Back to Journals » International Journal of General Medicine » Volume 15
CUL4B is a Potential Novel Prognostic Biomarker and is Correlated with Immune Infiltrates in Malignant Pleural Mesothelioma
Authors Liu L, Hui R, Zeng T, Yang X, Wu Q, Yang T
Received 10 January 2022
Accepted for publication 22 April 2022
Published 3 May 2022 Volume 2022:15 Pages 4613—4623
DOI https://doi.org/10.2147/IJGM.S355889
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Lu Liu,1 Ruting Hui,2 Tianyang Zeng,3 Xuetao Yang,3 Qingchen Wu,3 Tao Yang3,4
1Intensive Care Unit of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 2Department of Rehabilitation Medicine, Chengdu First People’s Hospital, Chengdu, 61007, People’s Republic of China; 3Department of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 4Department of Pulmonary, Critical Care and Sleep Medicine, Yale School of Medicine, New Haven, CT, 06520, USA
Correspondence: Tao Yang, Department of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China, Email [email protected]
Background: Malignant pleural mesothelioma (MPM) is a particularly fatal cancer with a median survival of less than one year. The value of single-agent checkpoint inhibitors is still obscure in MPM. We aim to reveal CUL4B prognostic role and immune infiltrates in MPM patients.
Methods: CUL4B expression profile and clinical information of malignant pleura mesothelioma individuals were collected from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) dataset. Quantitative real-time PCR (qRT-PCR) analysis measured CUL4B mRNA expression in epithelioid, biphasic, sarcomatoid, and normal pleural cell lines.
Results: CUL4B expression elevated in MPM and had a high diagnostic value with AUC = 0.772. Additionally, our results showed that CUL4B high expression significantly correlated with poorer outcomes in MPM. Moreover, GSEA revealed 15 KEGG pathways enriched in the CUL4B high-expression group, and 22 were exhibited in the CUL4B low-expression group. Otherwise, our results showed that CUL4B was relevant to Wnt antagonistic factors (BARX2), Insulin-like growth factor binding protein 3 (IGFBP3), and Phosphatase and tensin homolog (PTEN). In addition, our results revealed that CUL4B expression was positively linked with four types of immune cells, whereas CUL4B expression was negatively linked with three types of immune cells. Additionally, our results showed that CUL4B expression regulates T helper cells, Tcm, and Th2 cells infiltration MPM microenvironment. Finally, our results identified CUL4B high expression in MPM cell line NCI-H2052 (epithelioid), MSTO-211H (biphasic), and NCI-H28 (sarcomatoid).
Conclusion: CUL4B is a valuable prognostic biomarker and a critical immune cell infiltration regulator in MPM.
Keywords: malignant pleural mesothelioma, CUL4B, prognosis, tumorigenesis, immune infiltrates
Introduction
Malignant pleural mesothelioma (MPM) is a fatal disorder with the tumor in the pleural mesothelial cells and a history of asbestos exposure.1,2 MPM median survival is lower than one year.3,4 The therapeutic choices for MPM are restricted, consisting of surgical procedures, intraperitoneal chemotherapy, and systematic chemotherapy.5,6 The first-line chemotherapy, which comprises a combination of pemetrexed and cisplatin, only extends survival by around three months. Thus, new therapeutic approaches are urgently demanded to improve the outcome of this carcinoma.
Cullin 4B (CUL4B) is a family member of cullin proteins, potentially assembling ROC1 and/or ROC2, forming Cullin-RING E3 ubiquitin ligases.7,8 CUL4B has a function in a wide array of biological processes, consisting of cell cycle progression, chromatin remodeling, DNA damage response, signaling transduction, and embryonic development. Recent studies showed that CUL4B participated in the tumorigenesis of several types of cancers, including cervical cancer, lung cancer, esophageal cancer, breast cancer, and colorectal cancer.9–16 CUL4B plays an essential role in the development of various tumors by inhibiting the expression of Wnt antagonistic factors (BARX2 and PAX7), insulin-like growth factor binding protein 3 (IGFBP3), and Phosphatase and tensin Homolog (PTEN) via histone H2A Monoubiquitylation (H2AK119) single ubiquitination.9,17–19 Mayura et al found CUL4B high expression and worse progression-free survival in MPM patients. Furthermore, their study illustrates that pevonedistat inhibited CUL4B to alleviate MPM progression. However, there is no study related to CUL4B, and immune infiltrates lead to MPM occur.
Some studies showed CUL4B played an essential role in immune regulation. Asif et al found that CD4+ T cells are more abundant in CUL4B than Cul4A, Cul4B-deficient CD4+ T cells showed DNA damage sensing. This study revealed Cul4B in promoting the repair of damaged DNA to allow survival and expansion of activated T cells.20 Hung et al found that myeloid-specific Cul4B deficiency worsens LPS-induced peritonitis, and Cul4B deficiency leads to enhanced DNA replication and proliferation, increased production of chemokines, and decreased proinflammatory production cytokines of macrophages.21 These results suggested that CUL4B played a crucial role in the immune system. Immunotherapies including programmed death-1 (PD-1) inhibitors, programmed death ligand-1 (PD-L1) inhibitors and cytotoxic T lymphocyte correlated antigen 4 (CTLA4) inhibitors have made remarkable strides in recent years.22–25 In MPM, PD-L1 inhibitors show 17.3 and 6.1 months in overall survival and Progression-free survival in small phase II trials.26 The phase 3 study (CheckMate 743) shows CTLA4 inhibitors plus PD-L1 inhibitors have better overall survival.27 This clinical trial supports the use of CTLA4 inhibitors plus PD-L1 inhibitors as first-in-class regimen that was approved in the USA in October 2020 for previously untreated unresectable MPM. In some trials, dendritic cell (DC) immunotherapy shows it obtains antitumor efficacy.28 The value of single-agent checkpoint inhibitors is still obscure in MPM. CUL4B could be a biomarker to select the optimal immune treatment among MPM patients based on efficacy and tolerance.
Here, we conduct a comprehensive study of the association between CUL4B and MPM prognosis. In addition, we investigate the relationship between CUL4B and immune cells infiltration and related genes leading to MPM tumorigeneses. This study provides a novel insight into the function of CUL4B in MPM. It suggests a potential mechanism whereby CUL4B serves as an immunotherapy biomarker via MPM infiltrating immune cells in the tumor microenvironment.
Materials and Methods
RNA-Sequencing (RNASeq) and Clinical Characteristics
The gene expression profiles and clinical characteristics of pleura mesothelioma patients were downloaded from the publicly available TCGA and GEO datasets. GSE2549 includes 40 mesothelioma and 10 normal pleural tissues, and the TCGA dataset consists of 0 normal tissues and 86 MPM tissues. Furthermore, insufficient or missing data were excluded. Subsequent processing transformed RNASeq data to values more like those resulting from microarrays. To explore the influences of CUL4B expression on the immune microenvironment, MPM tissue was divided to two groups based on the expression of CUL4B in further study.
Gene Set Enrichment Analysis (GSEA)
GSEA was performed via the R package clusterprofiler (3.6.3) to reveal the potentially significant pathways related to the list of the genes initiated by CUL4B expression. For each analysis, the number of permutations was 1000. The pathway set with an adjust p value < 0.05, false discovery rate (FDR) q-value < 0.25, and a |normalized enrichment score (NES)| > 1 were recognized as significantly enriched.
Immune Infiltration Analysis via Single-Sample GSEA (ssGSEA)
According to the signature gene lists of 24 types of immune cells, the relative tumor cell infiltration levels were performed by the ssGSEA method from the Gene Set Variation Analysis GSVA package in R 3.6.3.29 Furthermore, CUL4B expression and immunocytes correlation was analyzed by Spearman correlation. In addition, the immune infiltration differences among the different expression groups of CUL4B were analyzed by the Wilcoxon rank-sum test.
Cell Culture
MPM cell line NCI-H2052 (epithelioid), MSTO-211H (biphasic), NCI-H28 (sarcomatoid), and normal mesothelium cell line Met-5A were purchased from ATCC, US. Met-5A cells were cultured in Medium-199 (Gibco, US) with 10% fetal bovine serum (Gibco, US), and NCI-H2052, MSTO-211H cells NCI-H28 were cultured in RPMI-1640 (Gibco, US) with 10% fetal bovine serum (Gibco, US). Cells were cultured in an incubator under 5% CO2 at 37 °C.
Quantitative Real‑time Polymerase Chain Reaction (qRT‑PCR) Analysis
Cell total RNA were isolated by RNeasy Mini Kit (QIAGEN, Germany), iScript cDNA Synthesis Kits (BIO-RAD, US) was used to reverse transcription, and SsoFast EvaGreen Supermix With Low ROX (BIO-RAD, US) was utilized to qRT-PCR of CUL4B and 18S (as endogenous control). Thermal cycling procedure is as follows: denaturation (95 °C, 3min); 40 cycles of denaturation (95 °C, 3s) and elongation (60°C, 30s). Each gene was measured in triplicate. The primers were purchased from CUL4B forward 5’-ACTCCTCCTTTACAACCCAGG-3’, reverse 5’-TCTTCGCATCAAACCCTACAAAC-3’; 18S forward CCATCCAATCGGTAGTAGCG, reverse GTAACCCGTTGAACCCCATT.
Statistics Analysis
R 3.6.3 was performed for statistical analysis for the dataset acquired from GEO and TCGA. X2 test, fisher's exact test, and Wilcoxon rank-sum test were used to assess the link between CUL4B expression and patients’ clinicopathological characteristics. Survival curves were plotted via the kaplan–meier method and Log rank test, including HR and p-values. Moreover, univariate and multivariate Cox analysis were performed to assess the influence of CUL4B expression and clinicopathological characteristics on survival. Bivariate and Spearman rank correlations were used to evaluate the correlation between CUL4B expression and 24 types of immune cells. Pearson rank correlation was utilized to evaluate the correlation between CUL4B expression and expression of Treg regulator (FOXP3), Wnt antagonistic factors (BARX2 and PAX7), insulin-like growth factor binding protein 3 (IGFBP3). Student’s t-tests were conducted to identify the statistical difference in CUL4B mRNA expression. The relative expression of CUL4B mRNA in qRT-PCR was calculated by the 2(-ΔΔC(T)) method. All tests were set p < 0.05 as the cut-off criterion.
Results
Clinicopathological Characteristics
TCGA data of 86 malignant pleural mesothelioma (MPM) patients included their characteristics regarding the T, N, M, and pathological stages, radiation therapy, gender, race, histological type, history of asbestos exposure, laterality, and age. The CUL4B expression was not significantly associated with patients’ clinicopathological characteristics (Supplementary Table 1).
Assessment of CUL4B Expression in MPM and Normal Tissues
We first explored the CUL4B expression in the GEO database (GSE2549) and verified it was significantly elevated relative to normal controls in pleural mesothelioma (Figure 1A). Otherwise, CUL4B expression was performed in the ROC test revealing that CUL4B expression had an excellent predictive use in pleural mesothelioma (AUC=0.772) (Figure 1B). Moreover, we evaluated the relationship between CUL4B expression and pleural mesothelioma patients using the TCGA database. We noticed that CUL4B expression elevation was related to prognosis (Progression-free interval HR = 2.02, 95% CI = 1.19–3.44, p = 0.009) (Figure 1C).
Gene Sets Enriched in CUL4B Expression Phenotype
CUL4B-related signaling pathways depended on GSEA were used to identify signaling pathways involved in pleural mesothelioma between high and low CUL4B expression data sets and indicated significant differences (p-value < 0.05, FDR q < 0.25, |NES| > 1) in enrichment using the molecular signatures database collection (C2.cp.v7.2.symbols.gmt). Thirty-seven enriched KEGG pathways were identified, consisting of 15 pathways that demonstrated a significantly differential enrichment in the CUL4B high-expression group and 22 pathways exhibited in the low-expression group (Supplementary Table 2). In addition, the top nine KEGG items (Table 1) include focal adhesion, regulation of actin cytoskeleton, maturity-onset diabetes of the young, arrhythmogenic right ventricular cardiomyopathy, progesterone mediated oocyte maturation, calcium signaling pathway, extracellular matrix (ECM) receptor interaction, transforming growth factor β (TGF β) signaling pathway and cell cycle were indicated significantly differential enrichment in CUL4B high expression phenotype based on p-value, FDR q and NES (Figure 2).
 |
Table 1 KEGG Gene Sets Enriched in the CUL4B High-Expression Phenotype |
Tumorigenesis Genes Related to CUL4B Expression
Wnt antagonistic factors, insulin-like growth factor binding protein 3 (IGFBP3), and Phosphatase and tensin Homolog (PTEN) are relevant to CUL4B tumorigenesis. We have checked the association between CUL4B expression and Wnt antagonistic factors BARX2 and PAX7, IGFBP3 and PTEN in mesothelioma, and there are significant association between CUL4B and BARX2; CUL4B and IGFBP3; and CUL4B and PTEN. However, there is no significant association between CUL4B and PAX7 (Figure 3).
Correlation Between CUL4B Expression and Immune Infiltration
We next further explored the correlation between CUL4B expression and levels of immune cell infiltration, which was quantified using ssGSEA in a pleural mesothelioma microenvironment (Figure 4A). CUL4B expression was positively linked with the abundance of Th2 cells, T helper cells, T central memory (Tcm) cells, and T effector memory (Tem) cells (Figure 4B–E). CUL4B expression was negatively linked with the abundance of dendritic cell (DC), plasmacytoid DC (pDC), and immature DC (iDC) (p < 0.05) (Figure 4F–H). The Wilcoxon rank sum test manifested that the infiltration levels of T helper cells, Tcm, and Th2 cells in the CUL4B high-expression group were significantly higher than those in the low-expression group (p < 0.05). In addition, FOXP3 plays a vital role in maintaining the homeostasis of the immune system by regulating the expansion and functions of regulatory T-cells (Treg). We have checked the correlation between FOXP3 and CUL4B expression in the TCGA-Meso dataset, and we found FOXP3 and CUL4B correlation has significance (p < 0.05) (Supplementary Figure 1).
CUL4B qRT-PCR Analysis
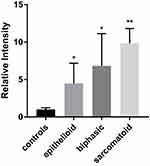
We confirmed the upregulation of CUL4B mRNA at the cellular level by qRT-PCR analysis. The average CUL4B/18S mRNA expression fold changes are 4.46, 6.50, and 9.60 in epithelioid, biphasic, and sarcomatoid MPM cell lines compared with normal mesothelium cell lines (Figure 5). The results above suggest that CUL4B may be a particular factor in MPM.
Discussion
There are no effective therapies for malignant pleural mesothelioma (MPM) due to the lack of defined targets. This study showed that CUL4B expression elevated in MPM and had a high diagnostic value with AUC = 0.772. Additionally, our results showed that CUL4B high expression significantly correlated with poorer outcomes in MPM. Moreover, GSEA revealed that 15 KEGG pathways enriched in the CUL4B high-expression group, and 22 pathways exhibited in the CUL4B low-expression group. Otherwise, our results showed that CUL4B was relevant to Wnt antagonistic factors (BARX2), insulin-like growth factor binding protein 3 (IGFBP3), and Phosphatase and tensin Homolog (PTEN). In addition, our results revealed that CUL4B expression positively correlated with the abundance of Th2 cells, T helper cells, T central memory (Tcm) cells, and T effector memory (Tem) cells. In contrast, CUL4B expression negatively correlated with the abundance of dendritic cells (DCs), plasmacytoid DCs (pDCs), and immature DCs (iDCs). Additionally, our results showed that CUL4B expression regulates T helper cells, Tcm, and Th2 cells infiltrate in the MPM microenvironment (Supplementary Figure 1A). Finally, our results identified CUL4B high expression in MPM cell line NCI-H2052 (epithelioid), MSTO-211H (biphasic), and NCI-H28 (sarcomatoid).
CUL4B is a scaffold protein of the CUL4B-RING E3 ubiquitin ligase complex.30,31 CUL4B plays a vital role in regulating cell-cycle progression, DNA replication, and DNA damage response. Recent studies revealed that CUL4B is overexpressed in various cancers and processes substantial oncogenic properties.32–34 However, the influence of CUL4B in MPM remains still poorly understood. In this study, CUL4B was upregulated in the GEO dataset and high expression in epithelioid, biphasic, and sarcomatoid cell lines. Furthermore, Mayura et al found CUL4B was upregulated in MPM tumor specimens compared to nonmalignant pleural tissues. We next found that CUL4B AUC is 0.772 via the ROC curve. To further understand the variation in CUL4B expression associated with prognosis in MPM, we used over survival (OS) and Progression-free survival (PFS) to evaluate the CUL4B expression role in MPM patients from the TCGA dataset. An elevated CUL4B expression was a prognostic factor for a negative prognosis. Those results suggest that CUL4B may play a key role in MPM prognostic biomarker.
In addition, we performed GSEA via TCGA data to investigate CUL4B functions in MPM. We found that focal adhesion, regulation of actin cytoskeleton, maturity-onset diabetes of the young, arrhythmogenic right ventricular cardiomyopathy, progesterone mediated oocyte maturation, calcium signaling pathway, ECM receptor interaction, TGF β signaling pathway, and cell cycle were enriched in CUL4B high expression phenotype. Otherwise, we found that CUL4B tumorigenesis was related to Wnt antagonistic factors, IGFBP3 and PTEN. Yang et al found that CUL4B led to a decrease in IGFBP3 and resulted in H2AK119 monoubiquitylation, H3K9 trimethylation, and DNA methylation. Fang et al found that CUL4B modulates the proliferation and invasion via the Wnt/β-catenin pathway, indicating that CUL4B is critical in controlling gastric cancer tumorigenesis17. Song et al found that CUL4B negatively repressed PTEN transcription by regulating Toll-like receptor-triggered proinflammatory responses.18 These suggest that CUL4B may be identified as a prognostic marker and therapeutic target in MPM.
Another important finding in this study was that CUL4B expression correlated with MPM diverse immune marker sets and immune infiltration levels. The results revealed that high CUL4B expression increased the immune infiltration in Th2 cells, T helper cells, T central memory (Tcm) cells, and T effector memory (Tem) cells and decreased immune infiltration in dendritic cells (DCs), plasmacytoid DCs (pDCs) and immature DCs (iDCs). T cells were typical tumor-infiltrating immune cells in different types of human cancers. In addition, we identified that CUL4B expression is related to T helper cells regulator FOXP3 (Supplementary Figure 1B). Several studies reported T cells’ response to immune checkpoint inhibition, drug survival, and predictors thereof.35–38 Here, this study appends the growing frame of literature revealing T cells as a positive prognostic aspect.
Conclusion
In conclusion, CUL4B was upregulated in MPM and was associated with poor prognostic factor. Moreover, higher CUL4B expression was involved in focal adhesion, regulation of actin cytoskeleton, maturity-onset diabetes of the young, arrhythmogenic right ventricular cardiomyopathy, progesterone-mediated oocyte maturation, calcium signaling pathway, ECM receptor interaction, TGF β signaling pathway, and cell cycle pathways. CUL4B tumorigenesis is related to Wnt antagonistic factors, IGFBP3 and PTEN. CUL4B is a paramount immune cell infiltration regulator in T lymphocytes for MPM.
Abbreviations
MPM, malignant pleural mesothelioma; CUL4B, Cullin 4B; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; qRT-PCR, quantitative real-time PCR; IGFBP3, insulin-like growth factor binding protein 3; PTEN, Phosphatase and tensin Homolog; H2AK119, histone H2A monoubiquitylation; H3K9, histone H3 lysine 9 trimethylation; PD-1, programmed death-1; PD-L1, programmed death ligand-1; CTLA4, cytotoxic T lymphocyte correlated antigen 4; DC, dendritic cell; pDC, plasmacytoid DC; iDC, immature DC; FDR, false discovery rate; NES, normalized enrichment score; OS, overall survival; PFS, progression-free survival; ECM, extracellular matrix; TGF β, transforming growth factor β; Tcm, T central memory cells; Tem, T effector memory cells; Th, T helper cells.
Data Sharing Statement
The datasets supporting the conclusions of this article are available in TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases.
Statement of Ethics
All data utilized in our study were obtained through the TCGA and GEO databases, which are freely accessible for research purposes. According to the ethics guidelines, informed consent and ethical approval are not required to use public and anonymized clinical data. Therefore, the medical ethics committee of the First Affiliated Hospital of Chongqing Medical University exempted the ethical approval.
Funding
This study was supported by Chongqing Natural Science Foundation (No. cstc2018jcyjAX0245).
Disclosure
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
1. Bibby AC, Tsim S, Kanellakis N, et al. Malignant pleural mesothelioma: an update on investigation, diagnosis and treatment. Eur Respir Rev. 2016;25(142):472–486. doi:10.1183/16000617.0063-2016
2. Fujii T, Itami H, Uchiyama T, et al. HEG1-responsive microRNA-23b regulates cell proliferation in malignant mesothelioma cells. Biochem Biophys Res Commun. 2020;526(4):927–933. doi:10.1016/j.bbrc.2020.03.172
3. Nicolini F, Bocchini M, Bronte G, et al. Malignant pleural mesothelioma: state-of-the-art on current therapies and promises for the future. Front Oncol. 2019;9:1519. doi:10.3389/fonc.2019.01519
4. Horio D, Minami T, Kitai H, et al. Tumor-associated macrophage-derived inflammatory cytokine enhances malignant potential of malignant pleural mesothelioma. Cancer Sci. 2020;111(8):2895–2906. doi:10.1111/cas.14523
5. Takuwa T, Hasegawa S. Current surgical strategies for malignant pleural mesothelioma. Surg Today. 2016;46(8):887–894. doi:10.1007/s00595-015-1275-3
6. Fiore D, Baggio V, Sotti G, Muzzio PC. Imaging before and after multimodal treatment for malignant pleural mesothelioma. Radiol Med. 2006;111(3):355–364. doi:10.1007/s11547-006-0034-3
7. Li Q, Cui M, Yang F, et al. A cullin 4B-RING E3 ligase complex fine-tunes pancreatic δ cell paracrine interactions. J Clin Invest. 2017;127(7):2631–2646. doi:10.1172/JCI91348
8. Yoshizawa T, Karim MF, Sato Y, et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19(4):712–721. doi:10.1016/j.cmet.2014.03.006
9. Yang Y, Liu R, Qiu R, et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene. 2015;34(1):104–118. doi:10.1038/onc.2013.522
10. Jia L, Yan F, Cao W, et al. Dysregulation of CUL4A and CUL4B ubiquitin ligases in lung cancer. J Biol Chem. 2017;292(7):2966–2978. doi:10.1074/jbc.M116.765230
11. Zhang H, Wang A, Tan Y, et al. NCBP1 promotes the development of lung adenocarcinoma through up-regulation of CUL4B. J Cell Mol Med. 2019;23(10):6965–6977. doi:10.1111/jcmm.14581
12. Wang X, Chen Z. Knockdown of CUL4B suppresses the proliferation and invasion in non-small cell lung cancer cells. Oncol Res. 2016;24(4):271–277. doi:10.3727/096504016X14666990347473
13. Li T, Wu S, Jia L, et al. CUL4 E3 ligase regulates the proliferation and apoptosis of lung squamous cell carcinoma and small cell lung carcinoma. Cancer Biol Med. 2020;17(2):357–370. doi:10.20892/j.issn.2095-3941.2019.0107
14. Huang H, Xu Y, Guo Z, Chen X, Ji S, Xu Z. MicroRNA-133b inhibits cell proliferation and promotes apoptosis by targeting cullin 4B in esophageal squamous cell carcinoma. Exp Ther Med. 2018;15(4):3743–3750. doi:10.3892/etm.2018.5906
15. Wang Y, Pan X, Li Y, et al. CUL4B renders breast cancer cells tamoxifen-resistant via miR-32-5p/ER-α36 axis. J Pathol. 2021;254(2):185–198. doi:10.1002/path.5657
16. Li Y, Hu H, Wang Y, et al. CUL4B contributes to cancer stemness by repressing tumor suppressor miR34a in colorectal cancer. Oncogenesis. 2020;9(2):20. doi:10.1038/s41389-020-0206-3
17. Fang Z, Zhong M, Wang Y, et al. miR‑381 and miR‑489 suppress cell proliferation and invasion by targeting CUL4B via the Wnt/β‑catenin pathway in gastric cancer. Int J Oncol. 2019;54(2):733–743. doi:10.3892/ijo.2018.4646
18. Song Y, Li P, Qin L, et al. CUL4B negatively regulates toll-like receptor-triggered proinflammatory responses by repressing PTEN transcription. Cell Mol Immunol. 2021;18(2):339–349. doi:10.1038/s41423-019-0323-0
19. Hu H, Yang Y, Ji Q, et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell. 2012;22(6):781–795. doi:10.1016/j.ccr.2012.10.024
20. Dar AA, Sawada K, Dybas JM, et al. The E3 ubiquitin ligase Cul4b promotes CD4+ T cell expansion by aiding the repair of damaged DNA. PLoS Biol. 2021;19(2):e3001041. doi:10.1371/journal.pbio.3001041
21. Hung MH, Jian YR, Tsao CC, Lin SW, Chuang YH, Enhanced LP. S-induced peritonitis in mice deficiency of cullin 4B in macrophages. Genes Immun. 2014;15(6):404–412. doi:10.1038/gene.2014.32
22. Akin Telli T, Bregni G, Camera S, Deleporte A, Hendlisz A, Sclafani F. PD-1 and PD-L1 inhibitors in oesophago-gastric cancers. Cancer Lett. 2020;469:142–150. doi:10.1016/j.canlet.2019.10.036
23. Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell Infiltration and PD-L1. Cancer Res. 2015;75(11):2139–2145. doi:10.1158/0008-5472.CAN-15-0255
24. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi:10.1038/nature21349
25. Tu L, Guan R, Yang H, et al. Assessment of the expression of the immune checkpoint molecules PD-1, CTLA4, TIM-3 and LAG-3 across different cancers in relation to treatment response, tumor-infiltrating immune cells and survival. Int J Cancer. 2020;147(2):423–439. doi:10.1002/ijc.32785
26. Okada M, Kijima T, Aoe K, et al. Clinical efficacy and safety of nivolumab: results of a multicenter, Open-label, single-arm, Japanese phase II study in malignant pleural mesothelioma (MERIT). Clin Cancer Res. 2019;25(18):5485–5492. doi:10.1158/1078-0432.CCR-19-0103
27. Baas P, Scherpereel A, Nowak AK, et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (checkmate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet. 2021;397(10272):375–386. doi:10.1016/S0140-6736(20)32714-8
28. de Gooijer CJ, Borm FJ, Scherpereel A, Baas P. Immunotherapy in malignant pleural mesothelioma. Front Oncol. 2020;10:187. doi:10.3389/fonc.2020.00187
29. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
30. Qi M, Jiao M, Li X, et al. CUL4B promotes gastric cancer invasion and metastasis-involvement of upregulation of HER2. Oncogene. 2018;37(8):1075–1085. doi:10.1038/onc.2017.380
31. Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12(4):220. doi:10.1186/gb-2011-12-4-220
32. Zhao M, Qi M, Li X, et al. CUL4B/miR-33b/C-MYC axis promotes prostate cancer progression. Prostate. 2019;79(5):480–488. doi:10.1002/pros.23754
33. Duan PJ, Zhao JH, Xie LL. Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. J Ovarian Res. 2020;13(1):76. doi:10.1186/s13048-020-00677-w
34. Wu P, Hu H, Li J, Gong W. Upregulation of cullin 4B promotes gastric cancer and predicts poor prognosis. Onco Targets Ther. 2020;13:1235–1243. doi:10.2147/OTT.S234706
35. Liu X, Bao X, Hu M, et al. Inhibition of PCSK9 potentiates immune checkpoint therapy for cancer. Nature. 2020;588(7839):693–698. doi:10.1038/s41586-020-2911-7
36. Khan M, Arooj S, Wang H. NK cell-based immune checkpoint inhibition. Front Immunol. 2020;11:167. doi:10.3389/fimmu.2020.00167
37. Kumagai S, Togashi Y, Kamada T, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat Immunol. 2020;21(11):1346–1358. doi:10.1038/s41590-020-0769-3
38. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi:10.1126/science.aaa8172
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.