Back to Journals » OncoTargets and Therapy » Volume 9
Correlations between EGFR gene polymorphisms and pleural metastasis of lung adenocarcinoma
Authors Guo H, Xing Y, Mu A, Li X, Li T, Bian X, Yang C, Zhang X, Liu Y, Wang X
Received 9 October 2015
Accepted for publication 7 May 2016
Published 25 August 2016 Volume 2016:9 Pages 5257—5270
DOI https://doi.org/10.2147/OTT.S97907
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Haisheng Guo,1,* Yunhui Xing,2,* Ailan Mu,1 Xia Li,3 Tingshan Li,4 Xia Bian,1 Chunmei Yang,1 Xiaolei Zhang,1 Yuefen Liu,1 Xunguo Wang1
1Department of Oncology, Dongying People’s Hospital, 2Department of Tuberculosis, Shengli Hospital of Shengli Oil Field, 3Department of Health, 4Personnel Department, Dongying People’s Hospital, Dongying, Shandong, People’s Republic of China
*These authors contributed equally to this work
Abstract: Proliferation, growth, and differentiation of cells are strictly controlled by the signal system of epidermal growth factor receptor (EGFR). If any link of the EGFR signals system is interfered with or damaged, the proliferation, growth, and differentiation of cells would become uncontrolled. EGFR is overexpressed in a variety of malignant tumors, such as non-small-cell lung cancer, colorectal cancer and breast cancer. Results of the study have proved that EGFR overexpression is closely associated with mutations and variants of the EGFR genes, whose mutations and variants are associated with occurrence, metastasis, and prognosis of different types of tumors, including lung cancer. This study is aimed at investigating whether the polymorphisms of CA simple sequence repeat in intron 1 (CA-SSR1), -216G/T, and R497K in the EGFR are able to induce EGFR activation and whether overexpression is associated with pleural metastasis of lung adenocarcinoma. A total of 432 lung adenocarcinoma patients with pleural metastasis (metastasis group) and 424 patients with lung adenocarcinoma but without pleural metastasis (nonmetastasis group) were enrolled in this study. For all patients, the CA-SSR1 genotypes were determined by capillary electrophoresis, polymerase chain reaction amplification, and direct DNA sequencing, and the R497K and -216G/T genotypes were determined by polymerase chain reaction amplification and direct DNA sequencing. EGFR expression was evaluated by immunohistochemical staining in primary tumor tissues with different -216G/T, R497K, and CA-SSR1 genotypes. Our results showed significant differences between pleural metastasis and nonmetastasis groups in the genotype and allele distribution of -216G/T, R497K, and CA-SSR1 polymorphisms of the EGFR gene. The -216T allele, Arg allele, and shorter CA-SSR1 (<17) had significantly increased risks of pleural metastasis compared with the -216G allele, Lys allele, and longer CA-SSR1 (≥17), respectively. The expression of EGFR was higher in patients with genotypes of -216T/T or -216G/T, Arg/Arg or Arg/Lys, and shorter CA-SSR1 (<17) than that in patients with genotypes of -216G/G, Lys/Lys, and longer CA-SSR1 (≥17), respectively. These results indicate that -216G/T, R497K, and CA-SSR1 polymorphisms are associated with the risk of pleural metastasis of lung adenocarcinoma, which may be related to the overexpression of EGFR protein induced by -216G/T, R497K, and CA-SSR1 polymorphisms.
Keywords: EGFR, polymorphism, pleural metastasis, lung adenocarcinoma
Introduction
At the advanced stage, patients with lung cancer, especially lung adenocarcinoma, frequently suffer from pleural metastasis,1 but the exact underlying mechanisms remain largely unclear.
Previous studies have shown that the occurrence, development, and distant metastasis of tumors were related to the overexpression of the EGFR gene. Some clinical models showed that the occurrence of pleural metastasis of lung adenocarcinoma was closely related to the abnormal excessive expression of the EGFR gene, but the specific mechanism was unclear.
EGFR, a transmembrane glycoprotein of 170 kDa whose gene is located at 7p12.1–12.3, is a major regulator of several signaling pathways, such as ras-raf-MEK-MAPK and PI3K-PKC-kB, and it could further activate transcription factors at the levels of transcription and translation, which mediate a series of processes, including cell differentiation, survival, migration, invasion, adhesion, and cell damage repair.2–4 Overexpression of EGFR often contributes to cell proliferation, angiogenesis, cancer invasion, and metastasis.5 Hemming et al6 reported that EGFR was associated with poor survival, more aggressive behavior, and increased risk of invasion/metastasis. Fontanini et al7,8 explored the relationship between the expression of EGFR and metastasis of lung cancer by immunohistochemical method. The result showed that the expression of EGFR was significantly higher in patients with hilar and mediastinal lymph node metastasis than those without lymph node involvement. Yudoh et al9 found that the expression of EGFR was higher in cell lines of high metastatic mouse sarcoma than that of low ones. Penetration and adhesion of cells increased when EGF was added to these cells with high expression of EGFR; lung metastasis rate was also greatly reduced when anti-EGFR monoclonal antibody was added to block EGF stimulation. Veale et al10 found that the expression of EGFR had close relation with tumor stage while detecting the expression of EGFR in non-small-cell lung cancer (NSCLC) by immunohistochemical method, since the staining was stronger in 30 cases of stage III cancer patients than that in 47 cases of stage I–II cancer patients. They observed that the average concentration of EGFR was 16.1 fmol/mg in longer survival groups and 68.6 fmol/mg in shorter survival groups.
Previous studies11–13 have shown that EGFR and its natural ligands, critical for the signal transduction pathway for cell proliferation, differentiation, metastasis, and survival, are overexpressed in most of the patients with NSCLC and can affect the prognosis of cancer patients.6,13
Experimental evidence indicates that polymorphisms of the gene may also regulate protein expression. Results of a previous study14 showed that the expression of EGFR is closely related to EGFR gene polymorphism.
EGFR was overexpressed in 50%–81% of patients with NSCLC, and such overexpression had a close bearing on cancer susceptibility and metastasis.8,15 Meanwhile, a large number of experimental results showed that EGFR gene polymorphism was closely related to the overexpression and activation of EGFR.11,16–18 Besides, several functional EGFR gene polymorphisms, such as CA simple sequence repeat in intron 1 (CA-SSR1), -216G/T, and R497K, have already been confirmed with higher frequency in lung cancer, and these polymorphisms often lead to increased activity and transcription of EGFR.19–21
One of the most common EGFR gene polymorphisms was CA-SSR1, and the relationship between CA-SSR1 repeat length and the expression of EGFR has been extensively studied in breast cancers.22–24 The experimental results showed that CA-SSR1 influenced transcription levels of EGFR obviously. The length of CA-SSR1 was closely related to the transcriptional level of the EGFR. Compared with the shorter CA-SSR1 repeats (<17), the transcriptional activity of EGFR decreased considerably in longer CA-SSR1 repeats (≥17).25
In addition to CA-SSR1, the other two kinds of single nucleotide polymorphisms, -216G/T and R497K, in the promoter region might correlate with increased promoter activity and expression of EGFR mRNA.26,27
-216G/T is located in the Sp1 transcriptional start site in the EGFR promoter, where multiple protein factors and transcriptional start sites have been identified.27,28 Since Sp1 binding site is a key area of EGFR gene transcription,29–32 the change (G→T) at site -216 resulted in an increase in the promoter activity by ~30%, which in turn led to an overexpression of EGFR.21,27,33
Another polymorphic locus in EGFR, R497K, a single nucleotide replacement (G→A), leads to an Arg→Lys substitution in exon 13. Experimental results have demonstrated that such a change influenced multiple aspects of biology, such as stimulation of cell growth, induction of proto-oncogenes, etc, compared with the “wild-type” 497R in vitro and in vivo.34
Based on the previous research results, we propose a hypothesis on whether the abovementioned polymorphisms of the EGFR gene might be associated with an increased risk of pleural metastasis of lung adenocarcinoma. Therefore, we performed the CA-SSR1, -216G/T, and R497K genotyping and immunohistochemical detection of EGFR expression in the pleural metastasis group and the nonmetastasis group, respectively, which aimed at determining the relationship between CA-SSR1, -216G/T, and R497K polymorphisms in EGFR gene and the risk of pleural metastasis of lung adenocarcinoma.
Patients and methods
Patient information
Patient recruitment was performed between May 2010 and April 2013. In total, 856 patients, including 432 lung adenocarcinoma patients with pleural metastasis and 424 patients with lung adenocarcinoma but no pleural metastasis, confirmed by pathology and/or cytology, were enrolled in this study. All patients were at stage IV according to the revised TNM staging system for NSCLC. The clinical characteristics, including age, sex, smoking history, and cancer stage, were recorded and the details are listed in Table 1.
The study was carried out with ethics committee approval from the Medical Ethics Committee of Dongying People’s Hospital and concerns no medical ethical issues, and written informed consent was obtained from all patients.
Sample preparation
Ten microliters of peripheral venous blood were collected from each patient enrolled in this study and stored in EDTA-coated tubes for the genotyping of genetic polymorphisms in EGFR gene. Meanwhile, tissue samples were also obtained from some of them and fixed in 10% buffered formalin, embedded in paraffin for evaluating EGFR expression.
CA-SSR1, -216G/T, and R497K genotyping
DNA was extracted using DNA extraction kit (Omega Bio-Tek, Inc., Beijing Hong YueChuangxin Technology Co., Ltd., Beijing, People’s Republic of China) according to the manufacturer’s instructions.
Agarose gel electrophoresis
Polymerase chain reaction (PCR) products were tested using 1.5% agarose gel electrophoresis and Alpha Imager TM 2200 Gel imaging system.
CA-SSR1, -216G/T, and R497K genotyping
CA-SSR1 genotyping was performed by capillary electrophoresis and direct sequencing. The primers were 5′-ACAAGCTTTTAAAGAGTTTCTTGT-3′ (forward) and 5′-AGGCAATGTGTTAGTACACA-3′ (reverse). A total of 80–100 ng of DNA was PCR amplified with a labeled forward primer and an unlabeled antisense primer. PCR conditions were as follows: initial denaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension for 7 minutes, with a total of 35 cycles. PCR products were purified using the MicroElute® Cycle-Pure Kit (Omega Bio-tek Inc., Guangzhou Feiyang Biological Engineering Co., Ltd, Guangzhou, People’s Republic of China). After PCR purification, the exact number of the (CA)n repeat was analyzed using the capillary electrophoresis system (instrument model: Agilent HP 3D; Agilent Technologies, Santa Clara, CA, USA). PCR products were directly sequenced with the same unlabeled forward primer to confirm the number of CA dinucleotide repeats.
-216G/T genotyping was performed by direct sequencing. The primers were 5′-GCTTGGTCCTCTTCGGCATCT-3′ (forward) and 5′-CCGTCTTGACCAGTCGCTTA-3′ (reverse). PCR conditions were as follows: predenaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds, annealing from 68°C to 60°C decreasing at 1°C/cycle for eight cycles and at 59°C for 22 cycles, extension at 72°C for 30 seconds, and a final extension for 7 minutes, with a total of 30 cycles. The PCR products were sequenced directly in the sense and antisense directions using an ABI373 instrument (Applied Biosystems, Foster City, CA, USA).
R497K genotyping was performed by direct sequencing. The primers were 5′-TTAGATGCAGCATTATTAGCC-3′ (forward) and 5′-CGGGTGATACATCATTAGAAA-3′ (reverse). PCR conditions were as follows: initial denaturation step at 94°C for 5 minutes followed by 35 cycles of 30 seconds at 94°C; 45 seconds at 60°C; 45 seconds at 72°C; and a final extension for 7 minutes.
PCR products were purified using MicroElute® Cycle-Pure Kit (Guangzhou Feiyang Biological Engineering Co., Ltd). After PCR purification, -216G/T and R497K polymorphisms were studied by direct sequencing from both ends using ABI PRISM 310 automatic DNA sequencer.
All primers and PCR conditions used are described in Table 2.
  | Table 2 Primers and PCR conditions used for polymorphisms amplification |
Immunohistochemical staining
The expression of EGFR protein was determined by immunohistochemistry. The tissue samples, fixed in 10% buffered formaldehyde solution and embedded in paraffin, were stained with EGFR antibody. In brief, dry frozen tissue sections of 5 μm thickness at room temperature for 2 hours. Fix sections with 4% paraformaldehyde for 15 minutes at room temperature. Wash slides two times, 4 minutes each, with tris-buffered saline (TBS)/0.1% saponin. Block endogenous peroxidase by incubating for 30 minutes in TBS/0.3% H2O2/0.1% saponin and 0.02% NaN3. Wash slides three times, 3 minutes each, with TBS/saponin. Block nonspecific binding sites with 1/100 diluted goat serum in TBS/saponin for 20 minutes. Incubate overnight at 4°C, with EGFR monoclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). Wash slides four times in TBS/saponin, and incubate the slides with biotinylated secondary antibody for 30 minutes. Add avidin–biotin–peroxidase reagents, and reveal the resulting peroxidase activity by incubating the slides with a 0.5 mg/mL HRP substrate solution (3, 3′-diaminobenzidine + H2O2 prepared in distilled water). Wash slides four times in TBS, counterstain for 1 minute with hematoxylin, dehydrate the slides with sequential ethanol washes for 1 minute each starting with 75%, followed by 80%, and finishing with a 100% ethanol wash and then seal slides. Staining without primary antibody was employed to create a negative control.
The level of EGFR expression was evaluated by multiplying the positive cell rate and staining intensity, as reported in previous studies.35,36 Briefly, the percentage of labeled cells was graded as follows: grade 0, no positive cells; grade 1, l%–10% labeled tumor cells; grade 2, 11%–50% labeled tumor cells; and grade 3, 51%–100% positive tumor cells. A composite score was obtained by multiplying the grade by the dominant intensity pattern of staining (0, negative; l, weak; 2, moderate; 3, intense) for EGFR, respectively, as described previously.37
Statistical analysis
Statistical analysis was carried out using commercial statistical software package SPSS 18 software for windows (SPSS 17.0, SPSS Inc., Chicago, IL, USA). The possible associations between EGFR gene polymorphisms and pleural metastasis of lung adenocarcinoma were assessed by computing the odds ratio (OR) and 95% confidence interval (CI). The categorical variables were analyzed using the χ2 test and Fisher’s exact test. EGFR expression data were analyzed statistically with the Mann–Whitney U-test. P<0.05 was considered significant.
Results
Agarose gel electrophoresis
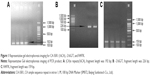
PCR products were tested using 1.5% agarose gel electrophoresis and Alpha Imager TM 2200 Gel imaging system. Representative gel electrophoresis images for CA-SSR1 (16CA), -216 G/T, and R497K are shown in Figure 1.
Genotype/allele frequencies of CA-SSR1
Twenty-four repeat genotypes were observed by capillary electrophoresis in the two groups, and the four most common genotypes are shown in Figure 2 (dark blue peak, as shown in Figure 2A–D).
According to the results of earlier wave figures of the nucleotide sequence by direct sequencing, the single dark blue peak (192 bp) in Figure 2A represents the homozygous genotype 16/16CA; the double peaks (192 bp and 196 bp) in Figure 2B represent the heterozygous genotype 16/18CA; the double peaks (192 bp and 200 bp) in Figure 2C represent the heterozygous genotype 16/20CA; and the single dark blue peak (200 bp) in Figure 2D represents the homozygous genotype 20/20CA, respectively.
The four PCR products mentioned above were further verified by direct sequencing. The results showed that the numbers of CA-SSR1 in products of 192 bp, 196 bp, and 200 bp were 16CA, 18CA, and 20CA repeats, respectively, and the corresponding waves are shown in Figure 3.
In total, nine alleles with 15–23CA repeats were found in the two groups, with a predominance of 16, 18, and 20CA repeats (Table 3), and the allelic distribution ranged from 1.62% to 35.42% in the metastasis group and from 1.42% to 39.15% in the nonmetastasis group, respectively.
16CA allele was significantly higher (P=2.425×10−7) while 20CA allele was much lower (P=0.002314) in the pleural metastasis group than that in the nonmetastasis group. Although 18CA allele was higher (17.92% [152/848]) in the nonmetastasis group than that in the pleural metastasis group (12.96% [112/864]), there was no obvious difference (P=0.05883).
The three main genotypes of CA-SSR1 polymorphism of the EGFR gene, namely, 16/16CA, 18/18CA, and 20/20CA, were detected among the patients. Each genotype is demonstrated by a representative sequencing wave in Figure 3. The genotype and allele frequencies of CA-SSR1 in the two groups are described in Table 4.
The distribution of genotype 16/16CA in the pleural metastasis group was significantly lower compared with that in the nonmetastasis group, with ORs of 1.43 (95% CI, 1.032–1.764) for 18/18CA and 1.94 (95% CI, 1.074–2.077) for 20/20CA, respectively. Following adjustment for the clinicopathological variables using logistic regression analysis, the adjusted ORs were 1.48 (95% CI, 1.415–1.845, adjusted P=0.132) for 18/18CA and 1.59 (95% CI, 1.075–3.002, adjusted P=0.034) for 20/20CA.
Patients with the 16/16CA genotype showed a higher risk of pleural metastasis. Compared with patients with the 16/16CA genotype, those with the 18/18CA and 20/20CA genotypes have less risk of pleural metastasis.
Genotype/allele frequencies of -216G/T
The genotype and allele frequencies of -216G/T in the nonmetastasis group and the metastasis group are shown in Table 4. The three genotypes, namely, G/G, G/T, and T/T, were detected among the patients, and each genotype is demonstrated by a representative sequencing wave in Figure 4. There were significant differences in genotype and allele frequency of the -216G/T polymorphism between the two groups.
  | Figure 4 Representative sequencing wave figures for -216G/T. |
The rate of allele T was 32.87% (284/864) in the metastasis group, being significantly higher than that in the nonmetastasis group (24.41% [207/848]) (OR =1.61, 95% CI: 1.228–1.916, adjusted P=0.021).
The distributions of G/T and T/T genotypes in the pleural metastasis group were much higher than that in the nonmetastasis group, with ORs of 1.42 (95% CI, 1.015–1.826) and 1.65 (95% CI, 1.026–3.015), respectively. Following adjustment for the clinicopathological variables by logistic regression analysis, the adjusted ORs were 1.57 (95% CI, 1.124–1.912, adjusted P=0.032) for G/T and 1.86 (95% CI, 1.043–3.225, adjusted P=0.004) for T/T (Table 4).
Genotype/allele frequencies of R497K in exon 13
The three genotypes of R497K in EGFR, namely, Arg/Arg, Arg/Lys, and Lys/Lys, were detected. Each genotype is demonstrated by a representative sequencing wave in Figure 5. The distributions of genotype and allele of R497K in the two groups are described in Table 4. The rate of allele Arg was 64.24% (555/864) in the metastasis group, which was significantly higher than that in the nonmetastasis group (50.94% (432/848); OR, 1.76; 95% CI, 1.237–1.982; P=0.016).
  | Figure 5 Three representative sequencing waves of genotypes of R497K in EGFR. |
The distribution of genotype Arg/Arg in the pleural metastasis group was obviously higher compared with that in the nonmetastasis group, with ORs of 1.14 (95% CI, 1.008–1.314) for Arg/Lys and 1.87 (95% CI, 1.074–3.086) for Lys/Lys, respectively. Following adjustment for the clinicopathological variables using logistic regression analysis, the adjusted ORs were 1.18 (95% CI, 1.014–1.363; adjusted P=0.087) for Arg/Lys and 1.94 (95% CI, 1.081–3.162; adjusted P=0.038) for Lys/Lys.
The genotype distributions of -216G/T, R497K, and CA-SSR1 were in agreement with the Hardy–Weinberg’s equilibrium in the two groups (P>0.05).
EGFR expression
Experiments have demonstrated that CA-SSR1, -216G/T, and R497K variants resulted in EGFR activation and thereby increased EGFR expression. In order to further reveal the internal contact between different genotypes and EGFR gene expression, EGFR expression was detected in lung adenocarcinoma tissues of various CA-SSR1, -216G/T, and R497K genotypes by immunohistochemical staining.
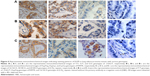
In total, 163 cases of lung adenocarcinoma tissues were detected, including 18 cases with the 16/16 CA genotype, 19 cases with the 18/18 genotype, and 20 cases with the 20/20CA genotype; 21 cases with the G/G genotype, 22 cases with the G/T genotype, and 19 cases with the T/T genotype; and 20 cases with the Arg/Arg genotype, 20 cases with the Arg/Lys genotype, and 22 cases with the Lys/Lys genotype, as shown in Figure 6; the diffuse/intense brown staining represented the positive expression of the EGFR protein. EGFR was mainly expressed on the membrane, but there were some expressions of EGFR in the cytoplasm. Weak positive to markedly positive staining for EGFR expression was detected.
The results of immunohistochemical staining showed that the average positive rates were 92%±3% in 16/16CA, 74%±6% in 16/18CA, 52%±4% in 18/18CA, and 39%±3% in 20/20CA genotype specimens, respectively. The expression of EGFR varied a lot in different genotype specimens (P<0.05). The rate of positive staining cells was significantly higher in subgroup of short alleles (<17) than that in subgroup of long alleles (≥17; 87%±2% vs 48%±5%, P=0.012).
The rates of positive staining cells were 88%±8% in T/T, 82%±5% in G/T, and 52%±5% in G/G genotype specimens, respectively, notably higher in genotypes of T/T or G/T than that in genotype of G/G (P=0.042).
The rates of positive staining cells were 81%±4% in Arg/Arg, 72%±8% in Arg/Lys, and 57%±6% in Lys/Lys genotype specimens, respectively, significantly lower in genotypes of Lys/Lys than that in genotypes of Arg/Arg or Arg/Lys (P=0.038).
Multifactor logistic regression analysis was applied to further investigate the correlation between CA-SSR1, -216G/T, and R497K polymorphisms. Common clinical features such as smoking, age, sex, and tumor differentiation degree and pleural metastasis are shown in Table 5.
The results of multifactor logistic regression analysis suggested that the factors including smoking, age, sex, and tumor differentiation degree were not closely associated with pleural metastasis (P>0.05), while the CA-SSR1, -216G/T, and R497K polymorphisms concerned the risk of the pleural metastasis of lung adenocarcinoma (P<0.05). The CA-SSR1, -216G/T, and R497K polymorphisms could be used as genetic susceptibility markers of pleural metastasis of lung adenocarcinoma.
Discussion
The results of clinical research showed that the overexpression of EGFR was closely related to the invasive and metastatic ability in NSCLC.38 EGFR overexpression resulting from genetic polymorphism has been found in other tumors such as breast cancer and stomach cancer,39–41 which induced and triggered a variety of cell transfers.42–44
EGFR inhibitors could inhibit tumor cells, including proliferation, differentiation, invasion, and metastasis of lung cancer cell. The process of cancer cell metastasis was associated with the activation of EGFR signal transduction pathways.45,46
The results of previous studies47–51 showed that single nucleotide polymorphisms usually modulated gene transcription and further affected the protein expression by interacting with trans-acting elements and cis-acting elements. These polymorphisms might modify the promoter activity. Previous studies have suggested that EGFR gene polymorphisms were associated with the development and metastasis of lung cancer.19 However, it remains largely unclear whether EGFR gene polymorphisms, such as CA-SSR1, -216G/T in the EGFR promoter, and R497K in exon 13 of the EGFR gene, play any critical roles in the pleural metastasis of lung adenocarcinoma, and the genetic mechanism by which the allele or genotype distribution of the EGFR gene polymorphism, including CA-SSR1, -216G/T, and R497K polymorphisms, affected the occurrence and development of pleural metastasis remains unclear.
Several studies have shown that the CA-SSR1, -216G/T, and R497K genotypes were detected in patients with pleural metastasis at different frequencies compared with cases of primary lung adenocarcinoma, and the expression of the EGFR protein also varied among different genotypes. All these results collectively indicated that CA-SSR1, -216G/T, and R497K were associated with the pleural metastasis of lung adenocarcinoma, possibly by affecting EGFR overexpression.
In our study, the results of genotyping of CA-SSR1, -216G/T, and R497K showed that patients with shorter CA-SSR1 (<17), the G/T or T/T genotypes, and the Arg/Lys or Arg/Arg genotypes had an increased risk of pleural metastasis of lung adenocarcinoma than those with longer length of CA-SSR1 (≥17), G/G, or Lys/Lys genotypes; CA-SSR1, -216G/T, and R497K polymorphisms were closely related to the pleural metastasis of lung adenocarcinoma. Our conclusion was consistent with the results of previous research.
EGFR was frequently overexpressed in a wide variety of solid tumors52 and thus represents an attractive target for novel treatments. The possibility that different genetic polymorphisms in the EGFR gene may regulate, at least in part, EGFR expression and/or activity, was an attractive hypothesis that could help identify the patients with tumors that were likely to concern pleural metastasis of lung adenocarcinoma.
The experimental results14 showed that the CA-SSR1 polymorphisms affected the activity of transcription factors in vivo and in vitro, and the number of CA-SSR1 was closely associated with the activity of gene transcription.
Buerger et al53 evaluated EGFR protein concentrations in specimens of 82 cases of patients with breast cancer using enzyme-linked immunosorbent assay quantitatively. All patients were divided into different subgroups according to different genotypes of the CA-SSR1 in EGFR. The results showed that the protein content was negatively correlated with CA repeat number: EGFR protein concentration was 127.5±47.4 fmol/mg in 15CA specimens, 52.0±59.1 fmol/mg in 18CA specimens, and varied a lot between short and long CA-SSR1 specimens (P<0.05).
Gebhardt et al14 and Tidow et al24 analyzed the mechanism behind CA-SSR1’s influence on protein expression. The research has found that CA-SSR1 was highly flexible, and the longer the repetitive sequence, the more the fold, and some of the biological molecules that possessed inhibitory activity for transcription or translation tended to be combined with CA-SSR1 that fold more than others, which in turn induced DNA structure change, inhibited the process of transcription and translation, and resulted in lower expression of EGFR protein and mRNA in patients with more CA-SSR1 repeat.
Gebhardt14 explored the gene transcriptional activity by applying seven kinds of cell lines in vivo and in vitro, respectively. These cell lines had no gene mutation but CA-SSR1 polymorphism. The experimental result showed that gene transcription activity was negatively related to the number of CA repeats, that is, more CA repeats would lead to lower transcriptional activity.
Previous research results25,53,54 also confirmed that EGFR protein was expressed more in tissues with CA homozygous genotype compared with CA heterozygote genotype. CA-SSR1 Polymorphism in Intron 1 of the EGFR Gene in Patients with Malignant Tumors Who Develop Acneiform Rash Associated with the Use of Cetuximab.55 Liu et al27 explored the correlation of EGFR gene transcription activity and CA-SSR1 polymorphisms in vitro. The results showed that the number of CA repeats increased from 16 to 21, and the transcription activity declined fivefold; the number of CA repeats decreased from 16 to 12, and the transcription activity increased fivefold. The experimental results fully confirmed that the CA-SSR1 polymorphism, namely, the number of CA repeats, influenced the expression of EGFR gene a lot.
CA-SSR1 might have a separate effect on gene regulation. CA-SSR1 polymorphism was associated with altering levels of EGFR transcription both in vitro and in vivo,42,52 and the length of CA-SSR1 inversely correlates with transcriptional activity of EGFR. The result of the previous study showed that CA-SSR1 affected the activity of gene transcription and translation. Compared with the shorter allele with 16 repeats, the longer allele with 21 repeats showed an 80% reduction of gene expression. Liu et al27 reported that transcriptional activity of EGFR gene decreased by fivefold when (CA)n repeats increased from 16/16CA to 21/21CA in vitro.
Previous studies32,56 have shown that 16 repeats and 20 repeats were the two most common alleles of CA-SSR1 of the EGFR gene. For the (CA)n repeat in intron 1 of EGFR polymorphism, alleles 20 (42.9%) and 16 (15.7%) were the two most common in a Chinese population in this study, which was similar to previous studies.57
In our study, the allele frequency distribution in the metastasis group ranged from 1.62% to 35.42% with a predominance of 16, 18, and 20 CA repeats, whereas in the nonmetastasis group it ranged from 1.42% to 39.15%, with the same predominance of 16, 18, and 20 CA repeats. Allele 16 frequency in the metastasis group was significantly higher than that in the nonmetastasis group (35.42% vs 19.58%, P=2.425×10−7); allele 20 CA frequency in the metastasis group was notably lower than that in the nonmetastasis group (28.94% vs 39.15%, P=0.002314). There were apparent differences between the two groups; allele 18 frequency in the metastasis group was lower than that in the nonmetastasis group (12.96% vs 17.92%), but there was no significant difference between the two groups (P=0.05883).
CA-SSR1 Polymorphism in Intron 1 of the EGFR Gene in Patients with Malignant Tumors Who Develop Acneiform Rash Associated with the Use of Cetuximab varied a lot in genotype of 16/16CA, 18/18CA, 16/20CA, and 20/20CA: 21.76%, 6.25%, 4.40%, and 17.59%; nonmetastasis group: 8.73%, 5.67%, 4.95% and 25.71%. The genotype frequency of 16/16CA in pleural metastasis was significantly lower compared with that in the nonmetastasis group, with ORs of 1.43 (95% CI, 1.032–1.764) for 18/18CA and 1.94 (95% CI, 1.074–2.077) for 20/20CA, respectively. Furthermore, following adjustment for the clinicopathological variables using logistic regression analysis, the adjusted ORs were 1.48 (95% CI, 1.415–1.845, adjusted P=0.132) for 18/18CA and 1.59 (95% CI, 1.075–3.002, adjusted P=0.034) for 20/20CA.
The result of immunohistochemistry showed that the average positive rate was 92%±3% in 16/16CA genotype specimens, 74%±6% in 16/18CA genotype specimens, 52%±4% in 18/18CA genotype specimens, and 39%±3% in 20/20CA genotype specimens, respectively. The expression of EGFR was significantly different in different polymorphism specimens (P<0.05). The rate of positive staining cells was much higher in subgroup of short alleles (<17) than that in subgroup of long alleles (≥17; 87%±2% vs 48%±5%, P=0.012).
Analyzing comprehensively the above results of our studies, we could conclude that the less the (CA)n repeated, the more the EGFR was expressed. It was the difference in genotype that resulted in differences in the expression of EGFR, which might cause pleural metastasis. CA-SSR1 polymorphism in EGFR was one of the risks for pleural metastasis.
This conclusion was basically consistent with that of the previous studies,58 which reported that expression of EGFR was significantly higher in primary lung adenocarcinoma patients with distant metastasis (M1) than those without distant metastasis (M0). This result showed that expression of EGFR played an important role in metastasis of lung cancer and other tumors.
Although there were enough data to show that CA-SSR1 had an important function of transcriptional regulation: mRNA and protein expressions decreased when the number of CA-SSR1 increased; mRNA and protein expression increased when CA-SSR1 decreased, the exact mechanism was not clear.
One possible mechanism was that the transcription activity of EGFR gene mainly depended on the combined effect of enhancer 2 and enhancer 3. As CA-SSR1 was located near the enhancer 2, there might be a regulating function of transcription in this region according to Helix theory. This needs further study in the near future.
The results of recent years’ research59 showed that -216G/T polymorphism, another polymorphism loci in the promoter region of EGFR, may independently regulate the activity and the process of gene transcription of EGFR promoter.
-216G/T, located in one of the four Sp1 recognition sites in the promoter region of EGFR, could adjust the promoter activity, influence the process of transcription and translation, and in turn affect the promoter activity and EGFR gene expression. Nomura et al21 found that average mRNA content was 1.0, average protein content was 0.97 in -216G/G genotype of tracheal epithelial cell lines, whereas average mRNA content was 1.2 and average protein content was 1.24 in -216G/T or -216T/T genotypes of tracheal epithelial cell lines. Although there was no significant difference in mRNA and protein contents between -216G/G genotype and -216G/T or -216T/T genotype, transcription activity tended to increase, that is, the change (G→T) at site -216 led to a high expression of EGFR.
In our study, the genotype and allele frequencies of -216G/T were detected, and each genotype was demonstrated by a representative sequencing wave. The rate of allele T was 32.87% in the metastasis group, much higher than that in the nonmetastasis group (24.41%; OR =1.61, 95% CI: 1.228–1.916; adjusted P=0.021). The distribution of G/T and T/T genotypes in the pleural metastasis group was significantly higher than that in the nonmetastasis group, with ORs of 1.42 (95% CI, 1.015–1.826) and 1.65 (95% CI, 1.026–3.015), respectively. Following adjustment for the clinicopathological variables using logistic regression analysis, the adjusted ORs were 1.57 (95% CI, 1.124–1.912; adjusted P=0.032) for G/T and 1.86 (95% CI, 1.043–3.225; adjusted P=0.004) for T/T. There were significant differences in genotype and allele frequency of the -216G/T between the nonmetastasis group and the pleural metastasis group.
The result of immunohistochemistry showed that the average positive rates were 88%±8% in T/T, 82%±5% in G/T, and 52%±5% in G/G genotype specimens, respectively, and the rate of positive staining cell was significantly higher in genotypes of T/T or G/T than that in genotype of G/G (P=0.042).
Based on the results of immunohistochemistry and different distributions of G/T and T/T genotypes in the nonmetastasis group and the metastasis group (41.28% vs 53.01%) in this study, it was reasonable to conclude that the mutations or variants in -216G/T in EGFR were closely associated with pleural metastasis of adenocarcinoma of lung, and the possible mechanism was that -216G/T variants promote EGFR overexpression, which in turn promote the pleural metastasis of lung adenocarcinoma.
In addition to CA-SSR1 and -216G/T, another polymorphism locus, namely, R497K in exon 13, with an Arg to Lys substitution at codon 497 of EGFR, was identified in 1993.60 R497K (Arg→Lys) was closely related to the occurrence, development, and transfer of cancer.60,61 Allele Arg in patients with distant metastasis was obviously higher than that in patients without distant metastasis. Compared with the allele Lys, allele Arg might be a higher risk factor for distant metastases.
Experiment showed that, compared with wild-type 497 R, mutation of 497 K could weaken the function of ligand binding, growth stimulation, glycine kinase activity and decrease the function of inducing proto-oncogene, such as myc, fos, and jun.34
Studies had shown that R497K polymorphism in exon 13 influenced some cancers.61,62 For example, Zhou et al63 reported in a study about R497K polymorphism in patients with dilated cardiomyopathy that the frequency of Lys allele in dilated cardiomyopathy patients was obviously higher than that in control subjects (64.4% and 53.8%, in dilated cardiomyopathy patients and control subjects, respectively). The frequency for Lys/Lys genotype was significantly overrepresented in dilated cardiomyopathy patients (P=0.020, for Lys/Lys vs Arg/Arg).
Our study detected that Arg allele in 64.24% of the patients with pleural metastasis was higher than that in 50.94% in patients with primary lung adenocarcinoma (OR, 1.76; 95% CI, 1.237–1.982; P=0.016).
For the R497K polymorphism, in the pleural metastasis group, 40.51% of the patients were homozygous for the Arg/Arg variant, 47.45% for Arg/Lys, and 12.04% for the Lys/Lys genotype. In the nonmetastasis group, 25.24% of the patients were homozygous for the Arg/Arg variant, 51.42% for Arg/Lys, and 23.35% for the Lys/Lys genotype. The distributions of genotypes and alleles of the R497K polymorphism were statistically different between the two groups. The genotype frequencies of Arg/Arg in the pleural metastasis group were higher than those in the nonpleural metastasis group, with ORs of 1.14 (95% CI, 1.008–1.314) for Arg/Lys and 1.87 (95% CI, 1.074–3.086) for Lys/Lys, respectively. Furthermore, following adjustment for the clinicopathological variables by logistic regression analysis, the adjusted ORs were 1.18 (95% CI, 1.014–1.363; adjusted P=0.087) for Arg/Lys and 1.94 (95% CI, 1.081–3.162; adjusted P=0.038) for Lys/Lys.
The result of immunohistochemistry in our study showed that the rates of positive staining cells were 81%±4% in Arg/Arg, 72%±8% in Arg/Lys, and 57%±6% in Lys/Lys genotype specimens, respectively, significantly lower in genotypes of Lys/Lys than that in genotypes of Arg/Arg or Arg/Ly (P=0.038).
The results mentioned above have demonstrated a clear trend that EGFR expression decreased when Arg allele was replaced by Lys allele at codon 497 of EGFR.
After comprehensive analysis of the previous studies, we could conclude that the Arg/Arg genotype was significantly higher in the pleural metastasis group than that in the nonmetastasis group, which resulted in a higher EGFR protein expression in the metastasis group than that in the nonmetastasis group. Compared with the patients with the Lys/Lys genotype, those with the Arg/Arg genotype suffered increased risk of pleural metastasis.
In summary, it was the difference in EGFR polymorphism that resulted in the difference of EGFR protein expression, which might cause pleural metastasis. EGFR gene polymorphism, especially -216G/T, R497K polymorphism in exon 13, and CA-SSR1 of the EGFR gene might be one of the molecular mechanisms of pleural metastasis of lung cancer.
Study limitations
First, a limited sample size should be considered; second, the molecular mechanism by which the EGFR gene polymorphisms were associated with pleural metastasis of lung cancer was unknown; third, in terms of EGFR gene expression, we had only discussed the impact of CA-SSR1, R497K, -216G/T without further exploration of the interaction among the three gene polymorphisms on the expression of EGFR protein; moreover, other polymorphisms of EGFR gene polymorphisms might also contribute to variability in EGFR expression. These issues need further study.
Conclusion
This study has demonstrated for the first time that the -216G/T polymorphism in the EGFR promoter, R497K polymorphism in exon 13, and CA-SSR1 of the EGFR gene were genetic susceptibility factors for the pleural metastasis of lung adenocarcinoma, with the -216T allele, Arg allele, and shorter CA-SSR1 (<17) genotypes being associated with increased metastatic risk.
Acknowledgments
This study was supported by all health care workers of Department of Cardiothoracic Surgery, Weixia Ma, Mingzhong Tian, Yanming Shen, Haimei Wang, Xiaojing Tan, and other physicians in Thoracoscopic Division and Bronchoscopy Room, Dongying People’s Hospital, who have provided selfless help to complete all specimen collection in this study. We would like to express our heartfelt thanks for their selfless help.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300. | ||
Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284(1):31–53. | ||
Murillo-Carretero M, Torroglosa A, Castro C, Villalobo A, Estrada C. S-Nitrosylation of the epidermal growth factor receptor: a regulatory mechanism of receptor tyrosine kinase activity. Free Radic Biol Med. 2009;46(4):471–479. | ||
Kerr KM. Pulmonary adenocarcinomas: classification and reporting. Histopathology. 2009;54(1):12–27. | ||
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. | ||
Hemming AW, Davis NL, Kluftinger A, et al. Prognostic markers of colorectal cancer: an evaluation of DNA content, epidermal growth factor receptor, and Ki-67. J Surg Oncol. 1992;51(3):147–152. | ||
Fontanini G, Vignati S, Bigini D, et al. Epidermal growth factor receptor (EGFR) expression in non-small cell lung carcinomas correlates with metastatic involvement of hilar and mediastinal lymph nodes in the squamous subtype. Eur J Cancer. 1995;3lA(2):178–183. | ||
Rao C, Hu Q, Ma J, et al. Comparison of the epidermal growth factor receptor protein expression between primary non-small cell lung cancer and paired lymph node metastases: implications for targeted nuclide radiotherapy. J Exp Clin Cancer Res. 2010;29(1):7. | ||
Yudoh K, Matsui H, Kanamori M, Maeda A, Ohmori K, Tsuji H. Effects of epidermal growth factor on invasiveness through the extracellular matrix in high- and low-metastatic clones of RCT sarcoma in vitro. Jpn J Cancer Res. 1994;85(1):63–71. | ||
Veale D, Ashcroft T, Marsh C, Gibson GJ, Harris AL. Epidermal growth factor receptor in non-small-cell lung cancer. Br J Cancer. 1997;55(5):513–516. | ||
Scagliotti GV, Selvaggi G, Novello S, Hirsch FR. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res. 2004;10(12 pt 2):4227s–4232s. | ||
Franklin WA, Veve R, Hirsch FR, Helfrich BA, Bunn PA Jr. Epidermal growth factor receptor family in lung cancer and premalignancy. Semin Oncol. 2002;29(1 suppl 4):3–14. | ||
Roskoski R Jr. The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem Biophys Res Commun. 2004;319(1):1–11. | ||
Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274(19):13176–l3l80. | ||
Selvaggi G, Novello S, Torri V, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15(1):28–32. | ||
Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005;23(14):3227–3234. | ||
Eberhard DA, Giaccone G, Johnson BE; Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in non-small-cell lung cancer working group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26(6):983–994. | ||
Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. | ||
Araújo A, Ribeiro R, Azevedo I, et al. Genetic polymorphisms of the epidermal growth factor and related receptor in non-small cell lung cancer – a review of the literature. Oncologist. 2007;12(2):201–210. | ||
Choi JE, Park SH, Kim KM, et al. Polymorphisms in the epidermal growth factor receptor gene and the risk of primary lung cancer: a case-control study. BMC Cancer. 2007;7:199. | ||
Nomura M, Shigematsu H, Li L, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4(4):e125. | ||
Buerger H, Packeisen J, Boecker A, Tidow N, Kersting C. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence-first results of an interethnic breast cancer study. J Pathol. 2004;203(1):545–550. | ||
Jami MS, Hemati S, Salehi Z, Tavassoli M. Association between the length of a CA dinucleotide repeat in the EGFR and risk of breast cancer. Cancer Invest. 2008;26(4):434–437. | ||
Tidow N, Boecker A, Schmidt H, et al. Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development. Cancer Res. 2003;63(6):1172–1178. | ||
Amador ML, Oppenheimer D, Perea S, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epider-mal growth factor receptor inhibitors. Cancer Res. 2004;64(24):9139–9143. | ||
Merlino GT, Ishii S, Whang-Peng J, et al. Structure and localization of genes encoding aberrant and normal epidermal growth factor receptor RNAs from A431 human carcinoma cells. Mol Cell Biol. 1985;5(7):1722–1734. | ||
Liu W, Innocenti F, Wu MH, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65(1):46–53. | ||
Kageyama R, Merlino GT, Pastan I. A transcription factor active on the epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1988;85(14):5016–5020. | ||
Chen LL, Clawson ML, Bilgrami S, Carmichael G. A sequence-specific single-stranded DNA-binding protein that is responsive to epidermal growth factor recognizes an S1 nuclease-sensitive region in the epidermal growth factor receptor promoter. Cell Growth Differ. 1993;4(12):975–983. | ||
Johnson AC, Ishii S, Jinno Y, Pastan I, Merlino GT. Epidermal growth factor receptor gene promoter. Deletion analysis and identification of nuclear protein binding sites. J Biol Chem. 1988;263(12):5693–5699. | ||
Kageyama R, Merlino GT, Pastan I. Epidermal growth factor (EGF) receptor gene transcription. Requirement for Sp1 and an EGF receptor-specific factor. J Biol Chem. 1988;263(13):6329–6336. | ||
Grinstein E, Jundt F, Weinert I, Wernet P, Royer HD. Sp1 as G1 cell cycle phase specific transcription factor in epithelial cells. Oncogene. 2002;21(10):1485–1492. | ||
McKibbin T, Zhao W, Tagen M, et al. Epidermal growth factor receptor polymorphisms and risk for toxicity in paediatric patients treated with gefitinib. Eur J Cancer. 2010;46(11):2045–2051. | ||
Moriai T, Kobrin MS, Hope C, Speck L, Korc M. A variant epidermal growth factor receptor exhibits altered type alpha transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci U S A. 1994;91(21):10217–10221. | ||
Nagashio R, Sato Y, Matsumoto T, et al. Expression of RACK1 is a novel biomarker in pulmonary adenocarcinomas. Lung Cancer. 2010;69(1):54–59. | ||
Kato T, Daigo Y, Aragaki M, et al. Overexpression of MAD2 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2011;74(1):124–131. | ||
Wu SG, Chang YL, Lin JW, et al. Including total EGFR staining in scoring improves EGFR mutations detection by mutation-specific antibodies and EGFR TKIs response prediction. PLoS One. 2011;6(8):e23303. | ||
Sasaki H, Yukiue H, Mizuno K, et al. Elevated serum epidermal growth factor receptor level is correlated with lymph node metastasis in lung cancer. Int J Clin Oncol. 2003;8(2):79–82. | ||
Han HS, Eom DW, Kim JH, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer. 2011;12(6):380–386. | ||
Wang H, Zhou M, Shi B, et al. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia. 2011;13(5):461–471. | ||
Gregorc V, Hidalgo M, Spreafico A, et al. Germline polymorphisms in EGFR and survival in patients with lung cancer receiving gefitinib. Clin Pharmacol Ther. 2008;83(3):477–484. | ||
Xue C, Wyckoff J, Liang F, et al. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66(1):192–197. | ||
Price JT, Wilson HM, Haites NE. Epidermal growth factor (EGF) increases the in vitro invasion, motility and adhesion interactions of the primary renal carcinoma cell line, A704. Eur J Cancer. 2008;32A(11):924–930. | ||
Turner T, Chen P, Goodly LJ, Wells A. EGF receptor signaling enhances in vivo invasiveness of DU-145 human prostate carcinoma cells. Clin Exp Metastasis. 1996;14(4):409–418. | ||
Yang Z, Bagheri-Yarmand R, Wang RA, et al. The epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) suppresses c-Src and Pak1 pathways and invasiveness of human cancer cells. Clin Cancer Res. 2004;10(2):658–667. | ||
Ueno S, Mojic M, Ohashi Y, Higashi N, Hayakawa Y, Irimura T. Asialoglycoprotein receptor promotes cancer metastasis by activating the EGFR-ERK pathway. Cancer Res. 2011;71(20):6419–6427. | ||
Baseggio L, Bartholin L, Chantome A, Charlot C, Rimokh R, Salles G. Allele-specific binding to the-308 single nucleotide polymorphism site in the tumour necrosis factor-a promoter. Eur J Immunogenet. 2004;31(1):15–19. | ||
Gazzoli I, Kolodner RD. Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol Cell Biol. 2003;23(22):7992–8007. | ||
Harendza S, Lovett DH, Panzer U, Lukacs Z, Kuhnl P, Stahl RA. Linked common polymorphisms in the gelatinase a promoter are associated with diminished transcriptional response to estrogen and genetic fitness. J Biol Chem. 2003;278(23):20490–20499. | ||
Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular helial growth factor gene promoter. Cancer Res. 2003;63(4):812–816. | ||
Mann V, Hobson EE, Li B, et al. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest. 2001;107(7):899–907. | ||
Shin DM, Ro JY, Hong WK, Hittelman WN. Dysregulation of epidermal growth factor receptor expression in premalignant lesions during head and neck tumorigenesis. Cancer Res. 1994;54(12):3153–3159. | ||
Buerger H, Gebhardt F, Schmidt H, et al. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 2000;60(4):854–857. | ||
Gebhardt F, Bürger H, Brandt B. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations – a link between genetics and epigenetics. Histol Histopathol. 2000;15(3):929–936. | ||
Jarząbek T, Rucińska M, Rogowski W, et al. CA-SSR1 polymorphism in intron 1 of the EGFR gene in patients with malignant tumors who develop acneiform rash associated with the use of cetuximab. Mol Diagn Ther. 2015;19(2):79–89. | ||
Yusoff P, Lao DH, Ong SH, et al. Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J Biol Chem. 2002;277(5):3195–3201. | ||
Lin-Bo Gao, Bin Zhou, Lin Zhang, et al. R497K polymorphism in epidermal growth factor receptor gene is associated with the risk of acute coronary syndrome. BMC Medical Genetics. 2008;9:74. | ||
Tateishi M, Ishida T, Kohdono S, Hamatake M, Fukuyama Y, Sugimachi K. Prognostic influence of the co-expression of epidermal growth factor receptor and c-erbB-2 protein in human lung adenocarcinoma. Surg Oncol. 1994;3(2):109–113. | ||
Wu SG, Gow CH, Yu CJ, et al. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J. 2008;32(4):924–930. | ||
Moriai T, Kobrin MS, Korc M. Cloning of a variant epidermal growth factor receptor. Biochem Biophys Res Commun. 1993;191(3):1034–1039. | ||
Sasaki H, Okuda K, Shimizu S. EGFR R497K polymorphism is a favorable prognostic factor for advanced lung cancer. J Cancer Res Clin Oncol. 2009;135(2):313–318. | ||
Wang WS, Chen PM, Chiou TJ, et al. Epidermal growth factor receptor R497K polymorphism is a favorable prognostic factor for patients with colorectal carcinoma. Clin Cancer Res. 2007;13(12):3597–3604. | ||
Zhou B, Rao L, Peng Y, Zhang Q, Zhang L. Epidermal growth factor receptor gene polymorphisms, R497K, but not (CA)n repeat, is associated with dilated cardiomyopathy. Clin Chim Acta. 2009;403(1–2):184–187. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.