Back to Journals » Cancer Management and Research » Volume 12
Curcumin Modifies Epithelial–Mesenchymal Transition in Colorectal Cancer Through Regulation of miR-200c/EPM5
Received 4 May 2020
Accepted for publication 21 August 2020
Published 1 October 2020 Volume 2020:12 Pages 9405—9415
DOI https://doi.org/10.2147/CMAR.S260129
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Hui Wang, Xiaolong Cai, Longyang Ma
Department of Emergency Surgery, Shaanxi Provincial People’s Hospital, Xi’an, Shaanxi Province, People’s Republic of China
Correspondence: Longyang Ma Email [email protected]
Background: The serious side effect of current conventional treatments for patients with metastatic colorectal cancer (CRC) highlights the requirement of an alternative treatment strategy. Natural compounds, such as curcumin, have been gained much attention due to its low toxicity and anti-tumor effect.
Methods: qPCR and Western blot were used to measure the molecular changes induced by curcumin. Wound-healing assay and transwell assay were conducted to study the effect on cell migration and invasion. RT1 PCR array was performed to identify the miRNAs involved in curcumin-repressed EMT. Three algorithms and luciferase reporter assay were used to identify EPM5 as a target of miR-200c. The bioinformatical analysis of TCGA-COAD and other CRC cohorts were used to examine the association of EPM5 with EMT signatures and clinical relevance. The ectopic expression or siRNA-mediated knockdown of EPM5 was applied to study the role of EPM5 in CRC.
Results: Treatment with curcumin changed the epithelial–mesenchymal transition (EMT)-related gene expression, repressed cell migration and invasion in CRC cells. Its anti-tumor capability required the upregulation of miR-200c. EPM5 was a direct target of miR-200c and enriched in the consensus molecular subtype (CMS) 4 of CRC. Ectopic expression of EPM5 alone was sufficient to induce EMT in CRC. Downregulation of EPM5 was necessary for curcumin-repressed EMT, migration, and invasion. Higher expression of EPM5 was associated with the advanced TNM stages and poor survival in CRC.
Conclusion: Our data provide the first evidence that the curcumin inhibits EMT in CRC by upregulation of miR-200c and downregulation of EPM5, and the use of curcumin might be able to prevent or delay CRC progression.
Keywords: curcumin, EMT, miR-200c, EPM5, CRC
Introduction
Metastasis, which is defined as the spread of tumor cells from the primary sites to a second part within the patients, is the major cause of death in colorectal cancer (CRC).1,2 Despite the tremendous advances achieved in the past years, there is a paucity of effective treatment for patients with metastatic CRC tumors.3 Thus, a better understanding of the underlying metastasis mechanism and novel compounds was urgently needed for proposing an effective treatment strategy.4
Epithelial–mesenchymal transition (EMT) has been considered as a key metastatic step, which is associated with poor prognosis in CRC.5,6 EMT is a complex process including the dissolution of cell-cell junction and loss of apicobasolateral polarity, which resulted in the formation of migratory mesenchymal cells with invasive properties.7,8 The decrease of E-cadherin expression and increase of VIM is generally accepted as a hallmark of the EMT process regulated by various transcriptional factors, such as ZEB1, ZEB2, TWIST, SLUG and SNAIL.9–11 In addition to these EMT transcriptional factors (EMT-TFs), recent evidence showed that microRNAs (miRNAs) are also critical for EMT regulation.6,8 For instance, miRNAs from the miR-200 family repress EMT by directly targeting and downregulating ZEB1/ZEB1/SNAIL/SLUG via miR-200-binding sites located within their 3′UTRs, resulting in enhanced E-cadherin expression and inhibition of cancer cell migration and motility.12–14 On the contrary, theses EMT-TFs bind directly to the common promoter regions of the miR-200c family and cause transcriptional repression.15 Therefore, the balance between miR-200c and EMT-TFs determines, at least in part, the status of epithelial or mesenchymal phenotype.16
Curcumin, a naturally occurring polyphenolic compound from Curcuma longa, has long been expected to be a therapeutic or preventive agent for several major human diseases due to its non-toxic and various therapeutic properties including anti-tumor activity.17,18 Curcumin exhibits anticancer activities through its effects on different biological pathways involved in the repression of oncogene expression and induction of cell cycle arrest.19,20 Recently, Ni et al reported that a curcumin analog demethoxycurcumin inhibited EMT in prostate cancer cells,21 which raised our concern that whether the use of curcumin alone will suppress EMT in CRC.
In the present study, we showed that the curcumin repressed EMT, migration, and invasion in CRC through induction of tumor suppressor miR-200c. Moreover, we identified that the EPM5 was a direct target of miR-200c. The dysregulation of miR-200c/EPM5 was required for curcumin-repressed EMT. The elevated expression of EPM5 was associated with the advanced TNM stage and poor survival rate of CRC patients.
Materials and Methods
Cell Culture and Materials
Human-derived colorectal cancer cell lines SW620 and HT29 were obtained from the American Type Culture Collection (ATCC). Both cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (GIBCO, Gaithersburg, MD, USA) and penicillin/streptomycin (Sigma-Aldrich) at 37°C with an atmosphere of 5% CO2 in humidified air. Curcumin was purchased from Sigma-Aldrich and dissolved in DMSO. The concentration of DMSO in the final working solution was below 0.05%. HMI0353-MISSION® microRNA Mimic from Sigma-Aldrich was used as a miR-200c mimic.
Western Blot
Cells were harvested by RIPA buffer and the whole-cell protein was subsequently extracted by centrifugation at 4°C for 30 minutes. BCA Protein Assay Kit (ab102536, ABCAM) was used to measure the concentration of protein in each sample. 50 µg of protein was loaded in the 10% or 12% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes. After blocking in 5% milk in Tris-buffered saline–Tween (TBST), membranes were incubated with primary antibodies overnight at 4°C. The following antibodies were used in this study: ZEB1 (D80D3) Rabbit mAb #3396; ZEB2 (E6U7Z) Rabbit mAb #97885; Snail (C15D3) Rabbit mAb #3879; Slug (C19G7) Rabbit mAb #9585; E-Cadherin (24E10) Rabbit mAb #3195; Vimentin (D21H3) XP® Rabbit mAb #5741; β-Actin (13E5) Rabbit mAb #4970.
RNA Extraction and qPCR
Total RNA was isolated from cells using the RNeasy Mini Kit (Cat No./ID: 74104, QIAGEN) and miRNA kit (Omega Bio-tek, Inc, Guangzhou, China) respectively followed by complementary DNA synthesis with 2 ng of total RNA. Real-time quantitative PCR (qRT-PCR) was performed in triplicate in a 96-well plate containing 1 µL of synthesized cDNA by the use of a QuantiNova™ SYBR Green PCR Kit (Qiagen, Germany). The primer sequences used for PCR were as follows: VIM, forward: TACAGGAAGCTGCTGGAAGG, reverse: ACCAGAGGGAGTGAATCCAG; CDH1, forward: CCCGGGACAACGTTTATTAC, reverse: GCTGGCTCAAGTCAAAGTCC; SNAIL, forward: GCACATCCGAAGCCACAC, reverse: GGAGAAGGTCCGAGCACAC; ZEB1, forward: TCAAAAGGAAGTCAATGGACAA, reverse: GTGCAGGAGGGACCTCTTTA. The relative gene expression was analyzed by the 2(-Delta Delta C(T)) Method.22
RT2 Polymerase Chain Reaction (PCR) Arrays
miRNA profiling was performed with miScript miRNA PCR array human miRNome from Qiagen (Qiagen, Valencia, CA). Total RNA was extracted from cells with or without curcumin treatment. cDNA was prepared as mentioned in RNA extraction and qPCR. The cDNA was mixed with RT1 qPCR master mix supplied by the manufacturer and real-time PCR was performed in a 96-well plate format. The reaction cycle as follows: 15 min at 95 °C and 40 cycles of 15 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C. Data were automatically analyzed by the online RT1 Profiler PCR Array Data Analysis Software version 3.5.
Bioinformatic Analysis
The potential targets of miR-200c were identified by 3 algorithms (TargetScan, Diana Tools, miRDB). TCGA data (gene expression data, miRNA expression data, and clinical data) was downloaded from The Cancer Genome Atlas Program (https://portal.gdc.cancer.gov/).23 GSE17536 and GSE37892 were obtained from NCBI GEO (www.ncbi.nlm.nih.gov/geo). The R package (CMScaller) was applied for CMS classification of TCGA data.24 Pearson was performed for analysis of the correlation coefficient between EPM5 and EMT transcription factors. Gene Set Enrichment Analysis (GSEA) was performed on pre-ranked gene lists based on expression correlation coefficients (Pearson) with EPM5 by the GSEA software.25
Wound-Healing Assay
The Culture-Insert (IBIDI) was used to generated scratch 2 hours after the addition of Mitomycin C (10 ng/mL) in the cell medium. To remove Mitomycin C and detached cells, cells were washed twice with cold PBS and culture medium was added. Subsequently, cells were cultured and allowed to close the wound for 24 hours and images were captured on a microscope. Rate of migration was calculated by the following formula: RM= (Wi-Wf)/t (RM: Rate of cell migration; Wi: Initial wound width; Wf: Final wound width; t = duration of migration).
Migration and Invasion Assay by Transwell
Cells were starved with serum-free medium 1 day before. For migration assay, 3x105 cells were seeded directly in the upper chamber in a serum-free medium. For invasion assay, the upper chambers of the transwell were coated with Matrigel, and 5x105 cells were seeded on the Matrigel. 10% FBS was used as a chemoattractant in the lower chamber. After culture for 24 hours, cells at the top of the upper chamber were removed and cells in the lower chamber were fixed with 4% PFA, followed by staining with crystal violet. The stained cells were counted with microscopy, at least 6 fields each well were analyzed.
Mice Xenograft
SW620 cells were treated with DMSO or curcumin for 72 hours, subsequently were injected to severe combined immunodeficient (SCID) mice through the tail vein (1 × 106 cells per mouse, 6 mice each group). After 6 weeks of the injection, mice were sacrificed and the lungs were taken out. The lungs were fixed in formaldehyde and stained by hematoxylin-eosin. The micro-metastasis tumors were counted by microscopy.
Luciferase Reporter Assay
The Dual-Luciferase Reporter Assay System (Promega) was used for the reporter activation determination strictly according to the manufacture’s instruction.26 Briefly. Cells were harvested and lysed for 20 minutes at room temperature by passive lysis buffer. The cell debris was removed after centrifugation, while the supernatant was further used. For analysis of the reporter activity of miR-200c mimic on the wild type or mutated EPM5 3ʹUTR, both were cloned into a Luc-expressing pGL3 vector. The activity of luciferase was normalized to the Renilla signal to control for differences in transfection efficiency.
Statistical Analysis
The statistics for Kaplan-Meier survival curves were calculated by the Log rank test. A two-tailed Student’s t-test was performed for the calculation of the difference between two groups. A one-way analysis of variance followed by a Tukey multiple comparisons post hoc test was used for comparison of multiple groups. Pearson’s correlation was applied to the mRNA expression correlation analysis. qPCR, migration, and invasion data were expressed as mean ± SD. Calculations were performed using Prism8. p-values ≤ 0.05 were considered as significant. * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001.
Results
Curcumin Suppresses EMT, Migration, and Invasion in CRC
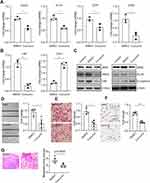
To investigate the potential role of curcumin in colorectal cancer cell metastasis, the mesenchymal-like CRC cell line SW620 was treated with DMSO or curcumin for 48 or 72 hours, and the EMT-related genes were subsequently evaluated. The EMT-inducing transcriptional factor SNAIL, SLUG, ZEB1, and ZEB2 were significantly decreased, accompanied by a downregulation of mesenchymal marker gene Vimentin (Figure 1A and B). In contrast, the epithelial marker gene CDH1 was upregulated. Accordingly, the protein analysis revealed the same pattern (Figure 1C). However, another two EMT-TFs, TWIST1 and TWIST2, were not changed after curcumin treatment (data not shown). A control group without treatment excluded the possibility that DMSO influences EMT in CRC cells.
It has been well-accepted that EMT is related to cell migration and invasion, which directly contribute to tumor metastasis.27 To study the function of curcumin on cell migration and invasion, the “wound healing” and transwell assay were performed. Treatment with curcumin slowed the “wound” close compared with the DMSO group (Figure 1D). Transwell assay showed that the number of migrated cells was decreased after curcumin treatment (Figure 1E). By using transwell containing matrigel, a downregulation in the number of invasive cells was observed (Figure 1F). Moreover, mice injected with curcumin-treated SW620 cells had more tumor formations, compared with mice injected with DMSO-treated cells (Figure 1G). Taken together, these results indicated that the curcumin could suppress EMT, migration, and invasion in CRC cells.
miR-200c Was Induced by Curcumin
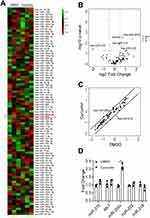
EMT is considered as a response to the combination of extracellular signaling, in which a number of molecules were involved, such as microRNA family.28,29 To address whether the curcumin-repressed EMT was due to the upregulation of tumor-suppressive miRNAs, a miRNA array containing 90 miRNAs was performed. Therefore, three miRNAs were upregulated, whereas one miRNA was downregulated in SW620 cells after curcumin treatment (Figure 2A-C). To verify the observation of miRNA array, the mature miR-210, let-7, miR-218, and miR-200c were analyzed by qPCR (Figure 2D). The miR-200c, which represents a tumor suppressor, was significantly upregulated by curcumin, suggesting that the miR-200c was a potential regulator in curcumin-repressed EMT. Since miR-200c belongs to the miR-200 family, we further checked the other members (miR-200a, −200b, −200c, −141, −429) of the miR-200 family in SW620 cells after curcumin treatments. However, these four miRNAs were not induced by curcumin (Supplementary Figure 1a). Therefore, we chose miR-200c for further study.
Upregulation of miR-200c Was Required for Curcumin-Repressed EMT
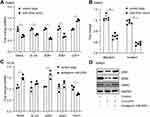
In order to further study the role of miR-200c in the curcumin-regulated EMT, a miR-200c mimic was transfected to SW620 cells. As expected, SNAIL, SLUG, ZEB1, and VIM were largely downregulated, whereas the CDH1 was upregulated by overexpression of miR-200c (Figure 3A). Accordingly, cell migration and invasion capabilities were decreased by ectopic miR-200c (Figure 3B). Moreover, after the endogenous miR-200c in an epithelial-like CRC cell line HT29 was neutralized by antagomiR miR-200c, an upregulation of SNAIL, SLUG, ZEB1, and VIM, as well as a downregulation of CDH1 (Figure 3C), were observed. Furthermore, the neutralization of miR-200c in curcumin-treated SW620 cells dramatically prevented the decrease of EMT markers repressed by curcumin, suggesting that the upregulation of miR-200c was required for curcumin-repressed EMT (Figure 3D).
EPM5 is a Direct Target of miR-200c
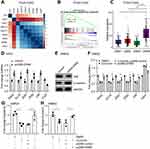
To figure out which gene was the downstream mediator or effector involved in the curcumin/miR-200c cascade, we identified EPM5 as a potential target of miR-200c. qPCR analysis showed that EPM5 was downregulated in SW620 cells after curcumin treatment (Figure 4A). Analysis of the TCGA database with 461 primary CRC tumor samples indicated an inverse correlation between EPM5 and mature miR-200c (Figure 4B). Moreover, the inspection of the EPM5 genomic region revealed a miR-200c binding site within the 3ʹ UTR of EPM5 (Figure 4C, Supplementary Figure 2a). Furthermore, the ectopic expression of miR-200c repressed EPM5 protein in SW620 cells (Figure 4D). The wild type of reporter containing full length of the 3ʹ UTR of EPM5 was repressed by miR-200c, whereas the mutated reporter decreased the responsiveness to miR-200c (Figure 4E). In line with the previous studies, a luciferase reporter of IL6 was also downregulated by miR-200c.30,31 Taken together, miR-200c directly repressed EPM5 through the binding site in its 3ʹ UTR.
EPM5 Induce EMT in CRC
Analysis of the TCGA-COAD database showed that the expression of EPM5 was positively correlated with EMT-inducing transcriptional factors, whereas inversely correlated with epithelial-related gene CDH1 (Figure 5A). Moreover, a similar correlation pattern was observed in another two cohorts (Supplementary Figure 3a). Gene Set Enrichment Analysis (GSEA) revealed that the mRNA of EPM5 was positively associated with EMT hallmark gene signature (Figure 5B, and Supplementary Figure 3b). To further explore the relationship between the EPM5 and tumor features, the association of EPM5 with consensus molecular subtypes (CMS) classification was analyzed. CMS was one of the most robust CRC classifications based on comprehensive gene expression profiles,32 which linked the gene expression to the cancer cell behavior and outcome. CMS4, which was characterized as EMT features and poor prognosis, showed a higher expression of EPM5 compare with the other three subtypes CMS1-3 (Figure 5C, and Supplementary Figure 3c). Taken together, these results implied the potential role of EPM5 in EMT.
Next, the function of EPM5 in curcumin-repressed EMT was experimentally investigated. EMT-inducing transcriptional factors were induced, whereas the epithelial related gene CDH1 was downregulated after the transfection of pcDNA-EPM5 in an epithelial-like CRC cell line HT29 (Figure 5D). Moreover, siRNA-mediated knockdown of EPM5 in mesenchymal-like CRC cell line SW620 increased the mesenchymal marker Vimentin and decreased the epithelial marker E-cadherin (Figure 5E), suggesting that EPM5 was an EMT inducer or mediator. Furthermore, overexpression of EPM5 in SW620 cells that were pre-treated with curcumin largely abolished the effect of curcumin on EMT repression (Figure 5F). Accordingly, curcumin-repressed migration and invasion were also prevented by overexpression of EPM5 (Figure 5G-H). Taken together, these results indicated that EPM5 induced EMT, and the downregulation of EPM5 was necessary for curcumin-repressed EMT in CRC cells.
EPM5 Was Involved in CRC Progress
To obtain further insight into the association between curcumin-repressed EPM5 and progression of CRC, the mRNA of EPM5 from TCGA-COAD database was examined in each of the T, N and M status that proposed by the American Joint Committee on Cancer (AJCC).33 In the T stage, the mRNA of EPM5 was upregulated as the T stage increased (Figure 6A). Moreover, the expression of EPM5 in M1 patients was higher than that of M0 patients (Figure 6B). Furthermore, the increased expression of EPM5 was associated with CRC patients’ lymph node metastasis, and the expression level of EPM5 was positively associated with the degree of lymph node metastasis (Figure 6C). CRC patients with higher expression of EPM5 had a short overall survival rate (Figure 6D). These results strongly suggested that the expression of EPM5 was associated with colorectal cancer progression, and curcumin had a potential protective role against CRC progression by repressing EPM5.
Discussion
Although chemotherapy and radiotherapy remain the most useful treatments for metastatic CRC, the serious side effects have been affecting the quality of patients’ life.34–36 Therefore, the use of natural compounds with low toxicity has gained much attention as an alternative treatment.34,37 In many Southeast Asian countries, the plant Curcuma longa from which the curcumin derived has been consumed as a daily condiment and traditional medicine for centuries.17,18,38 It has been reported as an effective medicine in the treatment of a variety of diseases, including CRC.17,39 Curcumin exerts its anti-tumor effect by downregulating multiple cell signaling pathways involved in the development and progression of cancer.40,41 miRNAs, which regulated the expression of about 60% of mammalian genes, were also regulated by curcumin.42,43 One of the first systematic studies to investigate the effect of curcumin on miRNA expression has been done in human pancreatic cancer cells, in which the authors found that the upregulation of miR-22 and downregulation of miRNA-199a* was the critical mediator for the effect of curcumin.44 Accordingly, it suggested that the modulation of miRNAs may be an important mechanism underlying the biological effect of curcumin.45 In the present study, we demonstrated that the curcumin inhibited the mesenchymal phenotype of CRC cells via the upregulation of miR-200c. Together with miR-200a, miR-200b, miR-141, and miR-429, miR-200c belongs to the miR-200 family.46 Previous studies have demonstrated a critical role of miR-200 family in the regulation of EMT through post-transcriptional repression of ZEB1, ZEB2, SNAIL, and SLUG.14,15 Additionally, theses EMT transcription factors bind directly to the common promoter regions of the miR-200 family to repress its expression. As a result of this bidirectional repression between the miR-200 family and EMT transcription factors, a feed-forward loop was established, resulting in the expression of an epithelial or mesenchymal phenotype depending on the initial signal.27,47 Here, we found that the expression of miR-200c in CRC cells was induced by curcumin, and thus repressed the mesenchymal phenotype of CRC cells. As a consequence of EMT repression, the cell migration and invasion were also inhibited by curcumin.
Besides the transcription factors, we found that the expression of EPM5 (also known as PRICKLE2) was downregulated by curcumin. Further investigation revealed that EPM5 acted as a downstream of curcumin/miR-200c cascade, and miR-200c mediated the curcumin-repressed EMT by direct repression of EPM5. EPM5, which encodes a homolog of Drosophila prickle, is associated with progressive myoclonic epilepsy type 5.48 In one previous study to examine the role of the transcriptional regulator NF-κB in EMT of primary human small airway epithelial cells, the authors found that EPM5 was highly induced by the well-known EMT inducer TGF-β.49 However, the function of EPM5 was not further studied. Here, we showed that the elevated expression of EPM5 was sufficient to induce EMT in CRC. Moreover, the downregulation of EPM5 was necessary for curcumin-repressed EMT. GSEA also revealed a strong positive correlation between EPM5 and TGF-β signaling gene signature (Supplementary Figure 4a-b), implying that TGF-β-induced EPM5 may be an effector of TGF-β signaling to induce EMT. However, this hypothesis needs further experimental evidence in the future.
Taken together, our results verified that curcumin repressed EMT, migration, and invasion in CRC through the upregulation of miR-200c. EPM5 was a direct target of miR-200c and involved in the curcumin-repressed EMT. Elevated expression of EPM5 was associated with the advanced TNM stages and poor prognosis. Despite the lack of in vivo evidence, our study makes it very likely that curcumin can contribute significantly to the improvement of cancer therapy and prevention.
Acknowledgments
We are grateful to all the members from the Department of Emergency Surgery, Shaanxi Provincial People’s Hospital.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
None of the authors has any financial or non-financial conflict of interest related to this study to disclose.
References
1. Seton-Rogers S. Surviving stress during metastasis. Nat Rev Cancer. 2020;20(3):139. doi:10.1038/s41568-020-0243-4
2. Esquerre N, Basso L, Dubuquoy C, et al. Aluminum ingestion promotes colorectal hypersensitivity in rodents. Cell Mol Gastroenterol Hepatol. 2019;7(1):185–196. doi:10.1016/j.jcmgh.2018.09.012
3. Hu Z, Ding J, Ma Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 2019;51(7):1113–1122. doi:10.1038/s41588-019-0423-x
4. Alhassan E, Yadav A, Kelly CP, Mukherjee R. Novel nondietary therapies for celiac disease. Cell Mol Gastroenterol Hepatol. 2019;8(3):335–345. doi:10.1016/j.jcmgh.2019.04.017
5. Zlobec I, Lugli A. Tumour budding in colorectal cancer: molecular rationale for clinical translation. Nat Rev Cancer. 2018;18(4):203–204. doi:10.1038/nrc.2018.1
6. Fennell L, Dumenil T, Wockner L, et al. Integrative Genome-Scale DNA methylation analysis of a large and unselected cohort reveals 5 distinct subtypes of colorectal adenocarcinomas. Cell Mol Gastroenterol Hepatol. 2019;8(2):269–290. doi:10.1016/j.jcmgh.2019.04.002
7. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196.
8. Rokavec M, Bouznad N, Hermeking H. Paracrine induction of epithelial-mesenchymal transition between colorectal cancer cells and its suppression by a p53/miR-192/215/NID1 axis. Cell Mol Gastroenterol Hepatol. 2019;7(4):783–802. doi:10.1016/j.jcmgh.2019.02.003
9. Molina-Castro SE, Tiffon C, Giraud J, et al. The Hippo Kinase LATS2 controls helicobacter pylori-induced epithelial-mesenchymal transition and intestinal metaplasia in gastric mucosa. Cell Mol Gastroenterol Hepatol. 2020;9(2):257–276. doi:10.1016/j.jcmgh.2019.10.007
10. Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat Rev Cancer. 2019;19(12):716–732.
11. Shi X, Kaller M, Rokavec M, Kirchner T, Horst D, Hermeking H. Characterization of a p53/miR-34a/CSF1R/STAT3 feedback loop in colorectal cancer. Cell Mol Gastroenterol Hepatol. 2020;10(2):391–418. doi:10.1016/j.jcmgh.2020.04.002
12. Franke FC, Muller J, Abal M, et al. The tumor suppressor SASH1 interacts with the signal adaptor CRKL to inhibit epithelial-mesenchymal transition and metastasis in colorectal cancer. Cell Mol Gastroenterol Hepatol. 2019;7(1):33–53. doi:10.1016/j.jcmgh.2018.08.007
13. Dinh TA, Jewell ML, Kanke M, et al. MicroRNA-375 suppresses the growth and invasion of fibrolamellar carcinoma. Cell Mol Gastroenterol Hepatol. 2019;7(4):803–817. doi:10.1016/j.jcmgh.2019.01.008
14. Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677.
15. Guan T, Dominguez CX, Amezquita RA, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J Exp Med. 2018;215(4):1153–1168. doi:10.1084/jem.20171352
16. Sciacovelli M, Goncalves E, Johnson TI, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537(7621):544–547. doi:10.1038/nature19353
17. Hsu CH, Cheng AL. Clinical studies with curcumin. Adv Exp Med Biol. 2007;595:471–480.
18. Wang Y, Yu J, Cui R, Lin J, Ding X. Curcumin in treating breast cancer: a review. J Lab Autom. 2016;21(6):723–731. doi:10.1177/2211068216655524
19. Wong S, Zhao J, Cao C, et al. Just add sugar for carbohydrate induced self-assembly of curcumin. Nat Commun. 2019;10(1):582. doi:10.1038/s41467-019-08402-y
20. Devassy JG, Nwachukwu ID, Jones PJ. Curcumin and cancer: barriers to obtaining a health claim. Nutr Rev. 2015;73(3):155–165. doi:10.1093/nutrit/nuu064
21. Ni X, Zhang A, Zhao Z, Shen Y, Wang S. Demethoxycurcumin inhibits cell proliferation, migration and invasion in prostate cancer cells. Oncol Rep. 2012;28(1):85–90.
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
23. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337.
24. Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7(1):16618.
25. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550.
26. Bamidele AO, Svingen PA, Sagstetter MR, et al. Disruption of FOXP3-EZH2 interaction represents a pathobiological mechanism in intestinal inflammation. Cell Mol Gastroenterol Hepatol. 2019;7(1):55–71. doi:10.1016/j.jcmgh.2018.08.009
27. Currey N, Jahan Z, Caldon CE, et al. Mouse model of mutated in colorectal cancer gene deletion reveals novel pathways in inflammation and cancer. Cell Mol Gastroenterol Hepatol. 2019;7(4):819–839. doi:10.1016/j.jcmgh.2019.01.009
28. Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31(3–4):653–662.
29. Suzuki K, Sentani K, Tanaka H, et al. Deficiency of Stomach-Type Claudin-18 in mice induces gastric tumor formation independent of H pylori infection. Cell Mol Gastroenterol Hepatol. 2019;8(1):119–142. doi:10.1016/j.jcmgh.2019.03.003
30. Rokavec M, Wu W, Luo JL. IL6-mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol Cell. 2012;45(6):777–789. doi:10.1016/j.molcel.2012.01.015
31. Zwarycz B, Gracz AD, Rivera KR, et al. IL22 inhibits epithelial stem cell expansion in an ileal organoid model. Cell Mol Gastroenterol Hepatol. 2019;7(1):1–17. doi:10.1016/j.jcmgh.2018.06.008
32. Okita A, Takahashi S, Ouchi K, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–18711. doi:10.18632/oncotarget.24617
33. Weiser MR. AJCC 8th Edition: colorectal Cancer. Ann Surg Oncol. 2018;25(6):1454–1455.
34. Huang XM, Yang ZJ, Xie Q, Zhang ZK, Zhang H, Ma JY. Natural products for treating colorectal cancer: A mechanistic review. Biomed Pharmacother. 2019;117:109142.
35. Kijima T, Nakagawa H, Shimonosono M, et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol Gastroenterol Hepatol. 2019;7(1):73–91. doi:10.1016/j.jcmgh.2018.09.003
36. Steele NG, Chakrabarti J, Wang J, et al. An organoid-based preclinical model of human gastric cancer. Cell Mol Gastroenterol Hepatol. 2019;7(1):161–184. doi:10.1016/j.jcmgh.2018.09.008
37. Sekiba K, Otsuka M, Ohno M, et al. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 Interaction. Cell Mol Gastroenterol Hepatol. 2019;7(2):297–312. doi:10.1016/j.jcmgh.2018.10.010
38. Prasad S, Aggarwal BB. Turmeric, the golden spice: from traditional medicine to modern medicine. In: Benzie IFF, Wachtel-Galor S, editors. Herbal Medicine: Biomolecular and Clinical Aspects. 2nd edition. Boca Raton (FL): CRC Press/Taylor & Francis; 2011. Chapter 13.PMID: 22593922.
39. Ravindran J, Prasad S, Aggarwal BB. Curcumin and cancer cells: how many ways can curry kill tumor cells selectively? AAPS J. 2009;11(3):495–510. doi:10.1208/s12248-009-9128-x
40. Park W, Amin AR, Chen ZG, Shin DM. New perspectives of curcumin in cancer prevention. Cancer Prev Res (Phila). 2013;6(5):387–400. doi:10.1158/1940-6207.CAPR-12-0410
41. Viennois E, Gewirtz AT, Chassaing B. Chronic inflammatory diseases: are we ready for microbiota-based dietary intervention? Cell Mol Gastroenterol Hepatol. 2019;8(1):61–71. doi:10.1016/j.jcmgh.2019.02.008
42. Mirzaei H, Masoudifar A, Sahebkar A, et al. MicroRNA: a novel target of curcumin in cancer therapy. J Cell Physiol. 2018;233(4):3004–3015. doi:10.1002/jcp.26055
43. Xu H, Li J, Chen H, Ghishan FK. NHE8 deficiency promotes colitis-associated cancer in mice via expansion of lgr5-expressing cells. Cell Mol Gastroenterol Hepatol. 2019;7(1):19–31. doi:10.1016/j.jcmgh.2018.08.005
44. Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–473.
45. Bachmeier BE, Killian PH, Melchart D. The role of curcumin in prevention and management of metastatic disease. Int J Mol Sci. 2018;19(6):6. doi:10.3390/ijms19061716
46. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi:10.1101/gad.1640608
47. O’Brien SJ, Carter JV, Burton JF, et al. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review. Int J Cancer. 2018;142(12):2501–2511. doi:10.1002/ijc.31282
48. Fox MH, Bassuk AG. PRICKLE1-Related Progressive Myoclonus Epilepsy with Ataxia. 2009 Sep 8 [Updated 2014 Apr 10]. In: Adam MP, Ardinger HH, Pagon RA, et al, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. Available from: https://www.ncbi.nlm.nih.gov/books/NBK9674/. Accessed September 17, 2020.
49. Martin-Mateos R, De Assuncao TM, Arab JP, et al. Enhancer of zeste homologue 2 inhibition attenuates TGF-beta dependent hepatic stellate cell activation and liver fibrosis. Cell Mol Gastroenterol Hepatol. 2019;7(1):197–209. doi:10.1016/j.jcmgh.2018.09.005
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.