Back to Journals » International Journal of Nanomedicine » Volume 17
Current Status, Opportunities, and Challenges of Exosomes in Oral Cancer Diagnosis and Treatment
Authors Liu H , Huang Y, Huang M, Huang Z, Wang Q, Qing L, Li L, Xu S, Jia B
Received 9 March 2022
Accepted for publication 1 June 2022
Published 16 June 2022 Volume 2022:17 Pages 2679—2705
DOI https://doi.org/10.2147/IJN.S365594
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Phong A Tran
Hongyu Liu,1 Yisheng Huang,1 Mingshu Huang,1 Zhijie Huang,1 Qin Wang,1 Ling Qing,1 Li Li,1 Shuaimei Xu,2 Bo Jia1
1Department of Oral Surgery, Stomatological Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 2Department of Endodontics, Stomatological Hospital, Southern Medical University, Guangzhou, People’s Republic of China
Correspondence: Shuaimei Xu; Bo Jia, Email [email protected]; [email protected]
Abstract: Oral cancer is one of the most common cancers in the world, with more than 300,000 cases diagnosed each year, of which oral squamous cell carcinoma accounts for more than 90%, with a 5-year survival rate of only 40– 60%, and poor prognosis. Exploring new strategies for the early diagnosis and treatment of oral cancer is key to improving the survival rate. Exosomes are nanoscale lipid bilayer membrane vesicles that are secreted by almost all cell types. During the development of oral cancer, exosomes can transport their contents (DNA, RNA, proteins, etc) to target cells and promote or inhibit the proliferation, invasion, and metastasis of oral cancer cells by influencing the host immune response, drug-resistant metastasis, and tumour angiogenesis. Therefore, exosomes have great potential and advantages as biomarkers for oral cancer diagnosis, and as drug delivery vehicles or targets for oral cancer therapy. In this review, we first describe the biogenesis, biological functions, and isolation methods of exosomes, followed by their relationship with oral cancer. Here, we focused on the potential of exosomes as oral cancer biomarkers, drug carriers, and therapeutic targets. Finally, we provide an insightful discussion of the opportunities and challenges of exosome application in oral cancer diagnosis and treatment, intending to offer new ideas for the clinical management of oral cancer.
Keywords: exosomes, oral cancer, biomarkers, drug delivery vehicles, therapeutic target
Introduction
As the first part of the upper gastrointestinal tract, the oral cavity is responsible for food intake, mastication, articulation, swallowing, sensation and expression.1 The normal performance of these physiological functions is inevitably affected to a greater or lesser extent by oral cancer, and the quality of life of most clinical patients is severely reduced in the late stage due to postoperative cosmetic defects and loss of oral and maxillofacial functions.2 Oral cancer, according to the International Classification of Diseases, Tenth Revision [ICD-10], includes cancers of the lip, cheek, tongue, gingiva, floor of the mouth, hard palate, and other unspecified parts of the oral cavity.3 Oral squamous cell carcinoma (OSCC) accounts for more than 90% of these cancers4–6 and is the most common malignancy of the head and neck. It has a high prevalence worldwide and is a major public health concern. In 2020, the number of new cases of OSCC worldwide was approximately 377,000, and the number of deaths in the same year was approximately 177,000.7 Early detection, even at the preclinical stage, is essential to reduce the morbidity and mortality of OSCC. Some studies have shown that patients with oral cancer can have a 5-year survival rate of more than 80% if diagnosed early (stages I and II). Unfortunately, up to 50% of oral cancers are diagnosed at an advanced stage (stages III and IV) after the onset of symptoms, such as pain, bleeding, or oral and maxillofacial masses. The risk of developing advanced oral cancer is significantly higher when the diagnosis is delayed for more than one month, and treatment for patients with advanced oral cancer is not only costly but also has a poor prognosis.8,9 Therefore, the early diagnosis of oral cancer is closely related to the prognosis, and it is urgent to develop methods for the early diagnosis thereof. OSCC has a high potential for local infiltration and lymph node metastasis,4–6 and once metastasis is present, conventional treatment approaches fail to achieve satisfactory results. Despite the development of multidisciplinary and comprehensive sequential treatment strategies and levels for OSCC, the 5-year survival rate has not improved significantly in the past decades (remaining around 50%).10–13 Therefore, in addition to focusing on early diagnosis of oral cancer, finding emerging strategies for targeted treatment is an effective way to reduce mortality and improve patients’ quality of life. The discovery of early diagnostic markers and targets for oral cancer treatment is the focus of current research.14
Extracellular vesicles are small membrane-derived vesicles secreted into the extracellular space. Based on the size of extracellular vesicles, they are classified into three main types: (1) apoptotic vesicles, (2) microvesicles, and (3) exosomes.15 Exosomes are lipid bilayer membrane vesicles with a diameter of approximately 30–150 nm16 and can be secreted by almost all cells (such as immune cells,17–19 tumour cells,20 and nerve cells)21. Exosomes contain a variety of molecules present in various body fluids, including saliva, blood, and urine. Exosomes have specific surface proteins that distinguish these nanoparticles from other microvesicles or apoptotic vesicles.22 These surface proteins (eg CD63, CD81, or CD9) allow for the screening, selective recruitment and analysis of exosomes of cancer cell origin.23 Previous studies have shown that the “cargo” loaded in exosomes can be used as a novel diagnostic biomarker for OSCC.24–27 Exosomal surface proteins or contents are diverse and dynamically altered, with significant differences between patients with oral cancer and healthy individuals.28,29 This provides a basis for circulating exosomes as a biomarker for the early diagnosis of oral cancer. In addition, exosomes are the “messengers” of intercellular communication,30,31 which can participate in intercellular information transfer, transmitting biological information, such as proteins, lipids and nucleic acids, from the parent cell to the recipient cell in a stable manner,27,32 causing a series of relevant reactions in the recipient cell, thus playing a regulatory role.33 Many studies have shown that after delivering exosomal contents to target cells, exosomes can influence the host immune response, drug-resistant metastasis, tumour angiogenesis and other processes by inducing phenotypic and functional changes in the recipient cells,34–36 thereby promoting or inhibiting the proliferation, invasion and metastasis of oral cancer. Moreover, exosomes have the characteristics required for easy uptake by cancer cells, small size, good biocompatibility, easy access, and high stability.37–39 This makes it possible to deliver drugs through exosomes or use engineered exosomes for targeted treatment of oral cancer. Therefore, exosomes have great potential and advantages for the diagnosis and treatment of oral cancer.
Research on exosomes for various cancers is increasing annually.37 The research progress and clinical application of exosomes in oral cancer diagnosis and treatment have also attracted much attention.40–42 This review takes a fresh perspective on the impact of exosomes on the development of oral cancer. It highlights the current status, opportunities, and challenges of exosomes in the diagnosis and treatment of oral cancer, providing new insights into the search for emerging biomarkers and potential targets for the diagnosis and treatment of oral cancer.
Biogenesis, Biological Functions, and Isolation Methods of Exosomes
The biogenesis of exosomes consists of different stages, including three main steps: biogenesis, loading and transport of related substances, and release.43 First, the plasma membrane bud inwards to form endocytic vesicles (ie early endosomes), and in some cases, early endosomes can be created by the trans-Golgi network (TGN). Early endosomes then form late endosomes through cargo selection and fusion, followed by invagination of the endosomal membrane to form intraluminal vesicles (ILVs) that promote the formation of multivesicular bodies (MVBs). Eventually, MVBs fuse with the plasma membrane, leading to the release of vesicle contents called exosomes.44–47 The biogenesis of exosomes is shown in Figure 1. In addition, MVBs can be degraded by lysosomes or autophagosomes.48,49 The released exosomes interact with target cells in three ways: (1) fusion of the plasma membrane between target cells and exosomes, (2) the binding of target cell membrane receptors with proteins on the membrane surface of exosomes, and (3) the in vivo transformation of exosomes by target cell endocytosis.50 The origin and biogenesis of exosomes are mediated by tumour susceptibility gene 101 (TSG101), apoptosis-linked gene 2-interacting protein X (ALIX), endosomal sorting complexes required for transport (ESCRT) protein, soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) complex proteins, and other related substances. However, the specific mechanism underlying the role and function of these related substances in exosome biogenesis requires further investigation.31,49,51,52
Exosome release can be achieved by plasma membrane fusion, extracellular action of MVB, or by small cytoplasmic protrusions directly from the cell surface.37,53 Exosome uptake is achieved by interacting with proteins on the surface of exosomes (such as CD9, CD63, and CD81)54–56 and recipient cells. Simultaneously, exosomes can transfer their contents from parent cells to nearby and distant recipient cells, thus mediating intercellular communication.27,32 Under physiological conditions, exosomes are important mediators of intercellular communication. Under pathological conditions (eg cancer, neurodegenerative diseases, cardiovascular diseases, infectious diseases, and respiratory diseases), exosomes play an essential role in regulating cellular activity.57–62 During oral cancer progression, exosomes produced by different cells (eg macrophages) can transfer nucleotides and proteins between cells and participate in the complex pathogenesis of oral cancer development and metastasis.63–67 Therefore, it is essential to explore the influence of exosomes on oral carcinogenesis and progression.
Therefore, obtaining high-purity and -quality exosomes is an essential condition and foundation for clinical and experimental purposes. Currently, based on the size, morphological structure, and specific surface proteins of exosomes, different methods of exosome isolation have been discovered, including ultra-high-speed centrifugation,56,68–71 polymer precipitation,70,72,73 chromatography including molecular sieve chromatography,56 density gradient centrifugation,74 ultrafiltration,75 immunoaffinity capture,56,75 and microfluidics-based separation techniques,76–78 among others. Because exosomes can be obtained in many ways and isolated by many methods, it provides the basis and feasibility to study the application of exosomes in oral cancer diagnosis and treatment. However, each of these methods has its own advantages and disadvantages, and further improvements are needed to promote the research and application of exosomes in oral cancer diagnosis and treatment (Table 1).
 |
Table 1 Exosome Isolation Methods |
Exosomes Associated with Oral Cancer
Tumour-derived exosomes can transmit their own biogenetic information to the surrounding normal or tumour cells79,80 to produce various substances that favour tumour proliferation, invasion, and metastasis, thus forming a microenvironment conducive to tumour metastasis.81 This microenvironment can induce the transformation of normal cells around the tumour to tumour cells,82–84 thus regulating the behaviour of recipient cells.32,85,86 Research related to exosomes and oral cancer is currently underway. Current studies on the effect of exosomes on oral cancer have focused on salivary exosomes from patients with oral cancer, exosomes of oral cancer cell origin, and exosomes obtained from circulating plasma or purified serum from patients with oral cancer.
The difference between oral cancer and other tumours is that the relationship between oral cancer and exosomes can be studied using salivary exosomes, in addition to tumour cell-, plasma-, or serum-derived exosomes. Therefore, it is hypothesized that there is still much room for discovering exosomes associated with oral cancer development as a possible way to find emerging oral cancer diagnostic methods and alternative treatment strategies.
Salivary Exosomes in Oral Cancer Patients
Different components of the salivary exosomes of patients with oral cancer may be directly related to oral cancer progression. The most studied oral cancer target is RNA (miRNAs account for the vast majority). miRNAs are small, non-coding RNA molecules consisting of 19–25 nucleotides that can regulate gene expression by degrading or inhibiting translation through complementary binding to a portion of the target mRNA sequence.87 Langevin et al88 performed miRNA microarray analysis of salivary exosomes from head and neck squamous cell carcinomas and the corresponding healthy controls. Compared with the corresponding controls, differences in miRNA expression levels were found in salivary exosomes from patients with head and neck squamous cell carcinoma. To obtain more specific results, He et al89 evaluated the miRNA microarray analysis of salivary exosomes from patients with OSCC compared to the corresponding control saliva samples, and the differences in miRNA expression levels were determined. In addition, Momen-Heravi et al90 provided a detailed review of the emerging role of non-coding RNAs in oral cancer. The RNAs associated with salivary exosomes in oral cancer and their differentially expressed salivary exosomal proteins suggest a link between exosomes and oral cancer. Some investigators have experimentally found a significant increase in the number and irregular morphology of salivary exosomes in oral cancer. These suggest that there are differences in morphology, number, surface markers and contents of saliva-derived exosomes between oral cancer patients and healthy controls and that exosomes may be the “key” to the early diagnosis of oral cancer.
Oral Cancer Cell-Derived Exosomes
Oral cancer cell-derived exosomes can promote oral cancer progression by modulating the immune response, promoting angiogenesis, and promoting aggressive metastasis.34–36,91 It has been shown that polarized tumour-associated macrophages (TAMs) can promote tumour growth and metastasis by producing large amounts of growth factors and cytokines, or by inhibiting the proliferation of effector T cells and directly suppressing the T cell immune response to cancer.63–65 Oral cancer cell-derived exosomes can influence oral cancer progression by affecting the phenotype of the host’s macrophages. It has been found that OSCC cell-derived exosomes can polarize macrophages into M1-like TAMs92 or modulate, enhance, and induce macrophage transition to the M2 phenotype93–96 which in turn affects the proliferation, invasion and migration of OSCC.
Blood supply plays a crucial role in cancer progression by providing nutrients for tumour growth and metabolism.97 The expression levels of exosomal miRNAs in OSCC cells were different compared to the corresponding controls, and differentially expressed exosomal miRNAs were associated with angiogenesis, migration of vascular endothelial cells, and microvessel density.98,99 It is hypothesized that OSCC cell-derived exosomes could also promote oral cancer progression by promoting angiogenesis.
In addition, highly invasive OSCC cell-derived exosomes can induce a phenotypic shift from low-invasive OSCC cells to highly invasive or promote the motility of low-invasive OSCC cells, promoting OSCC invasion and migration. Exosomes isolated from the highly metastatic human oral cancer cell line HOC313-LM can promote tumour cell growth by activating ERK and AKT, transferring biomarkers with highly aggressive characteristics to less aggressive cells, increasing the motility of low invasive OSCC cells, and leading to increased cancer cell motility and invasiveness.100 In contrast, exosomes from highly invasive OSCC cell lines can induce a shift in non-invasive OSCC cell lines to a highly invasive phenotype, which in turn promotes OSCC invasion and migration.101
Exosomes Obtained by Isolation and Purification of Circulating Plasma or Serum from Oral Cancer Patients
There is a large amount of research data on the relationship between exosomes and diseases related to liver,102 kidneys,103 lungs,104 cardiovascular diseases,105,106 and diseases affecting the central nervous system.107 In contrast, research data related to the relationship between circulating plasma and purified exosomes isolated from patients with oral cancer are scarce and need to be explored through continued research. It has been shown that upregulation of circ_0000199 in circulating plasma exosomes from patients with OSCC promotes cell proliferation and inhibits apoptosis, and is positively associated with poorer survival outcomes, whereas in knockdown experiments is the opposite was noted.108 In addition, Guo et al109 identified several differentially expressed proteins in serum exosomes of patients with OSCC, including C-reactive protein, vascular haemophilia factor, and leucine-rich alpha-2-glycoprotein, as potential biomarkers for the specific diagnosis of OSCC. They also found that OSCC-associated serum exosomes might promote the migration of oral cancer cells.
Current Status of Research on Exosomes in the Diagnosis and Treatment of Oral Cancer
Current Status of Exosomes in the Diagnosis of Oral Cancer
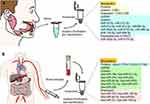
Surgical tissue biopsy is considered the gold standard for diagnosing solid tumours. Morphology-based biopsy is often physically invasive and time-consuming, adding excessive stress and pain to the patient. In addition, different sites of the primary tumour exhibit extensive inter-and intra-tumour variability, this tumour heterogeneity makes it difficult to determine an exact and effective treatment plan based on a single biopsy resulting in the need for another invasive biopsy.110–113 In these situations, this diagnostic approach not only raises the cost of treatment but may also result in potential complications, leading to reduced patient compliance. Over the past few decades, exosome-based liquid biopsies have received much attention from clinicians and scientists, with the advantages of easy access, convenience, non-invasiveness, time and effort savings, high reliability, reproducibility, ease of early detection, low cost, and high benefit.37 They can also track and monitor tumour progression and drug resistance in real time allowing for continuous sampling to provide information on tumour heterogeneity,114 making it an ideal method for early screening and diagnosis of oral cancer.Figure 2 provides a summary of exosomes as a source of biomarkers for oral cancer diagnosis.
Salivary Exosomes from Oral Cancer Patients as a Potential Tool for Oral Cancer Diagnosis
Saliva is an emerging biofluid for the early detection and diagnosis of diseases, reflecting the relevant conditions of oral and systemic health, and is a valuable source of clinically relevant information of these diseases.115 Saliva is secreted by three pairs of major salivary glands (parotid, submandibular, and sublingual) and numerous minor salivary glands throughout the oral cavity.116,117 The acquisition of salivary exosomes is not only painless, easy to obtain, cost-effective, and efficient, but also avoids unnecessary complications caused by invasive biopsies or blood collection, thus increasing patient compliance and motivation for treatment. The identification of alterations in salivary exosome production and abnormal expression of its contents may provide a new direction for oral cancer diagnosis. Recent studies have demonstrated that miRNAs in human salivary exosomes are potential biomarkers for oral cancer diagnosis. One researcher collected miRNA sequencing results from salivary exosomes of head and neck squamous cell carcinoma (HNSCC) patients and their corresponding healthy controls, and found that miR-486-3p, miR-10b-5p, and miR-486-5p were expressed at significantly higher levels in the salivary exosomes of HNSCC patients.88 The miRNA sequencing results of salivary exosome samples from OSCC patients showed that miR-24-3p, miR-412-3p, miR-512-3p, miR-27a-3p, and miR-494-3p expressions were significantly upregulated in salivary exosomes from patients with OSCC compared to that in their corresponding healthy controls, whereas miR-302b-3p, miR-517b-3p were only expressed in salivary exosomes from patients with OSCC.25,89 These studies suggest that miRNAs in salivary exosomes could be a potential resource for the diagnosis and differential diagnosis of oral cancer and may become a new oral cancer biomarker. However, more practical clinical data are required to confirm their diagnostic accuracy. Nevertheless, many differentially expressed RNAs still exist in the saliva of patients with oral cancer, and these RNAs hold great promise for research. These findings may aid in the diagnosis of oral cancer. In addition, exosomal protein markers are expected to be a new way to diagnose oral cancer by specifically identifying signature proteins that are differentially expressed on the surface of salivary exosomes in oral cancer patients to distinguish oral cancer patients from healthy individuals. In 2011, Sharma et al90 used high-resolution atomic force microscopy to show the differences between salivary exosomes from healthy individuals and patients with oral cancer. They found that exosomes in the saliva of oral cancer patients were irregular in shape and increased in size, number, and aggregation, especially oral cancer exosomes with a significantly higher surface density of CD63. Later, Zlotogorski-Hurvitz et al42 used nanoparticle tracking analysis to evaluate saliva samples from 36 patients with oral cancer and 25 healthy individuals, validating the claims of Sharma et al. They showed differences in the expression of exosome markers by ELISA and Western blotting between patients with oral cancer and healthy individuals showing higher expression of CD63 and lower expression of CD81 and CD9. Therefore, according to the morphological, molecular, and surface marker expression differences of salivary exosomes between patients with oral cancer and healthy individuals, salivary exosomes can be used as a potential tool for the early screening and auxiliary diagnosis of oral cancer patients. However, saliva is a complex fluid that is rich in a variety of proteins,118 and some salivary impurity protein components may be mixed with the salivary exosomes obtained by isolation. This may mask the presence of low levels of salivary exosomal proteins that may be important biomarkers119 and thus interfere with the accuracy of salivary exosome-based diagnosis of oral cancer. Therefore, studies on specific exosomal proteins that are directly related to oral cancer progression are scarce. At present, some scholars have obtained the proteome map of human parotid exosomes using multidimensional protein identification technology,120 which may be an auxiliary means to discover exosome protein markers related to oral cancer.121
Purified Exosomes from Plasma or Serum of Oral Cancer Patients as Potential Substances for Oral Cancer Diagnosis
The acquisition of salivary exosomes has significant advantages (eg the body cannot tolerate frequent blood collection) and provides a positive patient experience. Compared to salivary exosomes, exosomes obtained from plasma or serum purification may be more stable and less susceptible to interference. These may provide more accurate information on relevant biomarkers with differential expression. Therefore, differential expression of plasma or serum-purified exosome-associated biomarkers in oral cancer patients may be more representative, accurate, and suitable for real-time tracking and assessment of oral cancer progression. It has been shown that ApoA1, PF4V1, CXCL7, and F13A1 in serum-purified exosomes are valuable as potential markers for detecting metastasis of oral squamous cell carcinoma. Combining these biomarkers can improve the sensitivity of OSCC diagnosis and reduce clinical misdiagnosis in patients with OSCC with lymph node metastasis.122 Many differentially expressed miRNAs were also present in plasma or serum purified exosomes from patients with oral cancer compared to healthy individuals.
It was recently shown that circulating plasma exosome circ_0000199 expression levels were significantly higher in OSCC patients than healthy controls,108 and may be a potential biomarker for the diagnosis of oral cancer. Similarly, Rabinowits et al123 screened several exosomal miRNAs from tumour tissue samples and the plasma of patients with squamous cell carcinoma of the tongue. Sixteen of these miRNAs were differentially expressed between tumour tissues and matched benign tissues, while 15 of them had the same expression profile in plasma exosomes: nine were upregulated (hsa-miR-19a, hsa-miR-512-3p, hsa-miR-27b, hsa-miR-20a, hsa-miR-28- 3p, hsa-miR-200c, hsa-miR-151-3p, hsa-miR-223, and hsa-miR-20b), and six were downreguIated (hsa-miR-370, hsa-miR-139-5p, hsa-miR-let-7e, hsa-miR-30c, hsa-miR-22, and hsa-miR-145-3p). These differentially expressed miRNAs can be used as non-specific biomarkers of oral cancer. The combined application of these markers may aid in improving the sensitivity of oral cancer diagnosis and tracking dynamic changes in oral cancer. The application of relevant biomarkers in exosomes for oral cancer diagnosis has considerable research prospects and room for further exploration.
Current Status of Exosomes in Oral Cancer Treatment
The combination of surgical excision of lesions and radiotherapy remains the main treatment option for oral cancer; however, the results are unsatisfactory. Currently, the search for emerging therapeutic modalities to reduce the chemotherapy resistance rate, improve drug targeting, and improve patient survival rate remains a direction for future efforts. Recently, exosomes have received considerable attention as drug carriers and targets for the treatment of oral cancer (Table 2).
 |
Table 2 Status of Exosomes in Oral Cancer Treatment |
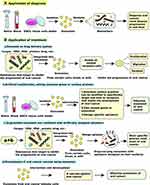
Exosomes as Natural Drug Delivery Vehicles for Oral Cancer
The interaction of therapeutic agents with non-targeted sites in the body (ie, off-target effects) can lead to harmful side effects, such as those observed in cancer chemotherapy. Tumours and health tissue can experience significant toxic side effects during chemotherapy, which is a major problem in oncology. The delivery of drugs to target organs and the prevention of toxic side effects remain the current focus of researchers. Precise targeted delivery of chemotherapeutic drugs may be a potential way to reduce the toxic side effects of chemotherapy.124 Recent advances in drug delivery methods aim to enhance targeting strategies, improve drug delivery rates, control drug release, and prolong drug action. The disadvantages of large materials, such as instability, low absorption rate, and low degradability in vivo, make them unsuitable for drug delivery applications. Based on the concept of targeted drug delivery, researchers have also worked on the development of non-toxic, stable, and biocompatible drug carriers.125 Therefore, nanomaterial-based drug delivery methods have been favoured. Nanoparticle carriers can be classified as inorganic materials (eg metal nanoparticles) or organic materials (eg liposomes and exosomes).126 Liposomes have been used as carriers for cancer therapy.127 Compared with liposomes, exosomes have superior drug delivery properties, such as good stability, allowing them to travel to distant target organs, a hydrophilic core that can encapsulate water-soluble drug molecules, and high biosafety, which does not induce an immune response in the body.128,129 In addition, exosomes can easily deliver their contents to targets by fusing with cell membranes, binding to membrane surface receptors, and cytocytosis. The nature of exosomes allows them to be loaded with many types of cargo (eg DNA, RNA, and proteins), and the loading of these cargoes can be achieved in vivo or in vitro.130 Therefore, exosomes can be used as natural drug delivery vehicles for oral cancer.
According to the different methods of exosomal drug loading, the two main types are passive and active drug loading. We next describe the characteristics, advantages, and disadvantages related to these two drug loading methods of exosomes. The passive drug loading method of exosomes includes three forms: the first is to separate and purify the exosomes from the donor cell and mix them with the drug to be loaded.131 This method can ensure the integrity of the exosomal membrane, but the drug loading rate is low. The second form is to transfect the drugs to be loaded into the donor cells, and then encapsulate these drugs into the exosomes inside the donor cells.132 This approach is safe and effective for immunotherapy and cancer treatment, but the drugs may have toxic effects on the donor cells. The third form is the fusion of plasmid DNA containing the therapeutic protein code with the coding peptide capable of targeting the therapeutic protein to the exosomes, followed by transfection into the nucleus of the donor cell. Plasmid DNA is then transcribed and translated to produce a drug that can be localized to the exosomes, ultimately allowing the drug to be integrated into the exosomes.133 This is a way to consistently produce exosomal drugs, but it is mechanistically complex, time-consuming, and costly. Because passive drug loading of exosomes does not use any external physical or chemical factors, this method does not affect the integrity of exosomes and is a technique that can be considered. Although the passive loading method for exosomes has the above advantages, this method has a low loading rate and is time-consuming. To increase the loading rate and shorten the loading time in case of exosomes, researchers normally use the active drug loading method.
There are four main approaches to active drug loading in exosomes. The first is the electroporation method, in which the exosomal membrane is pored by an electric field, allowing the therapeutic drug to penetrate the exosomes.134 This is a widely used method for transporting hydrophilic molecules to the hydrophilic core of exosomes. The second method is the extrusion method, in which isolated and purified exosomes are mixed with therapeutic drugs and passed simultaneously with an extruder through a multi-pore membrane with a pore size of approximately 100–400 nm.135 The drug loading rate of exosomes induced by the extrusion method is affected by the number of extrusions, type of exosomes, and type of transfected cells.136 This method has good drug loading capacity, but affects the integrity of the exosomal membrane, and the end product may be cytotoxic. The third form is the sonication method, in which the ultrasound energy provided by the probe sonicator stimulates the exosome and affects its integrity, allowing the therapeutic drug to penetrate the exosome during exosome deformation.137 This method increases the drug-carrying capacity in the hydrophobic bilayer, but increases the size of the exosome. The fourth method is the freeze-thaw cycle method, which involves freezing and then thawing a mixture of exosomes and drugs.138 However, both the electroporation and freeze-thaw cycling methods can cause aggregation of exosomes. There are reviews detailing the methods of exosomal drug loading and the use of exosomes in drug delivery and therapy.139 Based on the unique advantages of drug loading by exosomes and their diverse drug loading modes, a solid theoretical and methodological basis has been established for their use as natural drug delivery carriers for oral cancer.
In addition, studies have reported exosomes as carriers of therapeutic small molecules, proteins, and nucleic acids for therapeutic application in diseases. For example, a study reported that exosomes loaded with small molecules, such as adriamycin or paclitaxel, could easily cross the blood-brain barrier for the treatment of brain tumours.140 Similarly, exosomes loaded with curcumin, a small molecule drug, have inhibited the proliferation of breast cancer cells.141 Exosomes can also deliver therapeutic proteins (such as enzymes, cytoskeletal proteins, and transmembrane proteins) or therapeutic nucleic acids (such as miRNAs) to the corresponding recipient cells, resulting in phenotypic changes in the recipient cells and ultimately inhibiting tumour development as detailed in another review.62 For example, some researchers have successfully loaded proteins into the lumen of exosomes, which then efficiently deliver the loaded proteins to the target cells, leading to a significant increase in intracellular protein levels and their functions.142 Similarly, Zhang et al found that exosomes collected from transfected cells enriched with anti-c-Met siRNA significantly inhibited tumour growth in a mouse transplant tumour model and reversed cisplatin resistance in gastric cancer cells in vitro.143 Meanwhile, there are also reviews detailing new advances in exosome-based targeted drug delivery systems. Therefore, exosomes loaded with therapeutic small molecules, proteins, and nucleic acids are also a promising modality for the treatment of oral cancer (this will be explored in detail in later chapters).126 Using flow cytometry and in vivo uptake analysis, Wang et al144 confirmed that the uptake of exosomes by tumour cells was higher than that of macrophages and other immune cells at the single-cell level, suggesting that exosomes can inhibit oral cancer progression by loading drugs or carrying related substances (eg DNA, RNA, or protein), targeted delivery to the tumour site, and accumulation of anticancer treatment. In addition, exosomes can be modified through gene editing or protein modifications to increase their anti-tumour targeting.145 Several studies have shown that exosomes derived from bone marrow mesenchymal stem cells (MSC) and epithelial cells do not induce toxicity when repeatedly injected into mice.146,147 Repeated injections of MSC-derived exosomes are well tolerated without adverse or significant side effects. They can be used for their own treatment129 and for treating patients with immune rejection after transplantation.148 This indicates that exosomes have the advantage of good biocompatibility, almost nontoxic side effects, and can be used as a sound drug delivery system. In addition, the lipid bilayer membrane of exosomes protects their contents from degradation and destruction, making them highly stable in the circulation. Moreover, the small size of exosomes can easily cross various biological barriers (eg blood-brain barrier) and migrate to target cells or organs. This allows exosomes to maintain their potency and to target distant locations. The use of exosome-loaded drugs for targeted cancer treatment has been reported,149 revealing a new direction for using exosomes to load and deliver medicines to treat oral cancer.
Exosomes as a Target for Oral Cancer Treatment
Currently, the common treatment options for malignancies include surgery, radiotherapy, chemotherapy, targeted therapy, and adjuvant therapy,150 most of which can cause significant side effects and financial burden. The development of therapies with low side effects, high benefits, low cost, and high specificity is the goal of researchers. In recent years, there have been an increasing number of studies related to exosomal proteins, miRNAs, lncRNAs, and circRNAs as potential targets for treating oral cancer.62
Since exosomes contain numerous bioinformatic molecules related to the development of oral cancer, it may be possible to consider inhibiting the progression of oral cancer by suppressing the expression of these associated molecules in exosomes. To date, the field of oral cancer has been most studied for exosomal miRNAs, which can control cell division, differentiation, and death, and play an essential role in determining cell fate.151 The role of exosomal miRNAs in the proliferation, invasion, and migration of oral cancer cells has also been extensively studied. For example, the expression level of exosomal miR-1294 is low in OSCC tissue samples, and exosomal miR-1294 inhibits OSCC cell growth by regulating the c-Myc pathway. Upregulation of exosomal miR-1294 expression inhibited the proliferation and migration of OSCC cells, while inhibition of exosomal miR-1294 expression promoted the growth and migration of OSCC cells. Therefore, the expression of exosomal miR-1294 was negatively correlated with oral cancer progression and had an inhibitory effect on OSCC.152 Similarly, exosomal miR-6887-5p inhibited tumour growth and OSCC cell colony formation in vitro and in vivo. OSCC progression can be inhibited by upregulating the expression of exosomal miR-6887-5p in OSCC patients or by introducing miR-6887-5p-rich exosomes in vitro.153 In addition, exosomes from human bone marrow mesenchymal stem cells (HBMSCs) also contain miRNAs associated with oral cancer development. miR-101-3p overexpression in HBMSC-derived exosomes inhibits the proliferation, invasion, and migration of oral cancer cells by downregulating COL10A1, thus inhibiting oral cancer progression. Moreover, a tumorigenic assay in nude mice further confirmed the inhibitory effect of HBMSC-derived exosomes carrying miR-101-3p on oral cancer.154 This provides a basis for exosome-based targeted therapy for oral cancer, and exosomes are expected to be a potential therapeutic target for OSCC. When exosomes are used as a target for oral cancer therapy, the key is to determine how to increase the exosomes carrying capacity of miR-1294, miR-6887-5p, or miR-101-3p. As described above, exosomes are currently loaded with hydrophilic molecules, such as relevant miRNAs, mainly through the active loading method of electroporation, but this method leads to the aggregation of exosomes and thus a relative decrease in drug loading rate.155 Therefore, finding new methods to increase the drug loading of exosomes remains the key for efficient treatment of oral cancer. Recently it was reported that Fe3O4 nanoparticles and a constant magnetic field can induce exosomal miR-21-5p upregulation in bone marrow MSCs, and this extrinsic induction condition (nanoparticles and constant magnetic field) may be a promising approach to increase exosomes capacity to be loaded with more miRNAs relevant to the treatment of diseases.156 A related review has also described multifunctional nanomaterials with tunable physical, chemical and biological properties that may play a key role in exosome-based drug delivery.157 This combination of nanoparticles and exosomes allows for therapeutic implementation on the same platform and has the advantages of providing targeting, improving dispersion, and avoiding clearance by the immune system. This approach of targeted transport of therapeutic exosomes by the combined action of magnetic nanoparticles and external magnetic fields may not only improve the precise localization of the drug in the organism, but also improve drug retention, prolong drug half-life, reduce drug dose, and improve efficacy (even in high blood flow systems).158 This may be a promising approach to improve the effective drug loading rate and targeting of exosomes, which is another step forward in the clinical application of exosome-loaded drugs for the treatment of oral cancer.
Epithelial-mesenchymal transition (EMT) is closely associated with the invasion and migration of oral cancer cells, a dynamic process in which the migratory and invasive capacities of epithelial cells are enhanced by the loss of intercellular adhesion and polarity. EMT is considered the initiating step of the invasion-metastasis cascade.159 Inhibition of the EMT process in oral cancer is a new direction for the treatment of oral cancer. It can inhibit the progression of oral cancer by suppressing any response associated with the promotion of EMT. Fujiwara et al160 showed that OSCC cells secrete large amounts of exosomes highly expressing epidermal growth factor receptor (EGFR) in response to epidermal growth factor stimulation. Subsequently, EGFR-rich exosomes enter epithelial cells and promote EMT, thereby promoting OSCC invasion and migration. The internalization and pro-EMT effects of OSCC exosomes were blocked by cetuximab. The therapeutic anti-EGFR antibody cetuximab inhibits OSCC cell invasion and migration. Therefore, it is hypothesized that targeted inhibition of exosomal EGFR expression or other biomarkers that promote the EMT process could be a new therapeutic strategy to inhibit oral cancer progression.
The blood supply plays a crucial role in cancer progression by providing nutrients for tumour growth and metabolism. Therefore, angiogenesis is associated with tumour growth and metastasis, and OSCC cell-derived exosomes may have an inhibitory or proangiogenic effect on angiogenesis, thus affecting OSCC progression.161 Angiogenesis is a critical step in tumour development and metastasis, and blocking angiogenesis is a way to discover new therapeutic approaches for oral cancer. In recent years, many studies have elucidated the important role of OSCC cell-derived exosomes in oral cancer angiogenesis. For example, He et al98 found that OSCC-derived exosome miR-221 promotes human umbilical vein endothelial cell (HUVEC) migration and angiogenesis by targeting and negatively regulating PIK3R1. Similarly, OSCC-derived exosomal miR-210-3p levels were positively correlated with microvessel density. The expression of miR-210-3p could be upregulated by the PI3K/AKT pathway and downregulated by ephrinA3 expression to promote oral cancer angiogenesis.99 Therefore, exosomal miR-221 and miR-210-3p may be closely related to angiogenesis during OSCC proliferation and may become a new target for the clinical treatment of OSCC. Therefore, it is hypothesized that inhibiting angiogenesis-related functions in oral cancer may be a new strategy for the effective clinical treatment of oral cancer.
Resistance to chemotherapy remains a major obstacle in the effective treatment of OSCC and enhancing the specificity and targeting of chemotherapeutic agents remains a critical problem. By establishing cisplatin-resistant cell lines (HSC-3-R and SCC-9-R), Liu et al162 found that the conditioned medium of cisplatin-resistant OSCC cells enhanced drug resistance in parental OSCC cells. Further studies have revealed that exosomes are involved in resistance transfer. Cisplatin-resistant OSCC cell-derived exosomes can induce cisplatin resistance by targeting phosphatase and tensin homologue (PTEN) and programmed cell death 4 (PDCD4) and transferring miR-21 into OSCC parental cells. Additionally, they established a subcutaneous transplantation mouse model to confirm the role of cisplatin-resistant OSCC cell-derived exosomes in vivo. They concluded that exosomes released from cisplatin-resistant OSCC cells induce cisplatin resistance in OSCC cells through delivery. Therefore, inhibiting exosome miR-21 expression by targeting can enhance chemotherapy sensitivity in OSCC patients and may be a new therapeutic approach for OSCC cisplatin-resistant patients. In contrast, under hypoxic conditions, OSCC cell-derived exosome miR-21 is directly regulated by HIF-1a and HIF-2a and its expression level is upregulated, which increases the migration and invasion of OSCC cells.163 Therefore, blocking the pathway of exosome activation-related pathways, controlling in vivo environmental factors, or inhibiting the effect of related pathways by targeting may be a new approach to inhibit oral cancer progression. In addition, THP-1 cells and primary human macrophage (PHM)-derived exosomes can reduce the sensitivity of OSCC cells to chemotherapeutic drugs by activating the AKT/ GSK-3β signalling pathway.164 Therefore, targeted inhibition of the relevant signalling pathways may be a way to improve the sensitivity of oral cancer chemotherapy. In addition, Cui et al91 transfected docetaxel (DTX)-resistant HSC-3 cells (HSC-3DR) with lentivirus and co-cultured them with exosomes from normal tongue epithelial cells (NTECs) overexpressing miR-200c. Further studies revealed that NTECs transferred exosomal miR-200c into HSC-3DR cells and made HSC-3DR more sensitive to DTX by binding to TUBB3 and PPP2R1B. This suggests that exosomal miR-200c can enhance the sensitivity of patients with OSCC to chemotherapy, and exosomal miR-200c delivery may be a promising and effective strategy for treating drug resistance in tongue squamous cell carcinoma.
Exosomes also play important roles in regulating immune responses and coordinating immune system-related responses. Tumour-derived exosomes can activate or suppress the host immune system, suggesting that exosomes have potential therapeutic value in modulating immune responses against tumour progression and are promising candidates for immunomodulation in oral cancer.165,166 Recent studies have identified that THBS1 from OSCC cell-derived exosomes is involved in the polarization of macrophages to an M1-like phenotype, which polarizes macrophages to M1-like TAMs,92 and significantly promotes the migration of OSCC cells. In contrast, the OSCC cell-derived exosomes miR-23a-3p,93,94,96 CMTM6,94,95 and miR-29a-3p93,94 promote macrophage polarization toward the M2-like phenotype by activating related signalling pathways, which in turn promotes OSCC proliferation and invasion. Therefore, THBS1, miR-23a-3p, CMTM6, and miR-29a-3p may be potential immunomodulatory targets for the treatment of oral cancer. Additionally, exosomal PD-L1 in the plasma of patients with oral cancer can drive immunosuppression and contribute to the immune escape of oral cancer cells.167,168 Moreover, PD-L1-driven immunosuppression can be reversed by anti-PD-L1.169 Therefore, exosomal PD-L1 is a potential target for the treatment of oral cancer. Tumour-derived exosomes can not only mediate immunosuppression and thus promote tumour progression, but also interfere with immunotherapy.170 In addition, exosomes can also act as immunosuppressive “immune enhancers”. Wang et al171 analysed the effects of oral cancer-derived exosomes on natural killer (NK) cells and found that oral cancer-derived exosomes (NF-κB-activating kinase-associated protein 1) NAP1 enhanced the cytotoxicity of NK cells through the IRF-3 pathway and enhanced the tumour suppressor function of NK cells. Therefore, exosomes with high NAP1 expression can inhibit OSCC progression. In addition, oral cancer exosomal lncRNA can interact with the tumour microenvironment (TME) through a related mechanism, promoting oral cancer progression, and is a potential target for targeted treatment of oral cancer. Ding et al172 found that lncRNA FLJ22447 from OSCC exosomes significantly upregulated lncRNA-caf (Lnc-CAF) in tumour-associated fibroblasts. Lnc-CAFs activated by IL-33 can induce OSCC cell proliferation, and high Lnc-CAF/IL-33 expression correlates with high TNM (tumour, node, metastasis) stage, and high Lnc-CAF expression predicts poor prognosis. In vivo, Lnc-CAF or IL-33 knockdown reduced tumour cell proliferation and inhibited tumour growth. However, data from studies related to exosomal lncRNAs associated with oral cancer are lacking and need to be explored further.
Exosomes regulate intercellular communication between cancer cells and act in conjunction with the TME.173,174 Exosomes are important signalling mediators that regulate the TME and are closely related to its formation. Interfering with intercellular communication and TME formation by inhibiting the synthesis and release of exosomes may be an effective way to inhibit oral cancer progression. Sento et al175 showed that OSCC-derived exosomes are taken up by OSCC cells and significantly promote OSCC proliferation, migration, and invasion by activating the PI3K/Akt, MAPK/ERK, and JNK-1/2 pathways. The pro-tumorigenic effect of OSCC-derived exosomes can be blocked by inhibiting the uptake of OSCC-derived exosomes by OSCC cells. Figure 3 provides a comprehensive summary of the progress of research on exosomes as potential targets for the treatment of oral cancer.
Opportunities and Challenges of Exosomes in the Diagnosis and Treatment of Oral Cancer
Opportunities of Exosomes in the Diagnosis and Treatment of Oral Cancer
Opportunities for Exosomes in Oral Cancer Diagnosis
Exosomes have great potential for clinical application as biomarkers for early diagnosis of oral cancer. Momen-Heravi et al176 used a genome-wide high-throughput miRNA microarray assay and found that miRNA-27b was significantly upregulated in the saliva of patients with OSCC. Moreover, miRNA-27b had higher sensitivity and specificity for the detection of OSCC compared to the detection of other miRNAs. Similarly, miRNA-21 and miRNA-184 were significantly increased in the saliva of patients with OSCC, and miRNA-184 could distinguish OSCC from potential other oral malignant diseases and aid in the differential diagnosis.177 These studies suggest that the saliva of patients with oral cancer is rich in biomarkers related to oral cancer progression. Exosomes as intercellular messengers may be loaded with these related substances, revealing that there may be more biomarkers in salivary exosomes of patients with oral cancer that are worth exploring. Exosomes may be a potential aid for oral cancer diagnosis.
Several studies have shown that DNA mutations associated with cancer development can characterize the genetic material carried in circulating blood exosomes, suggesting that the detection of serum exosome-associated DNA is a potential method for diagnosing cancer.178–181 It is hypothesized that if exosomal DNA provides a larger fragment of DNA than circulating free DNA, this may help detect relevant DNA mutations in circulating serum exosomes of oral cancer patients to diagnose oral cancer, thus avoiding the trauma and pain associated with cancer tissue biopsy. The expression of carcinogenic and tumour suppressor miRNAs in exosomes differs between cancer and normal cells,182 and there is an increasing number of studies related to cancer diagnosis based on exosomal miRNAs183–186 as the detection of specific miRNAs or miRNA groups in exosomes may be a new approach for oral cancer diagnosis. It has been suggested that combining multiple miRNAs may improve the diagnostic potential of exosomal miRNAs,187 and the related mechanisms of occurrence need to be further investigated. In addition, there are many specific marker proteins on the surface of exosomes, which can specifically bond to these surface proteins by immunocapture for early diagnosis of cancer.187–190 According to the idea of previous studies, combining exosomal DNA, RNA and proteins for early diagnosis of cancer may improve the sensitivity and accuracy of exosomal marker-based diagnosis of oral cancer. In addition, more undiscovered or unstudied exosomal DNA, exosomal RNAs (such as miRNAs, lncRNAs, or circRNAs), and exosomal surface proteins can be explored for oral cancer progression, laying the foundation for emerging biomarkers for the clinical diagnosis of oral cancer. For example, Lu et al62 established a mouse model of chemically induced OSCC and found that some piRNAs were significantly altered. Recently, Wang et al191 identified piRNAs in exosomes of stem cells. However, the function of exosomal piRNAs in human oral cancer and their relationship with oral cancer need to be further investigated. In addition, because exosomes are widely found in various body fluids, exosome-based liquid biopsies can be used to monitor and track disease progression. This non-invasive method of monitoring disease progression not only benefits the development of optimal treatment strategies for the disease, but also reduces patient suffering. Based on exosomal liquid biopsy, it is also possible to develop intelligent devices for exosomal liquid biopsy to obtain test results and perform preliminary assessments in real time, making the diagnostic process more manageable, and less time-consuming and laborious.
Opportunities for Exosomes in Oral Cancer Treatment
Exosomes have considerable clinical potential for the treatment of oral cancers. The exosomes are small, biocompatible, almost non-toxic, highly permeable and induce less immune rejection,146,147 allowing them to be used as carriers for delivering drugs and related genetic material.192,193 Compared to conventional chemotherapeutic drugs, exosomes have the advantages of easy avoidance of first-pass metabolic effects, good stability and high targeting, making them an effective drug delivery system.128,194,195 Compared with the traditional, more desirable liposome or polymeric nanoparticle systems, exosomes have superior degradability, biosafety, and easy accessibility, effectively avoiding the degradation of autophagosomes and lysosomes, allowing direct delivery of cargo to the cytoplasm, which makes exosome-based targeted drug delivery possible for oral cancer treatment.128,195 In addition, CD47 on exosomes produce a signal that protects them from phagocytosis, and blood-derived ribonucleases may protect exosomal RNA from damage. These protective mechanisms may prolong the time for exosomes to remain and function in vivo, as well as allow exosomes to deliver cargo to more distant targets and perform their functional roles.196,197 Therefore, exosomes could be used as drug delivery vehicles to effectively deliver therapeutic agents to oral cancer lesions to inhibit the progression of oral cancer, which is good news in the search for new strategies to treat oral cancer.96 Kandimalla et al149 demonstrated that the use of colostrum-derived exosomes as a vehicle for the tumour-targeted oral agent paclitaxel not only improved the therapeutic efficacy against lung cancer in situ, but also reduced the systemic and immunotoxicity of conventional intravenous injections, providing a solid theoretical basis for the use of oral exosomal agents of paclitaxel as an alternative to current traditional lung cancer treatment regimens. On this basis, it is also important for researchers to find alternative options for oral cancer treatment using exosomes as carriers of oral cancer-targeted oral agents.
Since oral cancer-derived exosomes transport DNA, RNA, and surface proteins related to the development of oral cancer, their expression can be regulated by artificial intervention and targeting to inhibit the progression and metastasis of oral cancer. And they can exert a series of relevant effects on the receptor cells of the body.198 Wang et al199 found that the expression of miRNA-655-3p was significantly downregulated in OSCC tissues and cell lines. Moreover, miR-655-3p was found to inhibit OSCC proliferation and invasion by regulating the PTEN/AKT signalling pathway. Therefore, targeting specific receptor cells and acting through exosomes as messengers loaded with miR-655-3p or other relevant substances capable of inhibiting the development of oral cancer may be a new therapeutic strategy for patients with OSCC. The effective delivery of specific miRNAs or siRNAs by exosomes has been developed to treat central nervous system diseases and cancers.146,196,200,201 Therefore, delivering specific DNA, RNA, or proteins via exosomes has considerable research value as an entry point for treating oral cancer.
However, we can artificially modify or edit the expression of exosome-related genes or surface proteins to specifically target the surface receptors of the target cells and achieve precise blockage of the occurrence of related responses or activation of interconnected signalling pathways after binding, thus achieving the purpose of precise cancer treatment.202 For example, some researchers have used programmable genotypic fusion vesicles with high affinity for SIRPα variants and PD-1 to block the effects of both CD47 and PD-L1 mechanisms, significantly enhancing the phagocytosis of cancer cells by macrophages and promoting antigen presentation to activate anti-tumour T-cell immunity. This bispecific targeting design better targets tumour cells and improves therapeutic efficacy, while reducing systemic side effects. It has also been demonstrated in model experiments of malignant melanoma and breast cancer.145 Therefore, engineered exosomes through gene editing or protein modification are also very promising for research in the targeted treatment of oral cancer (eg through targeted inhibition of angiogenesis, invasive metastasis, and chemoresistant metastasis, enhancement of chemotherapy sensitivity, and modulation of the immune response).
In addition, specific aptamers (also known as chemoantibodies) designed on the surface of drug-laden exosomes can guide drug-loaded exosomes to the appropriate cell surface and bind to the corresponding target site for action.202,203 RNA aptamers targeting vascular endothelial growth factor were the first to enter the clinic as therapeutic agents,204 heralding the great potential of such drug-loaded exosomes with aptamers designed on their surface for targeted treatment of oral cancer.The aptamer binds to the target with high affinity through the same mechanism as antibody-antigen binding.205,206 Compared to antibodies, aptamers have advantages, such as higher stability, lower toxicity, more remarkable tissue penetration properties, and absence of immunogenicity, making them more promising for application in precision medicine.207,208 Because aptamers are chemically synthesized without animals or cultured cells, they are not limited to expansion or batch differences.209 Therefore, novel artificially designed affinity ligands combined with exosomes loaded with specific drugs are expected to be the next generation of intelligently engineered exosomes for precision medicine. Aptamer-mediated exosome drug delivery has the advantages of low cost, high benefit, simple operation, and a good safety profile and is a research hotspot for intelligent nano-drug delivery systems. This heralds a new era of targeted delivery of nanomedicine for oral cancer treatment.
In addition to targeted therapy for oral cancer, exosomes may be used as vaccines to prevent oral cancer. Ovarian cancer ascites-derived exosomes in combination with granulocyte-macrophage colony-stimulating factor for colorectal cancer and autologous dendritic cell derived exosomes for the treatment of patients with metastatic melanoma have been previously suggested in Phase I clinical trials to be feasible, safe, and well-tolerated. These two types of subcutaneous immunotherapies may be options for treating advanced colorectal cancer and metastatic melanoma.210,211 Many scholars have developed vaccines by applying tumour-derived exosomes to restore tumour-specific immunity and promote tumour clearance.212 Therefore, a thorough understanding of the boundaries between oral cancer-derived exosomes and specific and intrinsic immunity can help explore the pathways by which oral cancer-derived exosomes affect the immune system to facilitate tumour clearance. However, most studies are still in the preclinical stage, and more prospective clinical trials are needed to support the application of exosomes in oral cancer treatment practice. Figure 4 outlines the opportunities for exosomes to be used in the diagnosis and treatment of oral cancer.
The Challenge of Exosomes in the Diagnosis and Treatment of Oral Cancer
There are still many problems to overcome before exosomes can be successfully used in clinical diagnosis and treatment. The study of exosomes in oral cancer diagnosis focuses on saliva, cells, and exosomes isolated from circulating plasma or serum. First, saliva has a complex composition and ensuring the stability of samples during transportation and storage remains challenging. Ensuring the stability of exosomes during long-term storage is an issue that needs to be studied. Because of their low content of specific proteins, serum-derived exosomes require large amounts of serum or culture medium to obtain sufficient exosomes.198 At present, although there are many methods to extract exosomes, the purity and quality of the obtained exosomes are still not high. Obtaining a large number of ideal exosomes is time-consuming, laborious, costly, and inefficient. Combined application of multiple exosome extraction methods may solve this problem. In addition, some investigators have proposed the application of exosomes mixed with inorganic/organic nanoparticles to address the shortcomings of inefficient exosome isolation, low drug loading rate, difficulty in characterisation, and lack of specific biomarkers.158 Exosomal hybrid inorganic/organic nanoparticles may provide good diagnostic and therapeutic functions; however, more attention and evaluation of their safety, biocompatibility, and biodegradability are needed to ensure that inorganic/organic materials are safe, non-toxic, and do not accumulate in vivo.
Second, there is still a lack of unified standards for the acquisition, separation, and purification methods of exosomes, and the reference range of diagnostic content. The performance of exosomes is affected by many pre-analysis factors,213 which reduces the reliability of exosome-based diagnosis of oral cancer. To overcome these obstacles, some researchers have established novel, highly sensitive, and specific exosome assays,214–216 but these assays are costly, complex, and time-consuming. Therefore, there is still a need to develop more economical, efficient, time-saving, and feasible exosome-based assays for clinical use of such a method for oral cancer diagnosis.
The application of exosomes in oral cancer treatment includes two aspects: drug delivery and targeted therapy. Before applying exosome-targeted drug delivery in clinical trials, the shortcomings of existing exosome delivery methods (such as low drug delivery rate) must be overcome to achieve clinical efficacy with a small number of exosomes, thus reducing their toxicity and cost.194 Another important challenge for exosome-based targeted drug delivery for oral cancer is the need to meet the requirements of national regulatory agencies and obtain approval for exosome-based therapeutics. At present, some studies have shown that targeted treatment of oral cancer can be achieved by modifying exosome surface proteins or their contents.145 However, these research results are still in the stage of animal experiments. Much clinical research support is still needed before these research results can be stably and reliably applied to clinical patients with oral cancer. However, clinical studies on exosomes in oral cancer treatment are usually time-consuming, laborious, and expensive. At present, although we have made great achievements in the application of exosome drug delivery, many challenges still need to be overcome (such as improving pharmacokinetic effects, optimizing targeting, and clinical application of mass production). Some investigators have suggested that it is highly unlikely that high-quality exosomes can be produced on a large scale for sustained clinical administration to achieve therapeutic effects. Therefore, premature clearance of exosomes in vivo remains one of the current challenges in the use of exosome-loaded drugs for oral cancer treatment.217 The use of exosome-targeted therapy for oral cancer still needs to address the following issues (including determining the route of exosome delivery, the optimal dose of exosomes to be administered, the frequency of treatment, and the time interval between exosome deliveries) to ultimately achieve optimal clinical efficacy of exosomes without any adverse effects.194
The development of an exosome-based vaccine for oral cancer seems promising.218,219 However, due to the potential of oral cancer-derived exosomes to inhibit anti-tumour responses and promote metastasis, extensive studies are needed to address safety issues related to oral cancer-derived exosomes. The heterogeneous origin and differential effects of oral cancer-derived exosomes may be a major challenge in future vaccine development. Therefore, a better understanding of oral cancer-derived exosomes and their effects on cancer progression is a challenging new research area. A significant amount of basic and clinical research is required before the currently proposed exosome-based vaccine for oral cancer prevention can be translated into clinical practice.
Despite these challenges, exosomes hold revolutionary implications and great promise for the diagnosis and treatment of oral cancer. As research continues, we expect the application of exosomes in oral cancer practice to arrive in the near future. Figure 5 summarizes the challenges of exosomes in the diagnostic and therapeutic applications related to oral cancer.
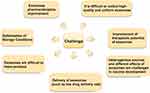 |
Figure 5 Challenges in the application of exosomes in the diagnosis and treatment of oral cancer. |
Summary
Exosomes are “messengers” that communicate between cells, transport their contents to recipient cells, and influence the progression of oral cancer by modulating host-related immune responses, tumour angiogenesis, drug resistance, or aggressive metastasis. The close association between exosomes and oral cancer has led to extensive studies of exosomes as oral cancer biomarkers, drug carriers, and therapeutic targets. Overall, exosome samples are obtained in a simple, low-cost, highly beneficial, less time-consuming, and non-invasive manner, which makes exosomes highly advantageous as biomarkers for oral cancer diagnosis and tracking disease progression, but the benefits and methods of exosome isolation and purification need to be improved. Exosomes have shown promise for clinical applications in cancer diagnosis.220,221 Based on the differentially expressed exosomal biomarkers between patients with oral cancer and healthy individuals and the biological functions and characteristics of exosomes, the use of exosomes to specifically diagnose oral cancer is almost within reach. Second, the advantages of exosomes, such as good biocompatibility, high cancer uptake, high stability, and ease of crossing various biological barriers, make them a suitable drug delivery system for targeted treatment of oral cancer. Currently, exosomal drug delivery has been reported in the treatment of cancer.149 However, there is a lack of research on exosomal drug delivery for oral cancer treatment, so it is worthy of further study in future work. Finally, initial results have been achieved in exosome-targeted therapy of oral cancer, and we can try to inhibit the progression of oral cancer by artificially integrating and wrapping relevant DNA, RNA, or proteins that can target and inhibit the progression of oral cancer into exosomes, so that they can interact with specific receptor cells of the body. Moreover, it can also influence the progression of oral cancer by regulating the expression of exosome-related DNA, RNA, and proteins, and intervene in the interaction between exosomes and receptor cells. Engineering smart exosomes to deliver drugs for precise targeting of oral cancer is currently a hot research topic that is receiving much attention. By artificially modifying the surface proteins of exosomes, the original biological functions of exosomes can be altered to specifically target oral cancer cells, thus achieving targeted drug delivery, or indirectly blocking the mechanism pathways related to oral cancer progression through aptamers, thereby inhibiting the progression of oral cancer. Notably, the development of exosome-based liquid biopsy diagnostic devices and vaccines for oral cancer prevention has also become a direction of research at this stage. Research on the role and mechanism of action of exosomes in oral cancer and the clinical application of diagnosis and treatment is still in its early stages. There are still many issues to be resolved, such as the timing of exosome collection, the quantity of sample required, the mode of obtaining exosomes, the preservation, isolation, and identification methods, the criteria of diagnosis, as well as the mode of administration, dose, frequency, and time window for drug administration. Therefore, further experimental studies and prospective clinical trials are needed to support the application of exosomes in clinical oral cancer diagnosis and treatment practice. Exosomes have shown application prospects in the diagnosis and treatment of oral cancer and further research on exosomes will provide new directions for these applications. We speculate that exosomes could be used to diagnose and treat patients with oral cancer in the future.
Abbreviations
ALIX, apoptosis-linked gene 2-interacting protein X; DTX, docetaxel; EGFR, epidermal growth factor receptor; EMT, Epithelial-mesenchymal transition; ESCRT, endosomal sorting complexes required for transport; HBMSCs, human bone marrow mesenchymal stem cells; HNSCC, head and neck squamous cell carcinoma; HSC-3DR, docetaxel resistant HSC-3 cells; HUVEC, human umbilical vein endothelial cell; ILVs, intraluminal vesicles; MSC, mesenchymal stem cells; MVBs, multivesicular bodies; NK, natural killer; NSF, N-ethylmaleimide-sensitive factor; NTECs, normal tongue epithelial cells; OSCC, oral squamous cell carcinoma; PDCD4, programmed cell death 4; PHM, primary human macrophage; PTEN, phosphatase and tensin homologue; SNARE, soluble N-ethylmaleimide-sensitive factor NSF attachment protein receptor; TAMs, tumour-associated macrophages; TGN, trans-Golgi network; TME, tumour microenvironment; TSG101, tumour susceptibility gene 101.
Acknowledgments
This work was supported by the Guangdong Science and Technology Program (2019A1515010408).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol. 2010;46:414–417. doi:10.1016/j.oraloncology.2010.03.009
2. Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 2017;67:51–64. doi:10.3322/caac.21384
3. Rengaswamy Sankaranarayanan KR, Amarasinghe H, Subramanian S, Johnson N. Cancer. editors, Gelband H, Jha P, Sankaranarayanan R, Horton S. In: Disease Control Priorities.
4. Liang D, Xiao-Feng H, Guan-Jun D, et al. Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim Biophys Acta. 2015;1852:2494–2503. doi:10.1016/j.bbadis.2015.08.011
5. Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8:11884–11894.
6. Su CC, Lee K-I, Chen M-K, et al. Cantharidin induced oral squamous cell carcinoma cell apoptosis via the JNK-regulated mitochondria and endoplasmic reticulum stress-related signaling pathways. PLoS One. 2016;11:e0168095. doi:10.1371/journal.pone.0168095
7. Vyhnalova T, Danek Z, Gachova D, Linhartova PB. The role of the oral microbiota in the etiopathogenesis of oral squamous cell carcinoma. Microorganisms. 2021;9:1549. doi:10.3390/microorganisms9081549
8. Jawert F, Nyman J, Olsson E, et al. Regular clinical follow-up of oral potentially malignant disorders results in improved survival for patients who develop oral cancer. Oral Oncol. 2021;121:105469. doi:10.1016/j.oraloncology.2021.105469
9. Liao CT, Chen H-N, Wen Y-W, et al. Association between the diagnosis-to-treatment interval and overall survival in Taiwanese patients with oral cavity squamous cell carcinoma. Eur J Cancer. 2017;72:226–234. doi:10.1016/j.ejca.2016.11.010
10. Bavle RM, Venugopal R, Konda P, Muniswamappa S, Makarla S. Molecular classification of oral squamous cell carcinoma. JCDR. 2016;10:Ze18–ze21. doi:10.7860/jcdr/2016/19967.8565
11. Brocklehurst PR, Baker SR, Speight PM. Oral cancer screening: what have we learnt and what is there still to achieve? Future Oncol. 2010;6:299–304. doi:10.2217/fon.09.163
12. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi:10.1002/ijc.29210
13. Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39:297–304. doi:10.1002/hed.24589
14. Sun X, Li P, Zhang M, Ma W. Research progress of exosomes in oral cancer. J Chin Oncol. 2020. doi:10.11735/j.issn.1671-170X.2020.06.B004
15. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi:10.1016/j.tcb.2016.11.003
16. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protocols Cell Biol. 2006;3:
17. Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi:10.1038/nm0598-594
18. Peters PJ, Geuze HJ, Van Donk HAD, et al. Molecules relevant for T cell-target cell interaction are present in cytolytic granules of human T lymphocytes. Eur J Immunol. 1989;19:1469–1475. doi:10.1002/eji.1830190819
19. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi:10.1084/jem.183.3.1161
20. Wolfers J, Lozier A, Raposo G, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi:10.1038/85438
21. Faure J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642–648. doi:10.1016/j.mcn.2005.12.003
22. Chung IM, Rajakumar G, Venkidasamy B, Subramanian U, Thiruvengadam M. Exosomes: current use and future applications. Clin Chim Acta. 2020;500:226–232. doi:10.1016/j.cca.2019.10.022
23. Khushman M, Bhardwaj A, Patel GK, et al. Exosomal markers (CD63 and CD9) expression pattern using immunohistochemistry in resected malignant and nonmalignant pancreatic specimens. Pancreas. 2017;46:782–788. doi:10.1097/MPA.0000000000000847
24. Pourhanifeh MH, Mahjoubin‐Tehran M, Shafiee A, et al. MicroRNAs and exosomes: small molecules with big actions in multiple myeloma pathogenesis. IUBMB Life. 2020;72:314–333. doi:10.1002/iub.2211
25. Gai C, Camussi F, Broccoletti R, et al. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer. 2018;18:439. doi:10.1186/s12885-018-4364-z
26. Abak A, Abhari A, Rahimzadeh S. Exosomes in cancer: small vesicular transporters for cancer progression and metastasis, biomarkers in cancer therapeutics. PeerJ. 2018;6:e4763. doi:10.7717/peerj.4763
27. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi:10.1083/jcb.201211138
28. Sohn W, Kim J, Kang SH, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. doi:10.1038/emm.2015.68
29. Wang H, Hou L, Li A, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. doi:10.1155/2014/864894
30. Pluchino S, Smith JA. Explicating exosomes: reclassifying the rising stars of intercellular communication. Cell. 2019;177:225–227. doi:10.1016/j.cell.2019.03.020
31. Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi:10.1038/s41556-018-0250-9
32. Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi:10.1038/ncb1596
33. Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi:10.1016/j.gpb.2015.02.001
34. Whiteside TL. Tumor-derived exosomes and their role in tumor-induced immune suppression. Vaccines. 2016;4:35. doi:10.3390/vaccines4040035
35. Ning Y, Shen K, Wu Q, et al. Tumor exosomes block dendritic cells maturation to decrease the T cell immune response. Immunol Lett. 2018;199:36–43. doi:10.1016/j.imlet.2018.05.002
36. Poggio M, Hu T, Pai -C-C, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177:414–427.e413. doi:10.1016/j.cell.2019.02.016
37. Yu W, Hurley J, Roberts D, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–477. doi:10.1016/j.annonc.2021.01.074
38. Nolte-’t Hoen EN, Buermans HPJ, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi:10.1093/nar/gks658
39. Ayyar KK, Moss AC. Exosomes in intestinal inflammation. Front Pharmacol. 2021;12:658505. doi:10.3389/fphar.2021.658505
40. Surman M, Drożdż A, Stępień E, Przybyło M. Extracellular vesicles as drug delivery systems - methods of production and potential therapeutic applications. Curr Pharm Des. 2019;25:132–154. doi:10.2174/1381612825666190306153318
41. de Jong OG, Kooijmans SAA, Murphy DE, et al. Drug delivery with extracellular vesicles: from imagination to innovation. Acc Chem Res. 2019;52:1761–1770. doi:10.1021/acs.accounts.9b00109
42. Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Salo T, Vered M. Morphological and molecular features of oral fluid-derived exosomes: oral cancer patients versus healthy individuals. J Cancer Res Clin Oncol. 2016;142:101–110. doi:10.1007/s00432-015-2005-3
43. Teng F, Fussenegger M. Shedding light on extracellular vesicle biogenesis and bioengineering. Adv Sci. 2020;8:2003505. doi:10.1002/advs.202003505
44. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev. 2002;2:569–579. doi:10.1038/nri855
45. Batista BS, Eng WS, Pilobello KT, Hendricks-Muñoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. J Proteome Res. 2011;10:4624–4633. doi:10.1021/pr200434y
46. Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. doi:10.1182/blood.V94.11.3791
47. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–312. doi:10.1007/s10571-016-0366-z
48. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev. 2018;19:213–228. doi:10.1038/nrm.2017.125
49. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. doi:10.1126/science.aau6977
50. Żbikowski A, Błachnio-Zabielska A, Galli M, Zabielski P. Adipose-derived exosomes as possible players in the development of insulin resistance. Int J Mol Sci. 2021;22:7427. doi:10.3390/ijms22147427
51. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1–11. doi:10.1016/j.pharmthera.2018.02.013
52. Ciardiello C, Cavallini L, Spinelli C, et al. Focus on extracellular vesicles: new frontiers of cell-to-cell communication in cancer. Int J Mol Sci. 2016;17:175. doi:10.3390/ijms17020175
53. Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34:214–219. doi:10.1016/j.bcmd.2005.03.002
54. Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. JoVE. 2012;e3037. doi:10.3791/3037
55. Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi:10.1002/pmic.200800109
56. Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8:307. doi:10.3390/cells8040307
57. Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. 2012;44:2060–2064. doi:10.1016/j.biocel.2012.08.007
58. Fleming A, Sampey G, Chung M-C, et al. The carrying pigeons of the cell: exosomes and their role in infectious diseases caused by human pathogens. Pathog Dis. 2014;71:109–120. doi:10.1111/2049-632X.12135
59. Gon Y, Shimizu T, Mizumura K, Maruoka S, Hikichi M. Molecular techniques for respiratory diseases: microRNA and extracellular vesicles. Respirology. 2020;25:149–160. doi:10.1111/resp.13756
60. Sasaki R, Kanda T, Yokosuka O, et al. Exosomes and hepatocellular carcinoma: from bench to bedside. Int J Mol Sci. 2019;20:1406. doi:10.3390/ijms20061406
61. Kimura K, Hohjoh H, Fukuoka M, et al. Circulating exosomes suppress the induction of regulatory T cells via let-7i in multiple sclerosis. Nat Commun. 2018;9:17. doi:10.1038/s41467-017-02406-2
62. Lu Y, Zheng Z, Yuan Y, et al. The emerging role of exosomes in oral squamous cell carcinoma. Front Cell Dev Biol. 2021;9:628103. doi:10.3389/fcell.2021.628103
63. Liu Y, Cao X. The origin and function of tumor-associated macrophages. Cell Mol Immunol. 2015;12:1–4. doi:10.1038/cmi.2014.83
64. Rhee I. Diverse macrophages polarization in tumor microenvironment. Arch Pharm Res. 2016;39:1588–1596. doi:10.1007/s12272-016-0820-y
65. Zhan C, Yang X, Yin X, Hou J. Exosomes and other extracellular vesicles in oral and salivary gland cancers. Oral Dis. 2020;26:865–875. doi:10.1111/odi.13172
66. You Y, Tian Z, Du Z, et al. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J Exp Clin Cancer Res. 2022;41:10. doi:10.1186/s13046-021-02222-z
67. Pang X, Wang S, Zhang M, et al. Correction to: OSCC cell‑secreted exosomal CMTM6 induced M2‑like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunol Immunother. 2022;71:505–506. doi:10.1007/s00262-021-03113-0
68. Jian, Y, Li, L, Wei-Dong T. Evaluation of isolation and identification methods of exosomes from different biofluids. 2021.
69. Gardiner C, Vizio DD, Sahoo S, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi:10.3402/jev.v5.32945
70. Helwa I, Cai J, Drewry MD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12(1):e0170628. doi:10.1371/journal.pone.0170628
71. Mol EA, Goumans MJ, Doevendans PA, Sluijter JPG, Vader P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine. 2017;13:2061–2065. doi:10.1016/j.nano.2017.03.011
72. Tian Y, Gong M, Hu Y, et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J Extracell Vesicles. 2020;9:1697028. doi:10.1080/20013078.2019.1697028
73. Gamez-Valero A, Monguió-Tortajada M, Carreras-Planella L, et al. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci Rep. 2016;6:33641. doi:10.1038/srep33641
74. Duong P, Chung A, Bouchareychas L, Raffai RL. Cushioned-Density Gradient Ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS One. 2019;14:e0215324. doi:10.1371/journal.pone.0215324
75. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi:10.7150/thno.18133
76. Szatanek R, Baj-Krzyworzeka M, Zimoch J, et al. The methods of choice for Extracellular Vesicles (EVs) characterization. Int J Mol Sci. 2017;18:1153. doi:10.3390/ijms18061153
77. Monguio-Tortajada M, Galvez-Monton C, Bayes-Genis A, Roura S, Borras FE. Extracellular vesicle isolation methods: rising impact of size-exclusion chromatography. Cell Mol Life Sci. 2019;76:2369–2382. doi:10.1007/s00018-019-03071-y
78. Nolan JP, Duggan E. Analysis of individual extracellular vesicles by flow cytometry. Methods Mol Biol. 2018;1678:79–92. doi:10.1007/978-1-4939-7346-0_5
79. Barros FM, Carneiro F, Machado JC, Melo SA. Exosomes and immune response in cancer: friends or foes? Front Immunol. 2018;9:730. doi:10.3389/fimmu.2018.00730
80. Wang J, Hendrix A, Hernot S, et al. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood. 2014;124:555–566. doi:10.1182/blood-2014-03-562439
81. Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi:10.1038/nrc2621
82. Jiang E, Xu Z, Wang M, et al. Tumoral microvesicle-activated glycometabolic reprogramming in fibroblasts promotes the progression of oral squamous cell carcinoma. FASEB J. 2019;33:5690–5703. doi:10.1096/fj.201802226R
83. Liu Y, Gu Y, Han Y, et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30:243–256. doi:10.1016/j.ccell.2016.06.021
84. Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21:139–146. doi:10.1016/j.semcancer.2011.01.002
85. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi:10.1146/annurev-cellbio-101512-122326
86. Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151–37168. doi:10.18632/oncotarget.6022
87. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi:10.1016/j.cell.2009.01.002
88. Langevin S, Kuhnell D, Parry T, et al. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget. 2017;8:82459–82474. doi:10.18632/oncotarget.19614
89. He L, Ping F, Fan Z, et al. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed Pharmacother. 2020;121:109553. doi:10.1016/j.biopha.2019.109553
90. Sharma S, Gillespie BM, Palanisamy V, Gimzewski JK. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir. 2011;27:14394–14400. doi:10.1021/la2038763
91. Cui J, Wang H, Zhang X, et al. Exosomal miR-200c suppresses chemoresistance of docetaxel in tongue squamous cell carcinoma by suppressing TUBB3 and PPP2R1B. Aging. 2020;12:6756–6773. doi:10.18632/aging.103036
92. Xiao M, Zhang J, Chen W, Chen W. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migration in oral squamous cell carcinoma. J Exp Clin Cancer Res. 2018;37:143. doi:10.1186/s13046-018-0815-2
93. Cai J, Qiao B, Gao N, Lin N, He W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Cell Physiol. 2019;316:C731–c740. doi:10.1152/ajpcell.00366.2018
94. Mori K, Hiroi M, Shimada J, Ohmori Y. Infiltration of m2 tumor-associated macrophages in oral squamous cell carcinoma correlates with tumor malignancy. Cancers. 2011;3:3726–3739. doi:10.3390/cancers3043726
95. Pang X, Wang -S-S, Zhang M, et al. OSCC cell-secreted exosomal CMTM6 induced M2-like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunol Immunother. 2021;70:1015–1029. doi:10.1007/s00262-020-02741-2
96. Baig MS, Roy A, Rajpoot S, et al. Tumor-derived exosomes in the regulation of macrophage polarization. Inflamm Res. 2020;69:435–451. doi:10.1007/s00011-020-01318-0
97. Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi:10.1038/nature04478
98. He S, Zhang W, Li X, et al. Oral squamous cell carcinoma (OSCC)-derived exosomal MiR-221 targets and regulates phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) to promote human umbilical vein endothelial cells migration and tube formation. Bioengineered. 2021;12:2164–2174. doi:10.1080/21655979.2021.1932222
99. Wang H, Wang L, Zhou X, et al. OSCC exosomes regulate miR-210-3p targeting EFNA3 to promote oral cancer angiogenesis through the PI3K/AKT pathway. Biomed Res Int. 2020;2020:2125656. doi:10.1155/2020/2125656
100. Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. doi:10.1038/srep38750
101. Kawakubo-Yasukochi T, Morioka M, Hazekawa M, et al. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol Carcinog. 2018;57:295–302. doi:10.1002/mc.22744
102. Masyuk AI, Masyuk TV, Larusso NF. Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J Hepatol. 2013;59:621–625. doi:10.1016/j.jhep.2013.03.028
103. Zhang W, Zhou X, Zhang H, et al. Extracellular vesicles in diagnosis and therapy of kidney diseases. Ren Physiol. 2016;311:F844–f851. doi:10.1152/ajprenal.00429.2016
104. Alipoor SD, Mortaz E, Garssen J, et al. Exosomes and exosomal miRNA in respiratory diseases. Mediators Inflamm. 2016;2016:5628404. doi:10.1155/2016/5628404
105. Jansen F, Li Q. Exosomes as diagnostic biomarkers in cardiovascular diseases. Adv Exp Med Biol. 2017;998:61–70. doi:10.1007/978-981-10-4397-0_4
106. Zhang Y, Hu YW, Zheng L, Wang Q. Characteristics and roles of exosomes in cardiovascular disease. DNA Cell Biol. 2017;36:202–211. doi:10.1089/dna.2016.3496
107. Kanninen KM, Bister N, Koistinaho J, Malm T. Exosomes as new diagnostic tools in CNS diseases. Biochim Biophys Acta. 2016;1862:403–410. doi:10.1016/j.bbadis.2015.09.020
108. Luo Y, Liu F, Guo J, Gui R. Upregulation of circ_0000199 in circulating exosomes is associated with survival outcome in OSCC. Sci Rep. 2020;10:13739. doi:10.1038/s41598-020-70747-y
109. Guo H, Jiang W, Huang S, Huang X, Li C. Serum exosome-derived biomarkers for the early detection of oral squamous cell carcinoma. Mol Cell Biochem. 2021;476:4435–4447. doi:10.1007/s11010-021-04254-7
110. Martinez P, Birkbak NJ, Gerlinger M, et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol. 2013;230:356–364. doi:10.1002/path.4214
111. Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol. 2017;3:740–741. doi:10.1001/jamaoncol.2016.2835
112. Tadimety A, Closson A, Li C, et al. Advances in liquid biopsy on-chip for cancer management: technologies, biomarkers, and clinical analysis. Crit Rev Clin Lab Sci. 2018;55:140–162. doi:10.1080/10408363.2018.1425976
113. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi:10.1038/nrclinonc.2013.110
114. Balasaheb Mali S, Dahivelkar S. Liquid biopsy = Individualized cancer management: diagnosis, monitoring treatment and checking recurrence and metastasis. Oral Oncol. 2021;123:105588. doi:10.1016/j.oraloncology.2021.105588
115. Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22:241–248.
116. Sas R, Dawes C. The intra-oral distribution of unstimulated and chewing-gum-stimulated parotid saliva. Arch Oral Biol. 1997;42:469–474. doi:10.1016/s0003-9969(97)00045-9
117. Sun QF, Sun QH, Du J, Wang S. Differential gene expression profiles of normal human parotid and submandibular glands. Oral Dis. 2008;14:500–509. doi:10.1111/j.1601-0825.2007.01408.x
118. Deutsch O, Fleissig Y, Zaks B, et al. An approach to remove alpha amylase for proteomic analysis of low abundance biomarkers in human saliva. Electrophoresis. 2008;29:4150–4157. doi:10.1002/elps.200800207
119. Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: emerging roles in systemic disease. Int J Biol Sci. 2018;14:633–643. doi:10.7150/ijbs.25018
120. Gonzalez-Begne M, Lu B, Han X, et al. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res. 2009;8:1304–1314. doi:10.1021/pr800658c
121. Principe S, Hui AB-Y, Bruce J, et al. Tumor-derived exosomes and microvesicles in head and neck cancer: implications for tumor biology and biomarker discovery. Proteomics. 2013;13:1608–1623. doi:10.1002/pmic.201200533
122. Li C, Zhou Y, Liu J, et al. Potential markers from serum-purified exosomes for detecting oral squamous cell carcinoma metastasis. Cancer Epidemiol Biomarkers Prev. 2019;28:1668–1681. doi:10.1158/1055-9965.Epi-18-1122
123. Rabinowits G, Bowden M, Flores LM, et al. Comparative analysis of microRNA expression among benign and malignant tongue tissue and plasma of patients with tongue cancer. Front Oncol. 2017;7:191. doi:10.3389/fonc.2017.00191
124. Sedykh S, Kuleshova A, Nevinsky G. Milk exosomes: perspective agents for anticancer drug delivery. Int J Mol Sci. 2020;21:6646. doi:10.3390/ijms21186646
125. Patra JK, Das G, Fraceto LF, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018;16:71. doi:10.1186/s12951-018-0392-8
126. Ferreira D, Moreira JN, Rodrigues LR. New advances in exosome-based targeted drug delivery systems. Crit Rev Oncol Hematol. 2022;172:103628. doi:10.1016/j.critrevonc.2022.103628
127. Perez AT, Domenech GH, Frankel C, Vogel CL. Pegylated liposomal doxorubicin (Doxil) for metastatic breast cancer: the Cancer Research Network, Inc., experience. Cancer Invest. 2002;20:22–29. doi:10.1081/CNV-120014883
128. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta pharmaceutica Sinica B. 2016;6:287–296. doi:10.1016/j.apsb.2016.02.001
129. Jiang X-C, Gao J-Q. Exosomes as novel bio-carriers for gene and drug delivery. Int J Pharm. 2017;521:167–175. doi:10.1016/j.ijpharm.2017.02.038
130. Bunggulawa EJ, Wang W, Yin T, et al. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnology. 2018;16:81. doi:10.1186/s12951-018-0403-9
131. Sun D, Zhuang X, Xiang X, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606–1614. doi:10.1038/mt.2010.105
132. Pascucci L, Coccè V, Bonomi A, et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J Control Release. 2014;192:262–270. doi:10.1016/j.jconrel.2014.07.042
133. Zeelenberg IS, Ostrowski M, Krumeich S, et al. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 2008;68:1228–1235. doi:10.1158/0008-5472.CAN-07-3163
134. Johnsen KB, Gudbergsson JM, Skov MN, et al. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68:2125–2138. doi:10.1007/s10616-016-9952-7
135. Fuhrmann G, Serio A, Mazo M, Nair R, Stevens MM. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J Control Release. 2015;205:35–44. doi:10.1016/j.jconrel.2014.11.029
136. Luan X, Sansanaphongpricha K, Myers I, et al. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754–763. doi:10.1038/aps.2017.12
137. Kim MS, Haney MJ, Zhao Y, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655–664. doi:10.1016/j.nano.2015.10.012
138. Sato YT, Umezaki K, Sawada S, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. 2016;6:21933. doi:10.1038/srep21933
139. Liao W, Du Y, Zhang C, et al. Exosomes: the next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta biomaterialia. 2019;86:1–14. doi:10.1016/j.actbio.2018.12.045
140. Yang Y, Chen Y, Zhang F, Zhao Q, Zhong H. Increased anti-tumour activity by exosomes derived from doxorubicin-treated tumour cells via heat stress. Int J Hyperthermia. 2015;31:498–506. doi:10.3109/02656736.2015.1036384
141. González-Sarrías A, Iglesias-Aguirre CE, Cortés-Martín A, et al. Milk-derived exosomes as nanocarriers to deliver curcumin and resveratrol in breast tissue and enhance their anticancer activity. Int J Mol Sci. 2022;23:2860. doi:10.3390/ijms23052860
142. Yim N, Ryu S-W, Choi K, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat Commun. 2016;7:12277. doi:10.1038/ncomms12277
143. Zhang Q, Zhang H, Ning T, et al. Exosome-delivered c-Met siRNA could reverse chemoresistance to cisplatin in gastric cancer. Int J Nanomedicine. 2020;15:2323–2335. doi:10.2147/IJN.S231214
144. Wang J, Li G, Tu C, et al. High-throughput single-cell analysis of exosome mediated dual drug delivery, in vivo fate and synergistic tumor therapy. Nanoscale. 2020;12:13742–13756. doi:10.1039/d0nr02344b
145. Meng QF, Zhao Y, Dong C, et al. Genetically programmable fusion cellular vesicles for cancer immunotherapy. Angewandte Chemie. 2021. doi:10.1002/anie.202108342
146. Mendt M, Kamerkar S, Sugimoto H, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3. doi:10.1172/jci.insight.99263
147. Zhu X, Badawi M, Pomeroy S, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6:1324730. doi:10.1080/20013078.2017.1324730
148. Kordelas L, Rebmann V, Ludwig A-K, et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973. doi:10.1038/leu.2014.41
149. Kandimalla R, Aqil F, Alhakeem SS, et al. Targeted oral delivery of paclitaxel using colostrum-derived exosomes. Cancers. 2021;13:3700. doi:10.3390/cancers13153700
150. Li L, Lu S, Liang X, et al. gammadeltaTDEs: an efficient delivery system for miR-138 with anti-tumoral and immunostimulatory roles on oral squamous cell carcinoma. Mol Ther Nucleic Acids. 2019;14:101–113. doi:10.1016/j.omtn.2018.11.009
151. Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi:10.1016/j.gde.2005.08.005
152. Wang Z, Yan J, Zou T, Gao H. MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc. Oncol Lett. 2018;16:2243–2250. doi:10.3892/ol.2018.8967
153. Higaki M, Shintani T, Hamada A, Rosli SNZ, Okamoto T. Eldecalcitol (ED-71)-induced exosomal miR-6887-5p suppresses squamous cell carcinoma cell growth by targeting heparin-binding protein 17/fibroblast growth factor-binding protein-1 (HBp17/FGFBP-1). In vitro cellular & developmental biology. Animal. 2020;56:222–233. doi:10.1007/s11626-020-00440-x
154. Xie C, Du LY, Guo F, Li X, Cheng B. Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Mol Cell Biochem. 2019;458:11–26. doi:10.1007/s11010-019-03526-7
155. Chan M-H, Chang Z-X, Huang C-YF, et al. Integrated therapy platform of exosomal system: hybrid inorganic/organic nanoparticles with exosomes for cancer treatment. Nanoscale Horiz. 2022;7:352–367. doi:10.1039/d1nh00637a
156. Wu D, Kang L, Tian J, et al. Exosomes derived from bone mesenchymal stem cells with the stimulation of FeO nanoparticles and static magnetic field enhance wound healing through upregulated miR-21-5p. Int J Nanomedicine. 2020;15:7979–7993. doi:10.2147/IJN.S275650
157. Yang L, Patel KD, Rathnam C, et al. Harnessing the therapeutic potential of extracellular vesicles for biomedical applications using multifunctional magnetic nanomaterials. Small. 2022;18:e2104783. doi:10.1002/smll.202104783
158. Chen Y, Hou S. Application of magnetic nanoparticles in cell therapy. Stem Cell Res Ther. 2022;13:135. doi:10.1186/s13287-022-02808-0
159. Vallina C, López-Pintor RM, González-Serrano J, et al. Genes involved in the epithelial-mesenchymal transition in oral cancer: a systematic review. Oral Oncol. 2021;117:105310. doi:10.1016/j.oraloncology.2021.105310
160. Fujiwara T, Eguchi T, Sogawa C, et al. Carcinogenic epithelial-mesenchymal transition initiated by oral cancer exosomes is inhibited by anti-EGFR antibody cetuximab. Oral Oncol. 2018;86:251–257. doi:10.1016/j.oraloncology.2018.09.030
161. Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi:10.1038/s41388-018-0619-z
162. Liu T, Chen G, Sun D, et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2017;49:808–816. doi:10.1093/abbs/gmx078
163. Li L, Li C, Wang S, et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi:10.1158/0008-5472.Can-15-1625
164. Tomita R, Sasabe E, Tomomura A, Yamamoto T. Macrophage‑derived exosomes attenuate the susceptibility of oral squamous cell carcinoma cells to chemotherapeutic drugs through the AKT/GSK‑3β pathway. Oncol Rep. 2020. doi:10.3892/or.2020.7748
165. Zhang B, Yin Y, Lai RC, Lim SK. Immunotherapeutic potential of extracellular vesicles. Front Immunol. 2014;5:518. doi:10.3389/fimmu.2014.00518
166. Li XB, Zhang ZR, Schluesener HJ, Xu SQ. Role of exosomes in immune regulation. J Cell Mol Med. 2006;10:364–375. doi:10.1111/j.1582-4934.2006.tb00405.x
167. Tekiki N, Fujita M, Okui T, et al. Dynamic contrast-enhanced MRI as a predictor of programmed death ligand-1 expression in patients with oral squamous cell carcinoma. Oncol Lett. 2021;22:778. doi:10.3892/ol.2021.13039
168. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi:10.1016/j.ccell.2015.03.001
169. Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi:10.1200/JCO.2009.26.7609
170. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi:10.1038/nature12634
171. Wang Y, Qin X, Zhu X, et al. Oral cancer-derived exosomal NAP1 enhances cytotoxicity of natural killer cells via the IRF-3 pathway. Oral Oncol. 2018;76:34–41. doi:10.1016/j.oraloncology.2017.11.024
172. Ding L, Ren J, Zhang D, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39:397–406. doi:10.1093/carcin/bgy006
173. Yang E, Wang X, Gong Z, et al. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther. 2020;5:242. doi:10.1038/s41392-020-00359-5
174. Wendler F, Favicchio R, Simon T, et al. Extracellular vesicles swarm the cancer microenvironment: from tumor-stroma communication to drug intervention. Oncogene. 2017;36:877–884. doi:10.1038/onc.2016.253
175. Sento S, Sasabe E, Yamamoto T, Tang C-H. Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell-derived exosomes. PLoS One. 2016;11:e0148454. doi:10.1371/journal.pone.0148454
176. Momen-Heravi F, Trachtenberg AJ, Kuo WP, Cheng YS. Genomewide study of salivary microRNAs for detection of oral cancer. J Dent Res. 2014;93:86s–93s. doi:10.1177/0022034514531018
177. Zahran F, Ghalwash D, Shaker O, Al-Johani K, Scully C. Salivary microRNAs in oral cancer. Oral Dis. 2015;21:739–747. doi:10.1111/odi.12340
178. Cai J, Han Y, Ren H, et al. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5:227–238. doi:10.1093/jmcb/mjt011
179. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi:10.1074/jbc.C113.532267
180. Kalluri R, LeBleu VS. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb Symp Quant Biol. 2016;81:275–280. doi:10.1101/sqb.2016.81.030932
181. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi:10.1038/cr.2014.44
182. Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: challenges and opportunities. J Cell Physiol. 2018;233:6370–6380. doi:10.1002/jcp.26481
183. Alhasan AH, Scott AW, Wu JJ, et al. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci USA. 2016;113:10655–10660. doi:10.1073/pnas.1611596113
184. Halvaei S, Daryani S, Eslami-S Z, et al. Exosomes in cancer liquid biopsy: a focus on breast cancer. Mol Ther. 2018;10:131–141. doi:10.1016/j.omtn.2017.11.014
185. Stevic I, Müller V, Weber K, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018;16:179. doi:10.1186/s12916-018-1163-y
186. Zhou X, Zhu W, Li H, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251. doi:10.1038/srep11251
187. Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. doi:10.3402/jev.v5.31292
188. Castillo J, Bernard V, San Lucas FA, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29:223–229. doi:10.1093/annonc/mdx542
189. Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. doi:10.1038/ncomms4591
190. Sharma R, Huang X, Brekken RA, Schroit AJ. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer. 2017;117:545–552. doi:10.1038/bjc.2017.183
191. Wang A, Liu J, Zhuang X, et al. Identification and comparison of piRNA expression profiles of exosomes derived from human stem cells from the apical papilla and bone marrow mesenchymal stem cells. Stem Cells Dev. 2020;29:511–520. doi:10.1089/scd.2019.0277
192. Arrighetti N, Corbo C, Evangelopoulos M, et al. Exosome-like nanovectors for drug delivery in cancer. Curr Med Chem. 2019;26:6132–6148. doi:10.2174/0929867325666180831150259
193. Pullan JE, Confeld MI, Osborn JK, et al. Exosomes as drug carriers for cancer therapy. Mol Pharm. 2019;16:1789–1798. doi:10.1021/acs.molpharmaceut.9b00104
194. Butreddy A, Kommineni N, Dudhipala N. Exosomes as naturally occurring vehicles for delivery of biopharmaceuticals: insights from drug delivery to clinical perspectives. Nanomaterials. 2021;11:1481. doi:10.3390/nano11061481
195. Yeo RW, Lai RC, Zhang B, et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi:10.1016/j.addr.2012.07.001
196. Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi:10.1038/nature22341
197. Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:23743. doi:10.3402/jev.v3.23743
198. Hu Y, Zhang R, Chen G. Exosome and secretion: action on? Adv Exp Med Biol. 2020;1248:455–483. doi:10.1007/978-981-15-3266-5_19
199. Wang Q, Lv L, Li Y, Ji H. MicroRNA‑655 suppresses cell proliferation and invasion in oral squamous cell carcinoma by directly targeting metadherin and regulating the PTEN/AKT pathway. Mol Med Rep. 2018;18:3106–3114. doi:10.3892/mmr.2018.9292
200. Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi:10.1016/j.canlet.2013.02.019
201. Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi:10.1038/mt.2012.180
202. Tran PHL, Xiang D, Tran TTD, et al. Exosomes and nanoengineering: a match made for precision therapeutics. Adv Mater. 2020;32:e1904040. doi:10.1002/adma.201904040
203. Kim MS, Haney MJ, Zhao Y, et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14:195–204. doi:10.1016/j.nano.2017.09.011
204. Ruckman J, Green LS, Beeson J, et al. 2’-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556–20567. doi:10.1074/jbc.273.32.20556
205. Zhao C, Busch DJ, Vershel CP, Stachowiak JC. Multifunctional transmembrane protein ligands for cell-specific targeting of plasma membrane-derived vesicles. Small. 2016;12:3837–3848. doi:10.1002/smll.201600493
206. Zhou G, Latchoumanin O, Hebbard L, et al. Aptamers as targeting ligands and therapeutic molecules for overcoming drug resistance in cancers. Adv Drug Deliv Rev. 2018;134:107–121. doi:10.1016/j.addr.2018.04.005
207. Gefen T, Castro I, Muharemagic D, et al. A TIM-3 oligonucleotide aptamer enhances T cell functions and potentiates tumor immunity in mice. Mol Ther. 2017;25:2280–2288. doi:10.1016/j.ymthe.2017.06.023
208. Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181–202. doi:10.1038/nrd.2016.199
209. Rahimizadeh K, AlShamaileh H, Fratini M, et al. Development of cell-specific aptamers: recent advances and insight into the selection procedures. Molecules. 2017;22:2070. doi:10.3390/molecules22122070
210. Dai S, Wei D, Wu Z, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–790. doi:10.1038/mt.2008.1
211. Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J Transl Med. 2005;3:10. doi:10.1186/1479-5876-3-10
212. Wan M, Ning B, Spiegel S, Lyon CJ, Hu TY. Tumor-derived exosomes (TDEs): how to avoid the sting in the tail. Med Res Rev. 2020;40:385–412. doi:10.1002/med.21623
213. Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3–10. doi:10.1016/j.ymeth.2015.02.019
214. Liu W, Li J, Wu Y, et al. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens Bioelectron. 2018;102:204–210. doi:10.1016/j.bios.2017.11.033
215. Xing S, Lu Z, Huang Q, et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics. 2020;10:10262–10273. doi:10.7150/thno.49047
216. Liu C, Xu X, Li B, et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018;18:4226–4232. doi:10.1021/acs.nanolett.8b01184
217. Ketabat F, Pundir M, Mohabatpour F, et al. Controlled drug delivery systems for oral cancer treatment-current status and future perspectives. Pharmaceutics. 2019;11:302. doi:10.3390/pharmaceutics11070302
218. Stremersch S, De Smedt SC, Raemdonck K. Therapeutic and diagnostic applications of extracellular vesicles. J Control Release. 2016;244:167–183. doi:10.1016/j.jconrel.2016.07.054
219. Chen T, Guo J, Yang M, Zhu X, Cao X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J Immunol. 2011;186:2219–2228. doi:10.4049/jimmunol.1002991
220. McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2:882–889. doi:10.1001/jamaoncol.2016.0097
221. McKiernan J, Donovan MJ, Margolis E, et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2–10 ng/mL at initial biopsy. Eur Urol. 2018;74:731–738. doi:10.1016/j.eururo.2018.08.019
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.