Back to Journals » OncoTargets and Therapy » Volume 11
CPEB4 promotes growth and metastasis of gastric cancer cells via ZEB1-mediated epithelial–mesenchymal transition
Authors Cao G, Chen D, Liu G, Pan Y, Liu Q
Received 26 May 2018
Accepted for publication 5 July 2018
Published 21 September 2018 Volume 2018:11 Pages 6153—6165
DOI https://doi.org/10.2147/OTT.S175428
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Gan Cao,* Dehu Chen,* Guiyuan Liu, Yan Pan, Qinghong Liu
Department of General Surgery, Taizhou People’s Hospital, The Fifth Affiliated Hospital of Nantong University, Taizhou 225300, People’s Republic of China
*These authors contributed equally to this work
Background: Cytoplasmic polyadenylation element-binding protein 4 (CPEB4) has previously been reported to be associated with biological malignancy in various cancers. However, its function in tumor growth and metastasis in gastric cancer (GC) remains obscure. Here, we explored the functional and molecular mechanisms by which CPEB4 influences GC.
Materials and methods: The expression of CPEB4 was assessed using Western blot and immunohistochemistry in GC specimens. The roles of CPEB4 in GC cell proliferation, migration, and invasion were investigated by cell-counting kit-8 (CCK-8), colony formation, and EdU assay; wound-healing assay; and transwell assay, respectively. Quantitative real-time PCR (qRT-PCR), Western blot, and immunofluorescence staining were performed to detect the expressions of CPEB4 and epithelial–mesenchymal transition (EMT)-related markers. The function of CPEB4 on GC cell growth and metastasis was also determined in vivo through establishing subcutaneous xenograft tumor and lung metastatic mice model.
Results: The results revealed that the expression of CPEB4 was increased in GC tissues compared with matched normal tissues. High expression level of CPEB4 was significantly associated with clinical metastasis and unfavorable prognosis in patients with GC. Furthermore, CPEB4 silencing remarkably inhibited GC cells’ proliferation, invasion, and metastasis in vitro and in vivo. Conversely, CPEB4 overexpression achieved the opposite effects. Mechanically, we proved that ZEB1-mediated EMT might be involved in CPEB4-facilitated GC cells’ proliferation, invasion, and metastasis.
Conclusion: Our findings implied that CPEB4 expression predicted a worse prognosis in patients with GC. Besides, CPEB4 contributed to GC cells’ proliferation, migration, and invasion via ZEB1-mediated EMT.
Keywords: CPEB4, gastric cancer, epithelial–mesenchymal transition
Introduction
Gastric cancer (GC), as the fifth most common malignancy and the third leading cause of cancer-associated deaths worldwide, is often diagnosed in the advanced stage, with a high tendency to metastasize and a poor prognosis.1–3 Despite advancements in a comprehensive therapy in recent decades, including the surgical treatment and chemotherapy, metastasis is still a major clinical challenge in the curative treatment of GC. Therefore, the investigation of the molecular mechanisms underlying GC progression and metastasis may provide potential therapeutic strategies for GC.
Recently, epithelial–mesenchymal transition (EMT) has emerged as a critical regulator in cancer cells’ invasion and metastasis.4 During EMT, cells shed their epithelial characteristics (such as cellular adherence and absence of motility) and acquire mesenchymal properties (such as motility and invasiveness), which are molecularly characterized by the loss of epithelial marker E-cadherin and the gain of mesenchymal markers N-cadherin and Vimentin.5 Additionally, the EMT process can be regulated by transcription factors (such as Snail, Slug, ZEB1, SIPI1, and Twist), as well as multiple complex signal pathways, including TGFβ, Notch, Wnt, and PI3K/AKT signaling cascade.4 Interestingly, increasing evidence reveals the potential clinical value of targeting EMT in cancer treatment.
Cytoplasmic polyadenylation element-binding protein 4 (CPEB4), a typical member of the CPEB family, is a sequence-specific RNA-binding protein and a translational regulator, which has been demonstrated to be selectively overexpressed in various malignancies.6 Notably particularly, recent studies have reported that CPEB4 functions importantly in cancer cells’ migration and invasion in certain types of cancer, such as glioma and breast cancers, and could be exploited as a target for cancer treatment.7–10 Nevertheless, to our knowledge, the clinical significance and biological function in GC remain undetermined and even less is known about the regulatory mechanism of CPEB4-mediated cancer progression.
Accordingly, we focused on the clinical significance of CPEB4 in GC tissues in this study, as well as the role and potential molecular mechanism of CPEB4 in GC cells’ growth, migration, and invasion.
Materials and methods
Patients, specimens, and cell lines
A total of 112 samples (tumor tissues and corresponding normal tissues) were collected from patients with gastric adenocarcinoma who underwent radical gastrectomy at our hospital. None of these patients received preoperative chemotherapy or radiotherapy. Among them, fresh tissues of 45 cases were evaluated by Western blot for CPEB4 protein and 112 cases were also embedded in paraffin blocks for immunohistochemical stainings. Preoperative written informed consent was obtained from each patient according to the Declaration of Helsinki, and this study was approved by the ethics committee of the Fifth Affiliated Hospital of Nantong University.
The human GC cell lines (AGS, BGC823, MGC803, MKN45, and SGC7901) and normal gastric epithelial GES-1 cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in 90% Roswell Park Memorial Institute-1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific). All cells were propagated at 37°C, in a humidified incubator containing 5% CO2.
Lentivirus infection
The sense sequence of short hairpin RNA (shRNA) oligonucleotides specifically targeting the CPEB4 transcripts was as follows: sh-CPEB4 sense, 5′-CUGCCUCAUUUGGCGAAUAC-3′ and antisense, 5′-UAUUCGCCAAAUGAGGCAGC-3′. A negative sequence was used as a control. Transfections were conducted using Lipofectamine 2000 following the manufacturer’s instructions. The full-length CPEB4 cDNA was inserted into the retroviral vector pBabe-puro and the expression vector HA-PMSCV. The empty vector was used as control. The CPEB4 expression was determined by Western blot and immunofluorescence.
Immunohistochemistry (IHC)
The approach for IHC was conducted as described previously.11 We used antibodies against CPEB4 (Abcam, Cambridge, UK), ZEB1 (CST, Massachusetts, USA), and Ki-67 (CST). Sections were observed under the microscope in a blinded manner. The immunoreactive score (IRS) was evaluated by the staining intensity and the percentage of positive cells. The staining intensity score was defined as follows: 0=negative, 1=weak intensity, 2=moderate intensity, and 3=strong intensity; and the percentage of staining cells was scored as follows: 0=0%–5%, 1=6%–25%, 2=26%–50%, and 3=>50%. Summation of intensity score and quantity score led to an IRS ranging from 0 to 6: the sample with IRS≥3 was considered positive, whereas the sample with IRS<3 was regarded as negative.
Western blot
Western blot was carried out using the standard protocol as described previously.12 Briefly, protein samples from tissues or cell lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). Membranes were incubated in 5% milk to block nonspecific binding, followed by incubation with a primary antibody at 4°C overnight. Afterward, membranes were incubated with a secondary antibody. Immunoreactivity was visualized by the enhanced chemiluminescence detection reagent (Thermo Fisher Scientific). Primary antibody against CPEB4 was obtained from Abcam. Primary antibodies against E-cadherin, Snail, Slug, ZEB1, SIP1, Twist, N-cadherin, and Vimentin were purchased from CST. GAPDH antibody, used as the loading control, was bought from Bioworld Technology (St Louis Park, MN, USA).
RNA isolation and real-time quantitative reverse-transcription PCR
Total RNA was isolated from cells using the TRIzol reagent (Thermo Fisher Scientific) and reversely transcribed to complementary DNA using the PrimeScript RT Reagent Kit (TaKaRa, Shanghai, China). Quantitative real-time PCR (qRT-PCR) was performed with the SYBR Green Assay kit (TaKaRa) on a 7500 RT-PCR System (Thermo Fisher Scientific). Relative quantification was calculated using the 2−ΔΔCt method. PCR primer sequences are listed in Table S1.
Immunofluorescence staining
Cells plated on coverslips were fixed with 4% paraformaldehyde for 15 min. Then, cells were blocked with Triton X-100 for 30 min. Subsequently, cells were blocked with 5% BSA for 30 min, followed by incubation with primary antibodies against CPEB4, E-cadherin, and N-cadherin at 4°C overnight. The coverslips were incubated with Alexa Fluor-conjugated secondary antibody (Bioworld Technology) for 60 min and stained with DAPI for 5 min. Finally, cells were observed under a fluorescence microscope.
Wound-healing assay
Cells were cultured to confluence in a six-well plate, and an artificial wound was gently created using a 10 μL plastic tip. Photomicrographs were taken at 0 and 48 h time points to monitor the wound-healing process. The wound coverage percentage=(0 h width−48 h width)/0 h width×100%.
Cells’ migration and invasion assays
For migration assay, cells were plated into the upper chamber of a 24-well 8 μm pore size transwell device (Corning Incorporated, Corning, NY, USA). For invasion assay, while the upper chamber was coated with a BD Matrigel™ Matrix Basement Membrane (BD Biosciences, San Jose, CA, USA). Medium containing 20% FBS as a chemoattractant was added to the lower chamber. After 24 h, cells on the upper surface of the chamber were removed by scraping and the chamber was fixed in 4% paraformaldehyde and stained with 0.05% crystal violet. Migrated and invaded cells were quantified under an inverted light microscope.
Cell proliferation assay
Cells at a density of 5×103/well were plated in 96-well plates. At the indicated times (0, 24, 48, 72, and 96 h after culture), 10 μL of cell-counting kit-8 (CCK-8; Shanghai, China) solution was added to each well and cultured for another 80 min. The absorbance value at 450 nm was detected to analyze cell viability.
Colony formation assay
Cells were seeded in a six-well plate and cultured for 2 weeks at 37°C in a humidified incubator containing 5% CO2. Subsequently, cells were fixed with crystal violet solution and counted under a microscope. Clone formation was defined as those containing ≥50 cells.
EdU assay
The proliferation potential of cells was assessed using the Cell-Light TM EdU imaging detecting kit (RiboBio, Shanghai, China). Briefly, cells (1×105/well) were seeded in each well of a 96-well plate. Cells within each well were cultured with 10 mM EdU for 2 h and then fixed with 4% paraformaldehyde, followed by permeabilization in 0.2% Triton X-100 and staining with Apollo solution for 30 min in the dark. Subsequently, DAPI solution was used to stain the cell nuclei. Finally, cells were imaged under a fluorescent microscope.
In vivo tumorigenesis
BALB/c nude mice (6-week old, male) were used to examine tumorigenicity. To determine the role of CPEB4 in tumor formation, SGC7901 cells (sh-NC/sh-CPEB4) or AGS cells (vector/CPEB4) were injected subcutaneously into nude mice followed by the measurement of tumor size at 5-day intervals using the following formula: volume=(short diameter)2× (long diameter)/2.13 The mice were sacrificed after 30 days, and tumor samples were resected for tumor mass, Western blot, and IHC staining. To ascertain the function of CPEB4 in tumor metastasis in vivo, cells mentioned earlier were injected intravenously into the tail vein of nude mice. At 30 days after the injection, the lungs of nude mice were surgically removed for the count of metastatic nodules and then processed for Western blot analysis. The assays were done using six nude mice per group. All animal procedures were approved by the Ethics Committee of the Fifth Affiliated Hospital of Nantong University and were performed according to the institutional guidelines.
Statistical analyses
Chi-squared test was performed to compare clinical features of patients between different groups. Kaplan–Meier survival analysis was done by the log-rank test. All of the data were expressed as the mean±SD and were analyzed by Student’s t-test. All the analyses were conducted using the SPSS 21.0 software (IBM Corporation, Armonk, NY, USA). Differences were considered statistically significant at P<0.05.
Results
Clinical significances of CPEB4 in patients with GC
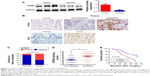
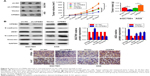
To investigate the function of CPEB4 in the progression of GC, we first detected CPEB4 protein expression in 45 pairs of tumor samples and matched normal tissues by Western blot. It was shown that CPEB4 expression was significantly higher in tumor samples compared with corresponding normal tissues (Figure 1A). Second, the expressions of CPEB4 and ZEB1 were determined in tumor tissues and matched normal tissues of 112 patients with GC by immunohistochemical staining (Figure 1B). As shown in Figure 1C, compared to normal tissues, CPEB4 expression was markedly increased in tumor tissues and its expression was significantly higher in metastatic tumor tissues (Figure 1B). Additionally, a clinicopathological association analysis demonstrated that CPEB4 was significantly correlated with tumor size, T stage, pTNM stage, and lymph node metastasis (P<0.05) (Table 1). Table 2 shows that the expression level of CPEB4 was positively related to that of ZEB1 (P<0.001, contingency coefficient [C]=0.491) in primary lesion. Additionally, compared with patients without lymph node metastasis, those with metastasis showed significantly higher staining scores for CPEB4 (P<0.05) (Figure 1D), indicating that CPEB4 might function importantly in metastasis. Furthermore, survival analysis revealed that patients with CPEB4 positive had poorer prognosis than those with CPEB4 negative (P<0.05) (Figure 1E).
  | Table 1 Relationship between CPEB4 expression and clinicopathological features in gastric cancer |
CPEB4 expression in GC cell lines
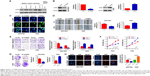
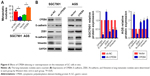
To identify the appropriate GC cell lines with low or high expression of CPEB4, Western blot was performed for protein samples derived from five GC cell lines (AGS, BGC823, MGC803, MKN45, and SGC7901) and normal gastric epithelial GES-1 cells. Among them, CPEB4 expression was the highest in SGC7901 cells and was the lowest in AGS cells and GES-1 cells (Figure 2A), which indicated that CPEB4 was overexpressed not only in primary tumors but also in GC cell lines compared with corresponding normal controls. Hence, SGC7901 cells were selected for stable transfection with shRNA lentivirus vectors toward CPEB4 and AGS cells were selected for stable transfection with CPEB4 expression vector. Obviously, CPEB4 expression was markedly decreased by shRNA-CPEB4 and significantly increased by CPEB4 overexpression via Western blot detection (Figure 2B) and immunofluorescence staining (Figure 2C).
CPEB4 promotes GC cells’ growth, migration, and invasion in vitro
To investigate the effects of CPEB4 on GC cell migration and invasion in vitro, we performed in vitro wound healing, transwell migration, and transwell invasion assays. Both wound healing and transwell migration assays indicated that the migratory capability of CPEB4-silencing SGC7901 cells was weaker than that of the corresponding sh-NC cells, whereas CPEB4-overexpressing AGS cells showed greater migration capability (Figure 2D and E). Consistently, the transwell invasion assay revealed the similar effect on GC cells’ invasion (Figure 2E). Subsequently, we examined the effect of CPEB4 expression on GC cells’ growth in vitro. Using the CCK-8 assay (Figure 2F), colony formation assay (Figure 2G), and EdU incorporation assay (Figure 2H), we revealed that CPEB4 knockdown markedly suppressed cell proliferation in SGC7901 cells, while CPEB4 overexpression in AGS cells exerted the opposite result. Taken together, these results indicated that CPEB4 could facilitate proliferation, motility, and invasiveness of GC cells in vitro.
ZEB1-mediated EMT is critical for CPEB4-induced GC cells’ growth, migration, and invasion
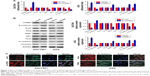
Accumulating evidence has demonstrated that EMT endows epithelial cells with their migratory and invasive abilities and functions importantly in many cancers’ metastasis process, including GC.4,14 To investigate the relationship between CPEB4 and EMT, qRT-PCR was performed to analyze the influence of CPEB4 on EMT-related molecules. We found that CPEB4 silencing resulted in higher level of the epithelial marker E-cadherin, lower levels of the mesenchymal markers N-cadherin and Vimentin, and transcription factor ZEB1 but did not significantly influence other transcription factors (Snail, Slug, SIP1, and Twist), whereas CPEB4 overexpression in AGS cells demonstrated the opposite effects (Figure 3A). Besides, Western blot and immunofluorescence staining further proved that CPEB4 negatively correlated with E-cadherin but positively correlated with ZEB1, N-cadherin, and Vimentin in GC cells (Figure 3B and C).
Next, to demonstrate the involvement of transcription factor ZEB1 in CPEB4-mediated GC cells’ growth, migration, invasion, as well as EMT, the ZEB1 expression was downregulated in GC cells. The results manifested that shRNA-mediated suppression of ZEB1 in AGS cells led to a noticeable reversal of CPEB4-induced EMT (Figure 4A). Besides, sh-ZEB1 expression in AGS cells observably suppressed CPEB4-facilitated cell migration and invasion (Figure 4B) as well as cell growth (Figure 4C–E). On the whole, these data implied that CPEB4 promoted GC cells’ growth, migration, and invasion via the activation of ZEB1-mediated EMT.
CPEB4 facilitates tumor formation and metastatic potential in vivo
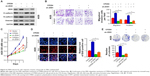
To further confirm the effect of CPEB4 on tumor growth and metastatic potential in vivo, we established subcutaneous xenograft tumor and lung metastasis model in nude mice. The result revealed that CPEB4 silencing in SGC7901 cells significantly hindered tumor growth, as evidenced by the reduced tumor volume and tumor mass (Figure 5A). Consistent with the results in vitro, CPEB4 silencing resulted in the higher level of E-cadherin and the lower level of ZEB1, N-cadherin, Vmentin, and Ki-67 by Western blot and IHC analysis in the xenograft specimens (Figure 5B and C). Inversely, CPEB4 overexpression in AGS cells led to the opposite effect (Figure 5A–C).
Additionally, the lung metastatic tissues indicated that CPEB4 knockdown had led to a significantly less number of lung metastatic nodules (Figure 6A) and a distinct reversal of EMT (Figure 6B). As expected, CPEB4 overexpression had imposed significantly opposite effects on GC cells’ metastasis in vivo (Figure 6A and B). Collectively, these results revealed that CPEB4 could promote GC cells’ growth and metastasis in vivo.
Discussion
Although great achievements have been made in surgical treatment and chemotherapy over the past few years, metastasis remains the major cause of death in cancer patients. Given that the spread of cancer cells to distant organs, from a relatively immobile type to a more invasive phenotype, is generally accepted as a leading function in tumor metastasis, novel therapeutic approaches are urgently required to prevent the metastatic dissemination of cancer cells.5,15 Hence, the identification of novel key targets and the potential mechanisms involved in cancer cells’ invasion and metastasis is promising. Interestingly, recent studies have revealed that CPEB4 was highly expressed in multiple tumor types and was involved in tumor progression and metastasis.6,7 For example, CPEB4 has been reported to be markedly increased in cancer tissues and be a promising therapeutic target in metastatic colorectal carcinoma.16 In breast cancer, CPEB4 was distinctly elevated in primary lesions compared with adjacent normal tissues and played a pivotal role in breast cancer invasion and metastasis.9 Ortiz-Zapater et al demonstrated that CPEB4 was overexpressed in pancreatic ductal adenocarcinoma and CPEB4 silencing in pancreatic cancer cells reduced the cell invasion ability, which described CPEB4 as a potential candidate target for cancer therapy.10 To data, however, little is known about the clinical significances of CPEB4 in GC tissues and the effect of CPEB4 expression on GC cells’ growth, invasion, and metastasis.
In this study, we provided evidence that CPEB4 might serve as a potential prognostic biomarker or therapeutic target for patients with GC. We found that CPEB4 expression was distinctly increased in GC primary lesion compared with matched normal tissues. Clinical significance analysis indicated that CPEB4 expression was positively correlated with clinical metastasis and patient survival, suggesting that CPEB4 might function crucially in GC evolution and metastasis. Also, we evaluated the biological roles of CPEB4 in GC cells. Our results revealed that the depletion of endogenous CPEB4 expression suppressed cell growth, migration, and invasion both in vitro and in vivo. Conversely, CPEB4 overexpression reversed these events. Subsequently, mechanical analysis demonstrated that CPEB4 controlled EMT in GC cells by regulating transcription factor ZEB1.
Recently, EMT has emerged as a key event of cancer cells’ invasion and metastasis through conferring an invasive phenotype in certain cancers.17,18 Noticeably, a very substantial effort has been made to discover novel targeted genes that accelerate cancer progression via stimulating EMT process.19–22 In this investigation, we revealed that CPEB4 silencing inhibited the EMT process, accompanied by downregulated expression of the mesenchymal markers N-cadherin and Vmentin and the upregulated expression of the epithelial marker E-cadherin, whereas CPEB4 overexpression showed the opposite effect. More interestingly, accumulating evidence indicated that transcription factor (such as Snail, Slug, ZEB1, SIP1, and Twist), as a crucial modulator of EMT, was an intracellular element relevant to cellular proliferation, longevity, and invasion.4 In many types of human cancer, transcription factor was overactivated, thus regulating cells’ proliferation, invasion, and metastasis.23 In our present study, we found that knockdown of CPEB4 expression could observably decrease ZEB1 expression and in turn suppressed EMT process, thereby reducing the growth, migration, and invasion abilities of GC cells. On the contrary, CPEB4 overexpression contributed to the opposite effects. However, no changes in the expressions of the other EMT transcription factors (Snail, Slug, SIP1, and Twist) were also observed when CPEB4 expression was changed. Next, we reduced the endogenous ZEB1 expression, which weakened not only CPEB4-induced EMT but also cells’ growth, invasion, and metastasis. Thus, we could reasonably assume that CPEB4 facilitated GC cells’ growth, invasion, and metastasis via ZEB1-regulated EMT.
Although ZEB1 acted as a downstream effector in CPEB4-induced migration and invasion, it is worth exploring the more in-depth mechanism(s) involved in the complex interaction of CPEB4 and ZEB1. As reported, ZEB1 was found to serve as a direct downstream transcriptional target involved in cancer cells’ invasion and metastasis. Besides, it demonstrated that the upstream target molecule and a binding partner constituted a transcriptional complex within ZEB1 promotor, thereby facilitating the ZEB1 expression.24 Clinically, CPEB4 expression was positively correlated with ZEB1 expression in this study. To gain more insights into CPEB4-mediated ZEB1 of GC cells, a luciferase reporter assay could be conducted to determine whether CPEB4 regulates ZEB1 promoter activity, which would prove a direct or indirect relationship between CPEB4 and ZEB1. Furthermore, in subsequent experiments, we evaluated whether other genes or certain signaling pathways are involved in CPEB4-mediated transactivation of ZEB1.
Conclusion
Our data demonstrated that high CPEB4 expression was positively associated with cancer metastasis and poor prognosis in patients with GC and CPEB4 could promote GC cells’ growth, invasion, and metastasis via activation of ZEB1-mediated EMT, thus possibly leading to the advancement of novel clinical targets for cancer patients. Nevertheless, further exploration of the complex interaction of CPEB4 and EMT is urgently required to enlarge the understanding of the mechanism.
Acknowledgments
This work was supported in part by funding from Taizhou Science and Technology Support Plan (Social Development) Project (TS201732) and Medical Science and Technology Development Foundation of Jiangsu University (JLY20160148 and JLY20160149).
Disclosure
The authors report no conflicts of interest in this work.
References
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. | ||
Fock KM. Review article: the epidemiology and prevention of gastric cancer. Aliment Pharmacol Ther. 2014;40(3):250–260. | ||
Catalano V, Labianca R, Beretta GD, et al. Gastric cancer. Crit Rev Oncol Hematol. 2009;71(2):127–164. | ||
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. | ||
Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci. 2014;35(9):479–488. | ||
Chen Y, Tsai YH, Tseng SH. Regulation of the expression of cytoplasmic polyadenylation element binding proteins for the treatment of cancer. Anticancer Res. 2016;36(11):5673–5680. | ||
Xu H, Liu B. CPEB4 is a candidate biomarker for defining metastatic cancers and directing personalized therapies. Med Hypotheses. 2013;81(5):875–877. | ||
Zhijun L, Dapeng W, Hong J, et al. Overexpression of CPEB4 in glioma indicates a poor prognosis by promoting cell migration and invasion. Tumour Biol. 2017;39(4):1010428317694538. | ||
Lu R, Zhou Z, Yu W, Xia Y, Zhi X. CPEB4 promotes cell migration and invasion via upregulating Vimentin expression in breast cancer. Biochem Biophys Res Commun. 2017;489(2):135–141. | ||
Ortiz-Zapater E, Pineda D, Martínez-Bosch N, et al. Key contribution of CPEB4-mediated translational control to cancer progression. Nat Med. 2012;18(1):83–90. | ||
Chen D, Liu G, Xu N, et al. Knockdown of ARK5 expression suppresses invasion and metastasis of gastric cancer. Cell Physiol Biochem. 2017;42(3):1025–1036. | ||
Chen DH, Yu JW, Wu JG, Wang SL, Jiang BJ. Significances of contactin-1 expression in human gastric cancer and knockdown of contactin-1 expression inhibits invasion and metastasis of MKN45 gastric cancer cells. J Cancer Res Clin Oncol. 2015;141(12):2109–2120. | ||
Chen D, Zhou H, Liu G, et al. SPOCK1 promotes the invasion and metastasis of gastric cancer through Slug-induced epithelial–mesenchymal transition. J Cell Mol Med. 2018;22(2):797–807. | ||
Li J, Zhen L, Zhang Y, et al. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. | ||
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. | ||
Cortés-Guiral D, Pastor-Iodate C, Díaz del Arco C, del Puerto-Nevado L, Fernández-Aceñero MJ. CPEB4 immunohistochemical expression is associated to prognosis in stage IV colorectal carcinoma. Pathol Res Pract. 2017;213(6):639–642. | ||
Kang HM, Park BS, Kang HK, et al. Delphinidin induces apoptosis and inhibits epithelial-to-mesenchymal transition via the ERK/p38 MAPK-signaling pathway in human osteosarcoma cell lines. Environ Toxicol. 2018;33(6):640–649. | ||
He SJ, Xiang CQ, Zhang Y, et al. Recent progress on the effects of microRNAs and natural products on tumor epithelial–mesenchymal transition. Onco Targets Ther. 2017;10:3435–3451. | ||
Wu K, Shen B, Jiang F, et al. TRPP2 enhances metastasis by regulating epithelial-mesenchymal transition in Laryngeal Squamous cell carcinoma. Cell Physiol Biochem. 2016;39(6):2203–2215. | ||
Chen Y, Peng Y, Xu Z, et al. LncROR promotes bladder cancer cell proliferation, migration, and epithelial-mesenchymal transition. Cell Physiol Biochem. 2017;41(6):2399–2410. | ||
Chen DH, Yu JW, Jiang BJ. Contactin 1: a potential therapeutic target and biomarker in gastric cancer. World J Gastroenterol. 2015;21(33):9707–9716. | ||
Chen D, Cao G, Qiao C. Alpha B-crystallin promotes the invasion and metastasis of gastric cancer via NF-κB-induced epithelial–mesenchymal transition. J Cell Mol Med. 2018. | ||
Goossens S, Vandamme N, van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta. 2017;1868(2):584–591. | ||
Guo X, Zhao L, Cheng D, Mu Q, Kuang H, Feng K. AKIP1 promoted epithelial–mesenchymal transition of non-small-cell lung cancer via transactivating ZEB1. Am J Cancer Res. 2017;7(11):2234–2244. |
Supplementary material
  | Table S1 Primers designed for qRT-PCR |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.