more than 150 inflammatory cytokines such as IFN-γ, IFN-α, IFN-β, TNF-α, IL-1, IL-6, IL-8, IL-12 and MCP-1 among others.27 An exaggerated and often imbalanced secretion of both pro-inflammatory and anti-inflammatory cytokines, found to be elevated in the serum, leads to a positive feedback cycle between cytokines and immune cells, culminating in a fierce and often lethal interplay between them. The unifying clinical feature of CS is a massive inflammatory reaction, which progresses to cause acute respiratory distress syndrome, multiple organ dysfunction and eventually death.43 The clinical symptoms, which develop due to these dysregulated pathways, are fever, redness, swelling, tachycardia, tachypnea, hypotension, extreme fatigue and nausea. Elevated cytokine production has been associated with poor clinical outcome and pathogenesis during respiratory corona viral infections in humans.31,39 The emergence of SARS-CoV-2, the most severe beta coronavirus so far and the causative agent of an acute and highly transmissible respiratory infection, even in the asymptomatic incubation period, has led to a worldwide pandemic. Covid-19 infections are generally associated with upper respiratory tract infections, the signs and symptoms of which commonly include fever, headache, and cough with some patients exhibiting lower respiratory tract infections.44 A significant percentage of cases present with acute respiratory distress syndrome that requires mechanical ventilation support and a subgroup of patients with severe Covid-19 can experience the “cytokine storm syndrome”, characterized by a fulminant and fatal hyper-cytokinemia associated with multi-organ failure.45
In SARS-CoV-2 infection, antigen-presenting cells, such as macrophages and dendritic cells process and present viral antigens to T cells, leading to T cell activation and differentiation, including the production of cytokines associated with the different T cell subsets CD8, CD4 and γδ-T cells. Moreover, Th17 cells may play a role in host defense against the pulmonary virus infection by mediating the recruitment of neutrophils and macrophages to infected tissues, significantly determining the pathogenesis of associated inflammation.46 Hence, these cells cause a massive release of cytokines for immune response amplification, promote accumulation of immune cells at inflammatory sites, and enhance tissue pathologies and pulmonary edema.
A study by Huang et al reported the level of inflammatory factors in a cluster of patients in Wuhan with Covid-19. They measured cytokine and chemokine levels in 41 in-patients (including 13 ICU patients and 28 non-ICU patients) and found that ICU patients had higher plasma levels of IL2, IL7, IL10, G-CSF, IP10, MCP1, MIP1A, and TNFα compared to non-ICU patients.47 Notably, the difference of serum IL-6 levels between ICU and non-ICU patients was not significant. However, in another retrospective, multicentre cohort study, a significant elevation of IL-6 levels was detected in the non-survival group of patients with Covid-19, as compared with that of the survivals.48 Another study analyzed the cytokines in 123 patients and found serum IL-6 levels in severe group were significantly higher than that in mild group.49 Other reports also confirmed the elevation of IL-6 in critically ill patients with Covid-19.50 et al also showed high levels of IL-6, TNFα, IL-1β, IL-8, IL2R, together with persistently elevated levels of erythrocyte sedimentation rate and CRP, associated with ARDS, hypercoagulation and disseminated intravascular coagulation, manifested as thrombosis, thrombocytopenia, gangrene of extremities.41 Cytokine storm amplifies immune cell recruitment at the site of infection causing tissue damage and in severe conditions may lead to multiple organ dysfunction and eventually death.
Cytokine Storm in the Elderly
Since the initial outbreak of infection in Wuhan, China, Covid-19 has claimed thousands of lives and steadily spread to almost every country across the world. Elderly people exhibit a higher risk for Covid-19 infection leading to serious illness and/or death. Increased cytokine production is a major contributory factor to age-related prognosis or death.
Poor Prognosis in Elder Covid-19 Patients
A study in China found the case fatality ratio to be 1·38% (1·23–1·53%). However, substantially higher ratios were recorded in older age groups (0·32% [0·27–0·38], where individuals aged <60 years had a ratio of 6·4% [5·7–7·2%] and those aged ≥60 years), up to 13·4% (11·2–15·9%).51 At the start of the pandemic, Italy was one of the countries with the highest number of fatalities. According to data from the website, “Statista. Health & Pharmaceuticals-State of Health”,52 by 30th June 2020, the mortality rate in Italy appeared to be higher for elderly patients, especially above 80 years. As per the statistics, the mortality rate increased significantly with increasing age, i.e. 10.1% in patients aged 60–69 years, 26.4% in 70–79 years and 58.9% in patients more than 80 years old.52 The median age of Italian patients who tested positive for SARS-CoV-2 and died in the first phase of the pandemic was 82 years.53 Similarly, in the United States of America, poor patient prognosis can also be linked to increase in patient age with 26.4% mortality in 65–74 years, 26.1% in 75–84 years and 33.16% in patients more than 85 years old recorded till 13th July 2020.54 The mortality rate trends compared between Italy and the US differ slightly with respect to the fact that more deaths have been reported in the US population in a relatively younger population aged 45 and above. This rate is found to increase with age and peak in individuals aged 80 years or more.53–55 Recent studies indicate that the infection fatality ratio (IFR) is the strongest predictor of a Covid-19 patient’s risk of dying. A study in England found that the IFR for age groups 15–44 years was close to zero. For 65–74 years, the IFR was 3.1%, which then increased substantially to 11.6% for patients ≥75 years.56 For Spain, the IFR was close to zero for patients under 50 years. This increased to 3.4% for infected individuals aged 70–79 years and then rose sharply to 7.2% for those aged ≥80 years old.56 Elderly patients with SARS also exhibited a poor prognosis with mortality rates of 50% in patients over the age of 65 years.57
Cytokine Secretion by Immune Cells in the Elderly
In SARS patients with ARDS progressed to late phase fibrosis, pneumonia, systemic inflammation responses and multiple organ failure.58 The lungs of SARS patients with ARDS were also associated with inflammatory cytokine induction including IL-1, IL-6, IL-8, CXCL-10, and TNFα.59 On recognizing infiltration by such coronaviruses, the innate immune system, initiates a rapid anti-viral signaling cascade, leading to the cytokine storm syndrome in severe patients, often progressing to the multi-organ dysfunction and eventually death. The reason behind the inequivalent cytokine secretions in different age groups although not clear, is statistically significant in elderly patients. Our immune system generates antibodies against viral infections throughout our lifetime. Depending upon prior exposures, some of these antibodies may potentially cross-react with SARS-CoV-2 and help in neutralizing the infection. Following immunization, decreased antibody production as well as shortened duration of protective immunity are characteristic features in the elderly.60 Also, the decrease in humoral immune response with increase in age is both quantitative and qualitative, as characteristics such as affinity, specificity and class of antibody produced are affected. Studies report a progressive decline in both numbers and size of germinal centers of B cell follicles.61 However, the effects of age on antibody affinity maturation are often conflicting. Studies show intrinsic B cell defects in aged mice and humans. These defects include a decreases in Ig class switch recombination (CSR), activation-induced cytidine deaminase (AID) and E47 transcription factor. Variations in the effects on somatic hypermutation (SHM) also depends on the system studied. For example, increase in AID in mice resulted in improved CSR but not SHM. Microarray analysis of human B cell subsets are now being used to link B cell defects with aging and such studies may help to select agents for an improved immune response in the aged population.62
T follicular helper (Tfh) cells, which are a subset of CD4+ T cells, are essential in directing B cell activities such as proliferation, isotype switching and antibody production.63 Tfh cells produce specific cytokines in response to a viral infection and can be found within germinal centers (GC). Intrinsic age-related defects in Tfh function can lead to reduced B cell expansion, differentiation, IgG production and affinity maturation.64 Also, various age-related changes in the secondary lymphoid organs microenvironment impact cell repertoire and protective capacity. For example, CCL21 expression which reduces CD4 T cell priming and antibodies generation post vaccination, is significantly impacted in intact aged mice leading to a reduction in protective capacity.65 These factors contribute to reduced Tfh differentiation and ability to generate a robust response to vaccination in aged individuals. Hence, elderly individuals have reduced protection against subsequent infection.
Li et al provided a detailed clinical and epidemiological description of the first 425 cases reported in the epicenter of the SARS-CoV-2 outbreak, the city of Wuhan in Hubei province, China.66 The median age of patients was 59 years. Higher morbidity and mortality rates were reported among the elderly and those suffering from coexisting conditions. The majority (56%) of infected patients were male. Notably, no cases were reported in children aged 15 years or younger, either suggesting that children were less likely to be infected or their symptoms were mild and escaped detection.67 According to a study by Qiu et al, 36 children (median age 8·3 [SD 3·5] years) infected with Covid-19 were found to have severe acute respiratory syndrome. However, the majority of children remained asymptomatic, making the identification of infections and determination of prevalence difficult and facilitating community transmission.68
Natural killer (NK) cells are effector cells of the innate immune system important for control of viral infections. Physiological ageing is linked to changes in the composition, phenotype and function of circulating NK cells, also known as NK cell immunesenescence.69 Virus-infected cells encounter NK cells as the first line of defense. Immunogerontological studies suggest that NK cell immunesenescence contributes to the higher incidence of viral infection in older adults.70 NK cell activation results in production of inflammatory cytokines, such as IFN-γ and TNF-α by cytotoxic degranulation.71 These cytokines in addition to having antiviral effects, also play a role in the activation and/or maturation of DCs,72 macrophages, and T cells.71 Additionally, NK cells exhibit regulatory functions such as the regulation of immune responses in viral infections.73 They are also known to produce granulocyte-macrophage colony-stimulating factor,74 IL-5, IL-13,75 and IL-10,73 thereby effecting downstream immune responses. NK cells can also affect the production of chemokines, such as CCL3, CCL4, and CCL5, which recruit other effector cells to sites of inflammation.76
The regulatory T cells (Tregs) are a subpopulation of T cells, which facilitate tissue repair through multiple mechanisms limiting inflammatory damage to tissues. Tregs also play an important role in resolution of acute lung damage.77 Tregs can produce IL-10, an anti-inflammatory cytokine to suppress excess immune responses78 and thus serve a protective function in lung injury by reducing TNF-α production and neutrophil activity.79 TGF-β is essential for the development of Tregs whereas, IL-6 and IL-2 obstruct Treg polarization.80 Studies in humans indicated that Tregs from both young and elderly individuals similarly inhibited the proliferation of responder cells. However, the production of IL-10 drops in cells in the elderly.81 Thus, in contrast to children and young adults, elderly individuals exhibit decreased NK and Treg cell activity in addition to limited humoral activity and function. This leads to excessive release of pro-inflammatory cytokines such as IL-6 and TNF-α which further tilt the balance of proinflammatory cytokine release. Such hyperinflammatory changes induce a cytokine storm leading to loss of immune regulatory control, resulting in mass recruitment of immune cells to the site of tissue injury (Figure 2). This is also associated with comorbidities.
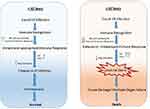 |
Figure 2 Comparison of immune responses in Covid-19 infection in individuals <40 years and >60 years old. |
Cytokine Storm Targeted Therapies
The inflammatory repertoire of Covid-19 remains unclear and hence, therapeutic options for prognosis and recovery of severe cases remain limited, with most treatment options focusing on supportive therapy. However, it is almost certain that anti-inflammatory therapies would play a preventative role in severe cases preventing exacerbation. Currently, a variety of anti-inflammatory agents such as steroids, glucocorticoids, chloroquine/hydroxychloroquine, immunosuppressive agents and inflammatory cytokines antagonists are being tested and trialed in severe patients with varying results.
Tocilizumab
A promising therapeutic strategy using inhibitory agents targeting key cytokines in Covid-19 patients is the use of tocilizumab, a recombinant humanised IL-6 blocking monoclonal antibody against the IL-6 receptor. Successful outcomes in some individuals were reported in a Chinese study using IL-6 blockade with anti-IL-6 antibody.82 A retrospective study in 20 patients with severe Covid-19, found that using tocilizumab, led to a reduction in fever and lung lesion opacity as well as recovery of lymphocytes in peripheral blood of patients.83 Another retrospective multicenter Italian cohort study of 544 adult patients with severe Covid-19 pneumonia found that administration of intravenous or subcutaneous tocilizumab prevented intubation or death in such patients.84 However, safety risks associated with the use of tocilizumab, especially with corticosteroids and increased risk of opportunistic infections need to be investigated further before more widespread use is possible.
Anakinra
A study of the use of the immunomodulatory agent Anakinra, a recombinant IL-1 receptor antagonist that blocks the activity of cytokines IL1α and IL1β, in Covid-19 patients with severe respiratory distress, found that the drug reduced need for invasive mechanical ventilation in ICU patients and reduced mortality in cases with severe complications, without serious side effects.85
Corticosteroids
Another therapeutic strategy used in severe cases of SARS-CoV-2, common with the treatment of other coronaviruses such as SARS and MERS, is the empirical administration of corticosteroids, which are known to inhibit and suppress inflammatory responses. A meta-analysis of the safety and efficacy of corticoid treatment in the treatment all three coronavirus infections found that corticosteroid use in subjects delayed virus clearing and did not convincingly improve survival, reduce hospitalization duration, ICU admission rate or use of mechanical ventilation.86 Another meta-analysis of 15 studies found that corticosteroid use to be associated with increased mortality in patients with coronavirus pneumonia.87 Currently, as per WHO guidelines, steroids are approved forms of therapy for Covid-19 treatment outside of clinical trials.
Chloroquine and Hydroxychloroquine
Traditionally known as anti-malarial drugs, chloroquine and hydroxychloroquine, are reported to have some anti-viral activity against viruses such as Ebola, dengue, H5N1 and SARS. These drugs have now been used in the treatment of Covid-19 in some countries.88 Both drugs accumulate in acidic organelles such as lysosomes, leading to an increase in endosomal pH, which interferes with viral replication and entry/exit from the infected host cell.89 Another, possible mechanism of action against SARS-CoV-2 is that both drugs reduce ACE2 receptor glycosylation, preventing the binding of virion particles with host cell.89,90 The anti-inflammatory effects of antimalarials are demonstrated by the fact that they are used to treat autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus. Chloroquine and hydroxychloroquine may block the events that lead to ARDS, such as blocking the production of IL-6 and other pro-inflammatory cytokines such as IL-1 and TNF-α involved in the cytokine storm.90 An early Chinese study of over 100 Chinese Covid-19 patients showed that chloroquine phosphate had good efficacy.91 Another study of a non-randomized clinical trial in France conducted on 20 Covid-19 patients who received hydroxychloroquine treatment reported good efficacy.92 However, more studies focusing on Covid-19 treatment with chloroquine and hydroxychloroquine are required to prove efficacy and beneficial effects.
Conclusion
Cytokine storm is one of the main factors associated with Covid-19 death in elderly individuals. Age-related immune imbalance with comorbidities leads to excessive release of pro-inflammatory cytokines. Resultant systemic hyper-inflammatory changes induce a CS leading to further loss of immune regulation, resulting in recruitment of excessive immune cells to the site of inflammation. Thus, identification of cytokine storm immunology and early-targeted therapies are important factors in patient prognosis and recovery in Covid-19.
Ethical Approval
Not required.
Funding
This paper was not supported by any grant or funding.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi:10.1056/NEJMoa2002032
2. World Health Organization, Director-General’s opening remarks at the media briefing on COVID-19 – 11 March 2020. World Health Organization. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020.
3. World Health Organization; WHO coronavirus disease (Covid-19) dashboard. Available from: https://covid19.who.int/.
4. Yang P, Wang X. COVID-19: a new challenge for human beings. Cell Mol Immunol. 2020;17:555–557. doi:10.1038/s41423-020-0407-x
5. Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J Heart Lung Transplant. 2020;39(5):405–407. doi:10.1016/j.healun.2020.03.012
6. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227(1):75–86. doi:10.1111/j.1600-065X.2008.00737.x
7. Petrilli V, Dostert C, Muruve DA, et al. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi:10.1016/j.coi.2007.09.002
8. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:727–733. doi:10.1056/NEJMoa2001017
9. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi:10.1016/S0140-6736(20)30251-8
10. Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eur Surveill. 2020;25(5):2000062. doi:10.2807/1560-7917.ES.2020.25.5.2000062
11. Li W, Moore M, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi:10.1038/nature02145
12. Hofmann H, Pyrc K, van der Hoek L, et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA. 2005;102:7988–7993. doi:10.1073/pnas.0409465102
13. Zhou P, Yang X, Wang X, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi:10.1038/s41586-020-2012-7
14. Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi:10.1007/s11427-020-1637-5
15. Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi:10.1007/s11684-020-0754-0
16. Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020. doi:10.1101/2020.01.30.927806
17. Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv. 2020. doi:10.1101/2020.02.03.931766
18. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi:10.1038/s41586-020-2179-y
19. Imai Y, Kuba K, Rao S, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi:10.1038/nature03712
20. Yu P, Zhu J, Zhang Z, et al. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J Infect Dis. 2020;221(11):1757–1761. doi:10.1093/infdis/jiaa077
21. Leonard WJ. Fundamental immunology. Chap. 1999;25:741–774.
22. Sblattero D, Berti I, Trevisiol C, et al. Human recombinant tissue transglutaminase ELISA: an innovative diagnostic assay for celiac disease. Am J Gastroenterol. 2000;95:1253–1257. doi:10.1111/j.1572-0241.2000.02018.x
23. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi:10.1038/nri3581
24. Zhang JM, An J. Cytokines, inflammation, and pain. Int Anesthesiol Clin. 2007;45(2):27–37. doi:10.1097/AIA.0b013e318034194e
25. Xu J, Zhang X, Pelayo R, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15:1318–1321. doi:10.1038/nm.2053
26. Young HA, Ortaldo J. Cytokines as critical co-stimulatory molecules in modulating the immune response of natural killer cells. Cell Res. 2006;16(1):20–24. doi:10.1038/sj.cr.7310004
27. Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79(12):7819–7826. doi:10.1128/JVI.79.12.7819-7826.2005
28. Law HK, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi:10.1182/blood-2004-10-4166
29. Yen YT, Liao F, Hsiao CH, et al. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J Virol. 2006;80(6):2684–2693. doi:10.1128/JVI.80.6.2684-2693.2006
30. Chien JY, Hsueh PR, Cheng WC, et al. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11(6):715–722. doi:10.1111/j.1440-1843.2006.00942.x
31. Zhang Y, Li J, Zhan Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72(8):4410–4415. doi:10.1128/IAI.72.8.4410-4415.2004
32. Cameron MJ, Bermejo-Martin JF, Danesh A, et al. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008;133(1):13–19. doi:10.1016/j.virusres.2007.02.014
33. Lau SKP, Lau CCY, Chan KH, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94(12):2679–2690. doi:10.1099/vir.0.055533-0
34. Chu H, Zhou J, Wong BHY, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2015;213(6):904–914. doi:10.1093/infdis/jiv380
35. Zhou J, Chu H, Li H, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209(9):1331–1342. doi:10.1093/infdis/jit504
36. Scheuplein VA, Seifried J, Malczyk AH, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(7):3859–3869. doi:10.1128/JVI.03607-14
37. Kim ES, Choe PG, Park WB, et al. Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. J Korean Med Sci. 2016;31(11):1717–1725. doi:10.3346/jkms.2016.31.11.1717
38. Ng DL, Hosani FA, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186(3):652–658. doi:10.1016/j.ajpath.2015.10.024
39. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539.
40. Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9(1):1123–1130. doi:10.1080/22221751.2020.1770129
41. Zhang W, Zhao Y, Zhang F, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the experience of clinical immunologists from China. Clin Immunol. 2020;214:108393. doi:10.1016/j.clim.2020.108393
42. Paules CI, Marston HD, Fauci AS. Coronavirus infections—more than just the common cold. JAMA. 2020;323(8):707–708. doi:10.1001/jama.2020.0757
43. Tisoncik JR, Korth MJ, Simmons CP, et al. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76(1):16–32. doi:10.1128/MMBR.05015-11
44. Coperchini F, Chiovato L, Croce L, et al. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi:10.1016/j.cytogfr.2020.05.003
45. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi:10.1002/jmv.25685
46. Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53(3):368–370. doi:10.1016/j.jmii.2020.03.005
47. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
48. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study [published correction appears in Lancet. 2020 Mar 28; 395(10229):1038][published correction appears in Lancet. 2020 Mar 28;395(10229):1038]. Lancet. 2020;395(10229):1054–1062.
49. Wan S, Yi Q, Fan S, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv. 2020. doi:10.1101/2020.02.10.20021832
50. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi:10.1016/S0140-6736(20)30211-7
51. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis [published correction appears in Lancet Infect Dis. 2020 Apr 15] [published correction appears in Lancet Infect Dis. 2020 May 4]. Lancet Infect Dis. 2020;20(6):669–677. doi:10.1016/S1473-3099(20)30243-7
52. Coronavirus (COVID-19) death rate in Italy as of June 30, 2020, by age group Statista. Health & Pharmaceuticals-State of Health. Available from: https://www.statista.com/statistics/1106372/coronavirus-death-rate-by-age-group-italy/.
53. Alicandro G, Remuzzi G, La Vecchia C. Italy’s first wave of the COVID-19 pandemic has ended: no excess mortality in May, 2020. Lancet. 2020;396:E27–E28. doi:10.1016/S0140-6736(20)31865-1
54. Provisional COVID-19 death counts by sex, age and state, provided by National Center for Health Statistics, Centre for Disease Control and Prevention. Available from: https://data.cdc.gov/NCHS/Provisional-COVID-19-Death-Counts-by-Sex-Age-and-S/9bhg-hcku.
55. Wortham JM, Lee JT, Althomsons S. Characteristics of persons who died with COVID-19 - United States, February 12-May 18, 2020, Centre for Disease Control and Prevention. Morb Mortal Wkly Rep. 2020;69(28):923–929. doi:10.15585/mmwr.mm6928e1
56. Mallapaty S. The coronavirus is most deadly if you are older and male – new data reveal the risks. Nature. 2020;585:16–17. doi:10.1038/d41586-020-02483-2
57. World Health Organization: SARS case fatality ratio, incubation period on World Wide Web URL. Available from: http://www.who.int/csr/sarsarchive/2003_05_07a/en/.
58. Tsushima K, King LS, Aggarwal NR, et al. Acute lung injury review. Int Med. 2009;48:621–630. doi:10.2169/internalmedicine.48.1741
59. Kong SL, Chui P, Lim B, et al. Elucidating the molecular physiopathology of acute respiratory distress syndrome in severe acute respiratory syndrome patients. Virus Res. 2009;145:260–269. doi:10.1016/j.virusres.2009.07.014
60. Saurwein-Teissl M, Lung TL, Marx F, et al. Lack of antibody production following immunization in old age: association with CD8CD28 T cell clonal expansions and an imbalance in the production of Th1 and Th2. J Immunol. 2002;168:5893–5899. doi:10.4049/jimmunol.168.11.5893
61. Zheng B, Han S, Takahashi Y, Kelsoe G. Immunosenescence and germinal center reaction. Immunol Rev. 1997;160:63–77. doi:10.1111/j.1600-065X.1997.tb01028.x
62. Frasca D, Blomberg BB. Effects of aging on B cell function. Curr Opin Immunol. 2009;21(4):425–430. doi:10.1016/j.coi.2009.06.001
63. Zhu J, Paul WE. CD4 T cells: fates, functions and faults. Blood. 2008;112:1557–1569. doi:10.1182/blood-2008-05-078154
64. Haynes L, Swain SL. Aged-related shifts in T cell homeostasis lead to intrinsic T cell defects. Semin Immunol. 2012;24(5):350–355. doi:10.1016/j.smim.2012.04.001
65. Swain SL, Kugler-Umana O, Kuang Y, et al. The properties of the unique age-associated B cell subset reveal a shift in strategy of immune response with age. Cell Immunol. 2017;321:52–60. doi:10.1016/j.cellimm.2017.05.009
66. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi:10.1056/NEJMoa2001316
67. Fauci AS, Lane HC, Redfield RR. Covid-19 - navigating the uncharted. N Engl J Med. 2020;382(13):1268–1269. doi:10.1056/NEJMe2002387
68. Qiu H, Wu J, Hong L, et al. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020;20(6):689–696. doi:10.1016/S1473-3099(20)30198-5
69. Solana R, Alonso MC, Pena J. Natural killer cells in healthy aging. Exp Gerontol. 1999;34:435–443. doi:10.1016/S0531-5565(99)00008-X
70. Hayhoe RP, Henson SM, Akbar AN, et al. Variation of human natural killer cell phenotypes with age: identification of a unique KLRG1-negative subset. Hum Immunol. 2010;71(7):676–681. doi:10.1016/j.humimm.2010.03.014
71. Vivier E, Tomasello E, Baratin M, et al. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510.
72. Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5(2):112–124. doi:10.1038/nri1549
73. Lee SH, Kim KS, Fodil-Cornu N, et al. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206(10):2235–2251. doi:10.1084/jem.20082387
74. Souza-Fonseca-Guimaraes F, Parlato M, de Oliveira RB, et al. Interferon-gamma and granulocyte/monocyte colony-stimulating factor production by natural killer cells involves different signaling pathways and the adaptor stimulator of interferon genes (STING). J Biol Chem. 2013;288(15):10715–10721. doi:10.1074/jbc.M112.435602
75. Hoshino T, Winkler-Pickett RT, Mason AT, et al. IL-13 production by NK cells: IL-13-producing NK and T cells are present in vivo in the absence of IFN-gamma. J Immunol. 1999;162(1):51–59.
76. Sawaki J, Tsutsui H, Hayashi N, et al. Type 1 cytokine/chemokine production by mouse NK cells following activation of their TLR/MyD88-mediated pathways. Int Immunol. 2007;19(3):311–320. doi:10.1093/intimm/dxl148
77. Lin S, Wu H, Wang C, et al. Regulatory T cells and acute lung injury: cytokines, uncontrolled inflammation, and therapeutic implications. Front Immunol. 2018;9:1545. doi:10.3389/fimmu.2018.01545
78. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi:10.1146/annurev-immunol-030409-101212
79. Inoue G. Effect of interleukin-10 (IL-10) on experimental LPS-induced acute lung injury. J Infect Chemother. 2000;6(1):51–60. doi:10.1007/s101560050050
80. Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol. 2010;40(7):1830–1835. doi:10.1002/eji.201040391
81. Fessler J, Ficjan A, Duftner C, et al. The impact of aging on regulatory T-cells. Front Immunol. 2013;4:231. doi:10.3389/fimmu.2013.00231
82. Xu X, Han M, Li T, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117(20):10970–10975. doi:10.1073/pnas.2005615117
83. Liu B, Li M, Zhou Z, et al. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020;111:102452.
84. Guaraldi G, Meschiari M, Cozzi-Lepri A, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2(8):e474–e484. doi:10.1016/S2665-9913(20)30173-9
85. Huet T, Beaussier H, Voisin O, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–400. doi:10.1016/S2665-9913(20)30164-8
86. Li H, Chen C, Hu F, et al. Impact of corticosteroid therapy on outcomes of persons with SARS-CoV-2, SARS-CoV, or MERS-CoV infection: a systematic review and meta-analysis. Leukemia. 2020;34:1503–1511. doi:10.1038/s41375-020-0848-3
87. Yang Z, Liu J, Zhou Y, et al. The effect of corticosteroid treatment on patients with coronavirus infection: a systematic review and meta-analysis. J Infect. 2020;81(1):e13–e20. doi:10.1016/j.jinf.2020.03.062
88. Soy M, Keser G, Atagündüz P, et al. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39(7):2085–2094. doi:10.1007/s10067-020-05190-5
89. Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi:10.1186/1743-422X-2-69
90. Qiu T, Liang S, Dabbous M, et al. Chinese guidelines related to novel coronavirus pneumonia. Preprints. 2020. doi:10.20944/preprints202004.0207.v1
91. Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi:10.5582/bst.2020.01047
92. Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56(1):105949. doi:10.1016/j.ijantimicag.2020.105949
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.