Back to Journals » Journal of Inflammation Research » Volume 14
Coronil, a Tri-Herbal Formulation, Attenuates Spike-Protein-Mediated SARS-CoV-2 Viral Entry into Human Alveolar Epithelial Cells and Pro-Inflammatory Cytokines Production by Inhibiting Spike Protein-ACE-2 Interaction
Authors Balkrishna A, Haldar S, Singh H, Roy P, Varshney A
Received 19 December 2020
Accepted for publication 29 January 2021
Published 16 March 2021 Volume 2021:14 Pages 869—884
DOI https://doi.org/10.2147/JIR.S298242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Video abstract of "Coronil: a potential inhibitor of SARS-CoV-2" [ID 298242].
Views: 1893
Acharya Balkrishna,1,2 Swati Haldar,1 Hoshiyar Singh,1 Partha Roy,3 Anurag Varshney1,2
1Drug Discovery and Development Division, Patanjali Research Institute, Haridwar, 249405, Uttarakhand, India; 2Department of Allied and Applied Sciences, University of Patanjali, Haridwar, 249405, Uttarakhand, India; 3Molecular Endocrinology Laboratory, Department of Biotechnology, Indian Institute of Technology Roorkee, Roorkee, 247667, Uttarakhand, India
Correspondence: Anurag Varshney
Drug Discovery and Development Division, Patanjali Research Institute, Roorkee-Haridwar Road, Haridwar, 249405, Uttarakhand, India
Tel +91-1334-244107, Ext.: 7458
Fax +91-1334-244805
Email [email protected]
Purpose: Coronil is a tri-herbal formulation containing extracts from Withania somnifera, Tinospora cordifolia, and Ocimum sanctum. Recently, it was shown that Coronil rescued humanized zebrafish from SARS-CoV-2 induced pathologies. Based on reported computational studies on the phytochemicals present in Coronil, it could be a potential inhibitor of SARS-CoV-2 entry into the host cell and associated cytokines’ production.
Methods: Through an ELISA-based biochemical assay, effects of Coronil on interaction between ACE-2 and different mutants of viral spike (S) protein, crucial for viral invasion of host cell, were evaluated. Additionally, using recombinant pseudoviruses having SARS-CoV-2 spike (S) protein in their envelopes and firefly luciferase reporter in their genomes, effects of Coronil on virus entry into human alveolar epithelial cells were evaluated through luciferase assay. UHPLC profiled Coronil also modulated S-protein mediated production of pro-inflammatory cytokines in A549 cells, like interleukin-6 (IL-6), interleukin-1β (IL-1β), and tumor necrosis factor-α (TNF-α), as evaluated through RT-qPCR and ELISA.
Results: Coronil effectively inhibited the interaction of ACE-2 not only with the wild-type S protein (SWT) but also with its currently prevalent and more infectious variant (SD614G) and another mutant (SW436R) with significantly higher affinity toward ACE-2. Treatment with Coronil significantly reduced the increased levels of IL-6, IL-1β, and TNF-α in A549 cells incubated with different S-protein variants in a dose-dependent manner. Likewise, it also prevented the SARS-CoV-2 S-protein pseudotyped vesicular stomatitis virus (VSVppSARS-2S) mediated cytokine response in these cells by reducing entry of pseudoviruses into host cells.
Conclusion: Coronil prevented SARS-CoV-2 S-protein mediated viral entry into A549 cells by inhibiting spike protein-ACE-2 interactions. SARS-CoV-2 S protein induced inflammatory cytokine response in these cells was also moderated by Coronil.
Keywords: Coronil, SARS-CoV-2, pseudovirus, spike protein, ACE-2, pro-inflammatory cytokines
Introduction
The COVID-19 pandemic is persistent and not showing any sign of fading away in the near future. Naturally, there is high demand toward finding a cure or a vaccine against it. The causative coronavirus, SARS-CoV-2, has originated from the same clade that produced SARS (SARS-CoV) and MERS (MERS-CoV), back in 2002 and 2012 respectively.1 Like SARS-CoV, this virus also enters host cells through ACE-2 receptors. Therefore, the alveolar type II epithelial cells with high level of expression of ACE-2 are the primary target of SARS-CoV-2 coronavirus.2 SARS-CoV-2 infection triggers an uncontrolled immune response directly responsible for severe COVID-19 prognosis.3 Therefore, the attempts toward therapeutic interventions are mainly focused on the development of drugs or vaccines that ultimately prevent the entry of SARS-CoV-2 into the host cells. It is presumed that this would automatically reduce the viral load and associated mal-adaptive immune responses leading to serious pathophysiological conditions.4 In order to control the sudden outbreak of the disease and rapid spread of the infection at the first instance, it has been attempted to repurpose several known drugs like hydroxychloroquine and remdesivir.5–7 However, with the outcomes of clinical trials using such repurposed drugs not being quite encouraging, there is a pressing need for the development of newer therapeutics which can specifically act against SARS-CoV-2. Among various modes of therapeutics, traditional systems of medicines which are mainly composed of herbal and other natural resources provide hope because of their multi-targeted functionality and minimal side effects.8
In this study, Coronil, a tri-herbal formulation, has been evaluated for its efficacy as an anti-viral therapy by inhibiting viral entry into host/target cells. Coronil is formulated based on the principles of Ayurveda, which has been used to treat respiratory ailments with symptoms overlapping with those of COVID-19. Coronil contains extracts of Withania somnifera, Tinospora cordifolia, and Ocimum sanctum. A large body of experimental evidence has demonstrated the anti-viral and immunomodulatory activities of the principal components present in Coronil. W. somnifera has been identified as one of the medicinal plants capable of boosting immunity among poultry infected with Newcastle Disease virus (NDV), belonging to Paramyxoviridae family.9 Recently root extract of W. somnifera has been reported to inhibit infection of chicken embryo fibroblasts by Bursal disease virus (BSV) in vitro.10 It was also reported to be effective in reducing human immunodeficiency virus (HIV)-induced neurotoxicity.11 Phytochemicals present in Withania somnifera have potential anti-viral activity against H1N1 influenza virus.12 Withaferin A, present in W. somnifera extract, was noted to effectively ameliorate inflammatory cytokine responses in pulmonary fibrosis and attenuate the expressions of fibrotic proteins, thus, proving to be a potential therapeutic agent against this lung disorder.13 Computational studies have shown that phytochemicals present in W. somnifera are capable of attenuating the interaction between host cell ACE-2, viral spike (S) protein, and main protease (Mpro), which is crucial for viral entry into host cells.14–16 Like W. somnifera, T. cordifolia is a potent immuno-stimulant which has been used as remedy against various microbial infections.17 According to a recent report, extracts from the stems of T. cordifolia inhibited the growth of Herpes Simplex virus (HSV)18 and other extracts of this plant have also been reported to be used in treating HIV patients.19,20 Extracts of O. sanctum are known for their anti-asthmatic and bronchodilation effects against histamine induced bronchospasm.21,22 Besides, like W. somnifera and T. cordifolia, O. sanctum is also a strong immunomodulant.22
In the current study, the ability of Coronil to inhibit ACE-2 and S protein interaction was assessed through an ELISA based assay. Once it was established that Coronil could remarkably inhibit the interaction of ACE-2 and S proteins biochemically, we analyzed its capacity to prohibit entry of SARS-CoV-2 S protein pseudotyped viruses into human alveolar epithelial A549 cells, expressing ACE-2. We also evaluated the effect of Coronil in ameliorating SARS-CoV-2 S protein- and pseudovirus-induced cytokine response in human alveolar epithelial cells.
Materials and Methods
Chemicals and Reagents
Cell culture media, namely RPMI1640, DMEM, and supplements like fetal bovine serum (FBS) and penicillin streptomycin mixture were obtained from Thermo Fisher Scientific (Waltham, MA, USA). The levels of cytokines such as interleukin-6 (IL-6) (Catalog number: 51–26451E, 51–26452E), interleukin-1 beta (IL-1β) (Catalog number: 51–9,002,512, 519,002,516), and tumor necrosis factor alpha (TNF α) (Catalog number: 51–26371E, 51–26372E) were quantified using respective ELISA kits from BD Biosciences (San Jose, CA, USA). Purified SARS-CoV-2 Spike (S) proteins and purified human ACE-2 protein were procured from Sino Biological US Inc. (Pennsylvania, USA). Lipofectamine 3000 (Catalog number L3000015) was obtained from Thermo Fisher Scientific (Waltham, MA, USA). All the other chemicals and reagents were from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. Coronil (Batch No. #A-CNT194, expiry June 2023) was obtained from Divya Pharmacy (Haridwar, India).
Compositional Analysis of Coronil
Compositional analysis was performed by Prominence-XR UHPLC system (Shimadzu, Japan) equipped with Quaternary pump (Nexera XR LC-20AD XR), DAD detector (SPD-M20 A), auto-sampler (Nexera XR SIL-20 AC XR), degassing unit (DGU-20A 5R), and column oven (CTO-10 AS VP). The separation was achieved using a Shimadzu Shim pack GIST-HP C18 (3 µm, 3 X 100 mm) column subjected to binary gradient elution. The two solvents used for the analysis consisted of water containing 0.1% orthophosphoric acid, pH adjusted to 2.5 with diethylamine (solvent A) and acetonitrile (solvent B). Gradient programming of the solvent system was initially at 5% B for 0–10 min, 5–15% B from 10–20 min, 15–25% B from 20–40 min, 25–65% B from 40–60 min, 65–90% B from 60–65 min, 90–95%B from 65–66 min, 5% B from 66–70 min, with a flow rate of 0.7 mL/min. Next 10 µL of standard and test solutions were injected and column temperature was maintained at 30°C. Wavelengths were set at 227 nm (for Withanone, Withaferin A, Withanoside IV, Withanoside V, Codifolioside A, and Magnoflorine), 325 nm (for Rosmarinic acid and Palmatine), and 210 nm (Ursolic acid and Betulinic acid).
The standards used in the compositional analysis of Coronil were prepared as follows:
Palmatine hydrochloride (purity-75.1%, Sigma Aldrich, Missouri, USA), Cordifolioside A (purity-98.0%, Chem Faces, Hubei, China), Magnoflorine (purity-99% Sigma Aldrich, Missouri, USA), Withanone (purity-93.6%, Natural Remedies, Noida, India), Withaferin A (purity-98%, Natural Remedies, Noida, India), Withanoside IV (Purity-96.2%, Natural Remedies, Noida, India), Withanoside V (purity-97.6%, Natural Remedies, Noida, India), Rosmarinic acid (Purity-98%, Sigma Aldrich, Missouri, USA), Ursolic acid (purity-97.3%, Tokyo Chemical Industries, Tokyo, Japan), and betulinic acid (purity-98.7%, Natural Remedies, Natural Remedies, Noida, India) were dissolved in analytical grade methanol (Sigma Aldrich, Missouri, USA) to prepare the appropriate concentrations of standard stock solutions.
For preparing the test sample, 0.5 g of powdered Coronil sample (Batch No. #A-CNT194, expiry June 2023) was diluted with 10 mL water: methanol (20:80) and then sonicated for 30 min. This was then centrifuged at 10,000 rpm for 5 min at room temperature and then filtered using 0.45 µm nylon filter. The filtrate thus obtained was used for the analysis.
ELISA-Based Assay to Determine Interaction Between Human ACE-2 and SARS-CoV-2 Spike (S) Proteins
Interactions between ACE-2 and three different types of SARS-CoV-2 spike (S) proteins, namely, wild type (Catalog number: 40,592-V08B) and two naturally occurring variants, D614G mutant (Aspartic acid “D” at 614th position replaced with Glycine “G”) (Catalog number: 40,591-V08H3) and W436R mutant (Tryptophan at 436th position replaced with Arginine “R”) (Catalog number: 40,592-V08H9) were quantitatively evaluated through an ELISA based SARS-CoV-2 Inhibitor screening Kit (Catalog number: EP‐105) from AcroBiosytems, Newark, USA, according to manufacturer’s protocol. Briefly, the wells of ELISA plate were separately coated overnight with 25 ng of different S proteins as described previously. Following thorough washings, 62.5 ng of purified biotinylated human ACE-2 protein (Catalog number: 10,108-H08H-B) was added to each well in a total volume of 100 µL with different concentrations of Coronil (0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1.0, 3.0, 10, and 30 µg/mL) and incubated for 30 min. Negative control included only ACE-2 and S proteins without any inhibitor. Positive control was supplied along with the kit and included a chemical inhibitor against ACE2-S protein interaction. Interactions between ACE-2 and S proteins were detected by adding streptavidin conjugated horse-radish peroxidase (streptavidin-HRP) along with 3.3′,5,5′-Tetramethylbenzidine (TMB) (Catalog number: 555,214, BD bioscience, San Diego, USA) as substrate followed by measuring the absorbance at 450 nm using Envision microplate reader (Perkin Elmer, USA). Percent (%) inhibition of interaction between ACE-2 and S proteins was calculated considering the interaction to be 100% in negative control group.
Determination of Cytokine Response Elicited in Alveolar Epithelial A549 Cells by Different S Proteins
Human alveolar epithelial A549 cells were obtained from ATCC recognized cell repository at National Centre for Cell Sciences (NCCS, Pune, India). For this assay, A549 cells were seeded at a density of 5 X 105 cells/mL and incubated at 37°C in the presence of 5% CO2 in a humidified incubator until 50–60% confluency. Subsequently, the cells were incubated with 8000 ng/mL, for wild type and W436R mutants while 20,000 ng/mL was used for D614G (mutant) for 48 h, followed by the estimation of different secreted pro-inflammatory cytokines. IL-6, TNF-α, and IL-1β (BD bioscience, San Diego, USA) were estimated in the supernatants of cultured cells by using specific ELISA kits for each of these cytokines, as per the manufacturer’s protocol. Absorbance was measured at 450 nm using the Envision microplate reader (Perkin Elmer, USA).
Pseudotyping of Vesicular Stomatitis Virus (VSV) with SARS-CoV-2 S Protein
VSV pseudotyping with SARS-CoV-2 S protein was performed according to earlier reports.23,24 Briefly, human kidney epithelial (HEK293) cells (from NCCS, Pune, India) were co-transfected with helper plasmids expressing VSV N, P and L proteins and pCG1-SARS-2S plasmid expressing SARS-CoV-2S protein 24 h before transducing with VSV*ΔG-FLuc viruses for 1 h. VSV*ΔG-FLuc viruses, helper plasmids and pCG1-SARS-2S construct were kind gifts from Prof. Stefan Pӧhlmann (Deutsches Primatenzentrum GmbH, Leibniz-Institut für Primatenforschung, Kellnerweg 4, 37,077 Göttingen, Germany). The supernatant containing SARS-CoV-2S pseudotyped VSV viruses (VSVppSARS-2S) were collected after 36 h and stored at −80°C to be used in subsequent experiments.
Luciferase-Based Analysis to Check Entry of Pseudotyped Virus into Target Cells
Luciferase assay was conducted according to published protocols with slight modifications.23,25 Briefly, 70–80% confluent A549 cells pre-treated for 16 h with different concentrations of Coronil (1, 3, and 10 µg/mL), were transduced with VSVppSARS-2S [at a multiplicity of infection (m.o.i.) of 5] and incubated at 37°C for 6 h. Subsequently, the medium was aspirated and lysate prepared in 50 µL Cell Culture Lysis 5X Reagent (Promega, Catalog number: E153A). To 20 µL of this lysate, 30 µL of luciferase substrate (Promega, Catalog number: E151A) reconstituted in Luciferase Assay Buffer (Promega, Catalog number: E152B) was added and immediately, firefly luciferase activity was measured using Envision microplate reader (Perkin Elmer, USA). The luciferase activity was used to depict the extent of VSVppSARS-2S entry into the cells and was represented as x-fold over background. The cells without viral transduction and the cells transduced with VSVppSARS-2S but without any treatment, were considered as normal control and disease control respectively. Virus transduced cells treated with 10 µM of camostat mesylate were used as positive control.23
Determination of the Level of Transcription of ACE-2 and Cytokines in Alveolar Epithelial A549 Cells in Response to Transduction with VSVppSARS-2S
mRNA levels of ACE-2 and different pro-inflammatory cytokines, namely, IL-1β, IL-8, IL-6 and TNF-α in normal, VSVppSARS-2S transduced and Coronil treated A549 cells were determined through RT-qPCR. cDNA was prepared using Verso cDNA Synthesis Kit (Catalog number: AB1453/B, Thermo Scientific, USA) according to manufacturer’s protocol. cDNA, thus synthesized, was used in RT-qPCR reaction set up with primers for ACE-2 (5ʹ- GGGATCAGAGATCGGAAGAAGAAA-3ʹ and 5ʹ- AGGAGGTCTGAACATCATCAGTG-3ʹ), IL-1β (5ʹ- AAGCTGATGGCCCTAAACAG-3ʹ and 5ʹ- AGGTGCATCGTGCACATAAG-3ʹ), IL-8 (5ʹ- TTCTAGGACAAGAGCCAGGAAG-3ʹ and 5ʹ- GGGTGGAAAGGTTTGGAGTATG-3ʹ), IL-6 (5ʹ- TCGAGCCCACCGGGAACGAA-3ʹ and 5ʹ- GTGGCTGTCTGTGTGGGGCG-3ʹ) and TNF-α (5ʹ- ATGAGCACTGAAAGCATGATCG-3ʹ and 5ʹ- GAGGGCTGATTAGAGAGAGGTC-3ʹ) using PowerUp SYBR Green Master Mix (Catalog no. A25778, Applied Biosystems, USA) according to manufacturer’s guidelines in qTOWER3G real time PCR machine (Analytic Jena, Germany). mRNA levels of ACE-2 were expressed relative to the house keeping gene PPIA (5ʹ-CCCACCGTGTTCTTCGACATT-3ʹ and 5ʹ-GGACCCGTATGCTTTAGGATGA-3ʹ). Alterations in the mRNA levels of IL-1β, IL-8, IL-6, and TNF-α genes were expressed as fold changes with respect to their corresponding levels in normal untreated, non-transduced A549 cells.
Determination of Pseudotyped Virus (VSVppSARS-2S)-Induced Cytokine Response in A549 Cells
50–60% confluent A549 cells were transduced with VSVppSARS-2S pseudoviruses. The levels of secreted pro-inflammatory cytokines, IL-6, IL-1β, and TNF-α in the supernatant were measured after 48 h of incubation through ELISA as described earlier.
Statistical Analysis Used in This Study
All quantitative data were represented as mean ± SEM. Statistical significance of the variations observed between the means of different groups was determined through one-way ANOVA followed by Dunnett’s multiple comparisons and denoted either with # or * depending on whether the comparison was with the normal control or transduced cells using GraphPad Prism software (version 7) (California, USA).
Results
Coronil is Enriched with Steroidal Lactones, Terpenoids, Alkaloids, and Furan Glycosides
Basically, Coronil has been formulated by using the aqueous extracts of Withania somnifera, Tinospora cordifolia, and Ocimum sanctum. As shown in Figure 1 and Table 1, the UHPLC pattern depicted that chemically, Coronil is a rich blend of several phytochemicals; steroidal lactones, of which Withaferin A (1.752 µg/mg), Withanoside IV (2.673 µg/mg) and V (0.822 µg/mg) and Withanone (0.008 µg/mg) are the most predominant ones. All these steroidal lactones were mainly from W. somnifera. Among other phytochemicals the major alkaloids like Magnoflorine (1.478 µg/mg) and Palmatine (0.027 µg/mg) and furan glycoside, Cordifolioside A (0.181 µg/mg) were from T. cordifolia. Further, O. sanctum contributed polyphenolic compound like Rosmarinic acid (0.091 µg/mg) and terpenoids like Betulinic (0.193 µg/mg) and Ursolic (0.046 µg/mg) acids.
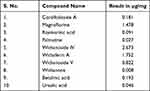 |
Table 1 Phytochemical Composition of Coronil |
Coronil Inhibits Interaction Between Human ACE-2 Receptor and Viral S Proteins
Coronil is a tri-herbal formulation containing phytochemicals from Withania somnifera, Tinospora cordifolia, and Ocimum sanctum. Based on our earlier computational analysis we could deduce that withanone and tinocordiside, the major phytochemicals present in Coronil as well, could impede the molecular interactions between human ACE-2 receptor and SARS-CoV-2 S protein.16,26 Therefore, as biochemical validation and the first step toward understanding the mechanism of action of Coronil as a potent anti-viral agent, we assessed its competency to interfere with ACE-2-S protein interaction through an ELISA based assay (Figure 2A). For this assay, essentially, biotinylated purified human ACE-2 protein was allowed to bind to the immobilized naturally occurring variations of SARS-CoV-2 S proteins, namely, SWT (the one identified at the beginning of the pandemic), SD614G (with the D at 614th position replaced by G) and SW436R (with R at 436th position instead of original W). A known inhibitor of ACE-2-S protein interaction provided with the kit was used as a positive control, while ACE-2 and S proteins without any inhibitor served as the negative control. The extent of interaction observed for the negative controls was considered as 100% and relative to this the inhibitions as displayed by positive control and different concentrations of Coronil, were calculated. Our data showed that Coronil exerted a dose-dependent inhibitory effect on the interactions between ACE-2 and all variants of S proteins (Figure 2B–D), albeit the magnitude of inhibition varied for each mutant. With our experimental set-up, we observed a maximum inhibition of ACE-2-S protein interactions ranging between 25–30%, of which the maximum response was observed with SW436R variant. Consequently, in order to perceive the comparative performance of Coronil with different variants of S proteins, IC20 values were calculated for each type and were found to be 19.08, 20.08, and 0.55 µg/mL for SWT (Figure 2B), SD614G (Figure 2C) and SW436R (Figure 2D), respectively, clearly suggesting better inhibitory potency of the formulation when SW436R mutant is the interacting partner for ACE-2 receptor. Therefore, from these observations, the potential of Coronil to impede the crucial molecular cross-talk between human ACE-2 receptor and viral S proteins becomes evident.
Coronil Attenuates Viral S Protein-Induced Cytokine Activation in A549 Cells
It has already been reported that SARS-CoV-2 interacts with ACE-2 receptors present on the alveolar type II (AT-II) epithelial cells invading host immune system. Therefore, we chose the pulmonary epithelial A549 cells, characterized as AT-II model, for all our further in vitro studies.27 Although, in case of SARS-CoV-2 infection, the main source of pathogenic IL-6 and pro-inflammatory cytokines (TNF-α and IL-1β) is myeloid cells,28 increased expressions of these cytokines are also reported in A549 cells due to Mycoplasma pneumoniae and Influenza A virus infections.29,30 Besides, A549 cells exhibit cytokine activation in response to inflammatory stimulus by lipopolysaccharides (LPS).31 Subsequent to our earlier observation that Coronil can inhibit ACE-2-S protein interaction, we evaluated the effect on cytokine activation in response to viral proteins by incubating A549 cells with S protein variants and measured the levels of immunoreactive pro-inflammatory cytokines in the presence of different concentrations (1, 3, 10, and 30 µg/mL) of Coronil. Cytosafety of Coronil in A549 cells has already been established up to 300 µg/mL (Figure 9a of reference 32).32 The cells incubated with S proteins but not treated with Coronil were included as disease control to measure the extent of respective cytokine activation evoked (Figure 3A). The cells without S proteins and Coronil treatment were taken as normal control. While in the presence of SWT and SW436R, we observed significant increases in the levels of immunoreactive cytokines, such reaction was not observed in case of SD614G variants for any of the cytokines tested by us (Figure 3B–D). Interestingly, treatment of cells with increasing concentrations of Coronil resulted in the reduction of these cytokines, although, dose-dependency was found only in some cases (Figure 3B–D). As shown in Figure 3B, the levels of IL-6 were elevated by about 4-fold in response to both SWT and SW436R proteins (p<0.05). Coronil was found to reduce the level of IL-6 close to normalcy in a dose dependent manner in SWT protein treated cells. A significant reduction in the level of IL-6 in SW436R protein treated cells was noted in response to Coronil, however, no dose dependent effect was observed in case of this mutant. The disease control groups of SWT and SW436R treated cells experienced almost 3.6- and 5.5-fold increases in TNF-α levels, respectively, as compared to vehicle treated control groups (p<0.05). However, treatment with Coronil demonstrated a dose-dependent inhibition of this elevated TNF-α level restoring it close to normal control (Figure 3C). When tested for IL-1β, as shown in Figure 3D, a 10-fold increase in the level of IL-1β was observed when the A549 cells were incubated with SWT protein which was subsequently attenuated almost close to normalcy upon treatment with Coronil (p<0.05). We did not notice any dose dependent effect of Coronil in this case, which needs further validation with lower doses of this drug. When analyzed for SW436R variant, it was found that the production of IL-1β increased only by ~3-fold which was significantly inhibited by Coronil close to the level of normal control, although here we also did not observe any dose dependency (Figure 3D). Taken together, it is evident that purified SARS-CoV-2 S proteins can elicit cytokine response in pulmonary epithelial cells in vitro and that Coronil can attenuate that response mostly restoring the cytokine levels to normalcy albeit its impacts vary based on the type of mutation that exists in the viral S proteins.
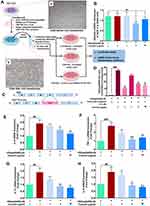
Coronil Inhibits Entry of VSVppSAR2S in Alveolar Epithelial Cells
The efficient management of S protein induced cytokine responses suggests that Coronil might act as entry inhibitor for SARS-CoV-2 virus. Vesicular stomatitis virus (VSV) pseudotyped with SARS-CoV-2 S protein was used to check this possibility. HEK293 cells were used to produce VSVpp2S viruses. Transduced VSV*ΔG fLuc viruses were repackaged with SARS-CoV-2 S glycoprotein in their capsid instead of its native G protein (Figure 4A). VSVpp2S viruses, thus produced, elicited cytopathic effects (Figure 4A, panel b) in HEK293 cells (Figure 4A, panel a) thereby confirming their release into the medium. The SARS-CoV-2 S protein pseudotyped VSV viruses (VSVppSARS-2S) were collected for transducing A549 cells for future experimentations. A549 cells pre-treated with different concentrations of Coronil (1, 3, and 10 µg/mL) were transduced with VSVppSARS-2S particles. Coronil treatment was continued in these cells. Pseudovirus transduced cells without any treatment and uninfected untreated cells were included as disease and normal controls, respectively. We did not observe any difference in the mRNA levels of ACE-2 across different groups (Figure 4B). The amount of VSVppSARS-2S entering the pulmonary epithelial A549 cells was quantified through luciferase activity produced by firefly luciferase gene (fLuc), stably integrated in the genome of VSVppSARS-2S25 (Figure 4C). The pseudovirus-transduced cells exhibited around 35-fold more virus entry when compared to uninfected cells (considered as background) (Figure 4D). The infected cells treated with camostat mesylate, a clinically proven serine protease effective against TMPRSS 2, showed almost 3.5-fold decrease in virus entry relative to the infected cells.23 Interestingly, virus entry observed for cells treated with Coronil (1, 3, and 10 µg/mL) was significantly reduced as compared to untreated cells and was almost comparable to that found in cells treated with camostat mesylate. Thus, based on these data it would be conceived that Coronil is capable of preventing entry of SARS-CoV-2 into host cells. The transcript levels of different pro-inflammatory cytokines namely, IL-6, TNF-α, IL-1β, and IL-8 were found to be significantly elevated in the cells transduced with pseudoviruses (Figure 4E–H). Upon Coronil treatment, the mRNA levels of these cytokines exhibited dose-dependent reduction, suggesting a cytokine-moderating effect of this medicine.
Coronil Rescues the Alveolar Epithelial Cells from VSVpp2S-Induced Cytokine Release
SARS-CoV-2 infection sets the immune system into an active mode thus triggering cytokine release syndrome which in general terms is also called “cytokine storm”. In a pragmatic context, mimicking SARS-CoV-2 virus induced cytokine surge, we used non-pathogenic, replication defective SARS-CoV-2 S protein pseudotyped vesicular stomatitis viruses (VSVpp2S) to transduce A549 cells and follow the levels of immunoreactive pro-inflammatory cytokines through ELISA with and without Coronil treatment. For this part of the experiment, VSVpp2S transduced A549 cells (without any Coronil treatment) were considered as disease control while uninfected cells served as normal control (Figure 5A). The infected cells were treated with different concentrations of Coronil (1, 3, and 10 µg/mL) and the subsequent effects on the levels of immunoreactive TNF-α, IL-1β, and IL-6 were assessed through ELISA as described earlier. VSVpp2S virus infected A549 cells experienced 4.5, 10, and 7-fold increases in IL-6, TNF-α, and IL-1β levels, respectively (Figure 5B–D). Treatment with Coronil significantly reduced their levels, close to the level shown by normal control (uninfected) cells. Surprisingly, the reduction in the level of the tested cytokines in response to Coronil was not found to be dose-dependent. This fact needs further validation with a wider range of doses for Coronil in our future studies. Taken together, these observations implicate the potential of Coronil as an antagonist of cytokine release syndrome.
Discussion
SARS-CoV-2 coronavirus infection engenders a strange sort of immune response, in which the initial phase resembles the regular innate anti-viral reaction, but soon a mal-responsive adaptive immune retaliation sets in that eventually leads to severe outcomes, like acute respiratory distress syndrome and multi-organ failure.33–36 Despite several studies, a clear understanding of COVID-19 associated immune response is still unmet and appears to be a combination of several pathological manifestations with several unresolved prognoses. Plausibly, these features of SARS-CoV-2 are responsible, to a great extent, for the continuing absence of specific COVID-19 treatment. Knowledge-base of traditional systems of medicines, particularly Traditional Chinese Medicine (TCM) has been successfully used against the SARS-CoV pandemic in 2003.37 On similar lines, TCM has been employed against the current pandemic as well, although, in both cases, its application has been restricted to China. Nevertheless, some of the reported information is encouraging enough to explore other ancient systems of medicine, Ayurveda being one of them. Ayurveda is a 5000-year old system of traditional Indian medicine worth considering as an option for treating COVID-19.8,38
The current study was designed with a straightforward objective of evaluating the ability of an Ayurvedic formulation, Coronil, to inhibit viral entry into the host cell. There are several independent computational studies, a few initial ones being from our group as well, that predicted anti-viral effects of the phytochemicals identified in Coronil, like Cordifolioside A, Magnoflorine, Rosmarinic acid, Palmatine, Withanoside IV, Withaferin A, Withanoside V, Withanone, Betulinic acid and Ursolic acid, against SARS-CoV-2. Earlier, we demonstrated through molecular docking and dynamic simulation that Withanone is capable of inhibiting the interaction between human host cell ACE-2 receptor and viral S protein.16,26 Similarly, the study by Kumar et al15 predicted that Withaferin A could bind to TMPRSS 2, the host serine protease important for SARS-CoV-2 entry.23 Shree et al39 observed through computational study that Withanoside V and Ursolic acid can bind SARS-CoV-2 MPro with high affinities. Likewise, an in-silico study by Tripathi et al40 determined that Withanoside IV and V can bind to MPro. Sagar and Kumar41 noted that Magnoflorine and Palmatine bind to SARS-CoV-2 MPro, surface glycoprotein and RNA polymerase. Chowdhury et al42 predicted that tetrahydropalmatine, an important phytoconstituent of T. cordifolia, can bind SARS-CoV-2 Mpro, although, this phytocompound was not detected in Coronil by UHPLC analysis. Carino et al43 showed biochemically that Betulinic acid disrupts ACE-2-RBD interaction. Betulinic acid, a naturally occurring pentacyclic triterpenoid and its derivatives are reported to be effective in inhibiting the growth of HSV, HIV, IFV, and ECHO6 viruses in vitro.44–46 Similarly, other phytochemicals like Ursolic have been found to be effective against rotavirus47 and Rosmarinic acid against enterovirus 71 in vitro48 and Japanese Encephalitis virus (JEV) in a mouse model.49 A study by Yan et al50 found that rosmarinic acid can also bind to SARS-CoV-2 MPro. Tinocordiside, a major phytochemical present in T. cordifolia has been reported by multiple independent computational studies to interact with SARS-CoV-2 surface glycoprotein and RNA polymerase,41 besides the viral MPro.39,41 In-silico study from our group also showed that tinocordiside binds at the interface of ACE-2-RBD complex and energetically weakens this interaction.26 Altogether, this evidence suggests that the herbal constituents present in Coronil have a proven record of anti-viral activities, albeit to a wide range of viral strains/species, which actually substantiates our current observations. Therefore, it was intriguing to experimentally evaluate the existing computational observations.
An ELISA-based protein-protein interaction assay was used in this study to assess the ability of Coronil to inhibit ACE-2-S protein interactions. Three different variants of S protein, namely, SWT (the original strain that appeared in the beginning of the pandemic), SD614G (Aspartic acid “D” at 614th position has been naturally replaced by a Glycine “G”), and SW436R (Tryptophan “W” at 436th position replaced by Arginine “R”) were used in this interaction study. Currently, SARS-CoV-2 with SD614G mutation is the predominantly circulating variety.51 D614G mutation does not affect the affinity of S protein for ACE-2 receptors. An indirect reflection of this fact was noticed in the comparable inhibitory effects that Coronil exerted on ACE-2 – SWT and ACE-2 – SD614G interactions, as evident from similar IC20 values of 19.08 and 20.08 µg/mL, respectively. However, it does increase the viral infectivity as evident from higher respiratory tract viral loads in case of infections caused by this variant of SARS-CoV-2.51 This mutation quantitatively improved S protein incorporation into the virions during packaging, which explains the observed increase in transmission and infectivity.52 However, D614G mutation was not associated with disease severity.51,52 Our observation that A549 cells exposed to SD614G protein did not exhibit increase in the levels of any of the secreted pro-inflammatory cytokines, namely IL-6, TNF-α, and IL-1β, corroborates the clinical report of no association of this mutation with disease severity.51 Being an RNA virus, SARS-CoV-2 genome is undergoing mutations albeit at a slower pace than reported for HIV.51 Nevertheless, based on the evolutionary advantage to be gained by the virus, residues included under hot and cold spots of positive selection pressure have been predicted. W436 is one such hot-spot residue and the W436R mutation displayed increased ACE-2 affinity.53,54 Complex formed due to ACE-2 - SW436R interaction is more rigid than ACE-2-SWT complex.54 We observed that Coronil inhibited ACE-2 - SW436R interaction with much greater efficiency than it did for ACE-2-SWT or ACE-2-SD614G interactions, as shown by IC20 value being almost 36 times less than those for the latter cases. This suggested that Coronil would be a promising viral entry antagonist for SARS-CoV-2 SW436R mutant with higher affinity for ACE-2, should it happen to make mass appearance. Until now, this mutation has been reported in only one isolate.54 The cytokine response evoked by SW436R protein in A549 cells was prominent and comparable to that observed for SWT protein. Our data showed that Coronil attenuated this response with equal efficiency in both the cases. Finally, infecting A549 cells with SARS-CoV-2 S protein pseudotyped vesicular stomatitis virus (VSV) elicited cytokine response as well as the expression of luciferase gene, both of which were efficiently attenuated by Coronil. Taken together, these observations can be attributed to the effective inhibition of virus entry into the host cell by Coronil.
Human lungs are the primary targets of SARS-CoV-2 infection. Therefore, we chose to conduct our studies with A549 cells of human alveolar epithelial origin which are also shown to express ACE2 (Figure 4B), the host cell receptor protein indispensable for SARS-CoV-2 entry. Vero E6 cells, on the other hand, are monkey kidney cells used for general cytotoxicity assessments, and also for the replication studies for certain viruses like SARS-CoV-2.55–57 In addition, A549 cells also elicit sizable cytokine response on stimulation by SARS-CoV-2 S proteins, therefore, these cells are an ideal model system to study SARS-CoV-2 entry and induced inflammatory responses. Although A549 cells express ACE2 receptors, the gaseous exchange and its concomitant effect on virus entry into host cells could not be simulated under these conditions. Moreover, our observations were based on near maximal bioavailability of Coronil, as under in-vitro conditions, which may not be the case in real life context, particularly when it is administered orally. Thus, the in-vitro experimental set-up cannot really be correlated with in-vivo conditions in terms of pharmacokinetics. Nevertheless, such a system seamlessly ensures the authenticity of the biochemical and cell biological studies, more importantly - in this context of understanding - the mode of action of Coronil as the SARS-CoV-2 entry inhibitor. In addition, we included only three SARS-CoV-2 variants in our current study, which might be seen as a limitation. However, when this study was being conducted, these mutations were the prominent ones, particularly the D614G variant.
According to Butterweck and Weber, severe COVID-19 prognosis can be an outcome of cross-reactivity of anti-SARS-CoV-2 nucleoprotein antibody with human interleukin-11 (IL-11), the anti-inflammatory cytokine responsible for tissue protection.58 Our current data showed that Coronil can effectively suppress the initial cytokine response resembling the innate anti-viral reaction. In fact, one of our recent studies showed that Coronil could rescue zebrafish with swim bladders xenotransplanted with human alveolar epithelial A549 cells from SARS-CoV-2 spike (S) protein induced pathologies.32 Subsequent to SARS-CoV-2 entry into the human body, cytokine responses are mounted in which pro-inflammatory cytokines, interleukin-6 (IL-6), interleukin-1β (IL-1β), and tumor necrosis factor-α (TNF-α) are highly elevated. These cytokines affect each other’s levels which is responsible for the complex cytokine response that follows SARS-CoV-2 infection in humans. In our recent study32 this situation was mimicked in vitro by treating pulmonary epithelial A549 cells with pro-inflammatory cytokine IL-1β to induce increase in IL-6 and TNF-α levels. The dose-dependent effect of Coronil was checked in this set-up. It was observed that the levels of IL-6 and TNF-α in IL-1β treated cells were reduced in a dose-dependent manner upon treatment with Coronil (Figures 9 of the reference 32).32 TNF-α is one of the master transcriptional regulators known to mediate cytokine response through a downstream effector, NF-κB, which is also a transcriptional regulator. So, the effect of Coronil on TNF-α induced NF-κB-mediated response was checked using a transactivation assay (Figure 9d of the reference 32).32 For this purpose, a stable cell line, HEK-Blue engineered to express secreted alkaline phosphatase (SEAP) enzyme from an NF-κB/AP-1 (co-factor Activator protein-1) regulatable promoter was used. When TNF-α was added to these cells, NF-κB expression was triggered which resulted in SEAP expression, quantifiable through an enzymatic reaction. This quantification indirectly reflected the amount of TNF-α to which the cells have been exposed. Secreted TNF-α harvested from Coronil treated A549 cells with induced inflammation showed lower SEAP activity than that collected from untreated cells (Figure 9d of the reference 32).32 This indicated that Coronil helped in reducing the inflammatory rise in TNF-α levels. All these observations suggested that Coronil can prevent viral entry and associated immunogenic responses, implying reduced chances of disease progression beyond the innate immune response phase into the adaptive response stage. Also, the chances of the adaptive response getting over-driven are also reduced. All these may finally eliminate the possibilities of anti-nucleoprotein antibody-mediated organ failure and severe disease outcomes.
Conclusion
In conclusion, this study provides evidence in support of a tri-herbal formulation based on Ayurvedic (Indian traditional medicinal) system, that can effectively ameliorate the cytokine response mounted during SARS-CoV-2 infection and thus inhibit the viral infection by acting at various levels of physiological systems (Figure 6). In light of the current evidence, Coronil has turned out to be a promising candidate, particularly with its similar effectivity against different variants of S protein. It is further considered, that being a purely tri-herbal formulation, and the long history of usage of its phytoconstituents for curing various ailments (including anti-viral), that all the anti-COVID-19 effects imparted by this formulation would have minimum adverse side effects. In fact, Kaplan-Meier survival analysis from our recent study32 (Figure 2C of reference 32) showed that Coronil did not have any adverse effects on the zebrafish. All these data thus warrant pre-clinical and clinical studies to develop Coronil into an anti-COVID-19 therapeutic agent, which is our future study and is in progress. For all future studies, the data obtained out of this initial but well-planned and consistent experimental set-up, have provided a strong base and strengthened the hope for development of a cure for COVID-19 using the knowledge of traditional healthcare systems like Ayurveda.
Abbreviations
ACE-2, Angiotensin Converting Enzyme 2; ATCC, American Type Culture Collection; COVID-19, Coronavirus Disease 2019; DMEM, Dulbecco’s Modified Eagle Medium; ELISA, Enzyme-linked Immunoassay; FBS, Fetal Bovine Serum; HRP, Horse Radish Peroxidase; IL-1β, Interleukin-1β; IL-6, Interleukin-6; MERS, Middle East Respiratory Syndrome; MERS-CoV, Middle East Respiratory Syndrome-related Coronavirus; Mpro, Main Protease; NCCS, National Centre for Cell Science; RBD, Receptor Binding Domain; RPMI, Roswell Park Memorial Institute Medium; RT-qPCR, Quantitative reverse transcription PCR; S protein, Spike protein; SARS, Severe Acute Respiratory Syndrome; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; TNF-α, Tumor Necrosis Factor-α; UHPLC, Ultra-high-performance liquid chromatography; VSV, Vesicular Stomatitis Virus; VSVppSARS-2S, SARS-CoV-2S pseudotyped VSV.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
We express our humble gratitude toward Prof. Stefan Pӧhlmann and Dr. Markus Hoffmann (Deutsches Primatenzentrum GmbH, Leibniz-Institut für Primatenforschung, Kellnerweg 4, 37077 Göttingen, Germany) for sharing VSV*ΔG-fLuc viruses and helper plasmids for pseudotyping with us. We thank Ms. Meenu Tomer, Mr. Sudeep Verma and Dr. Jyotish Srivastava for their help with HPLC analysis; Dr. Pratima Singh, Mr. Ajeet Chauhan, Mr. Shoor Singh and Mr. Arun Raturi for their help with visual graphics. We extend our gratitude to Ms. Priyanka Kandpal, Mr. Tarun Rajput, Mr. Gagan Kumar and Mr. Lalit Mohan for their swift administrative support.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This research received no external funding. This presented work has been conducted using internal research funds from Patanjali Research Foundation Trust, Hardwar, India.
Disclosure
The test article was provided by Divya Pharmacy, Haridwar, Uttarakhand, India. Acharya Balkrishna is an honorary trustee in Divya Yog Mandir Trust, in addition he holds an honorary managerial position in Patanjali Ayurved Ltd, Haridwar, India. Besides, providing the test article, Divya Pharmacy was not involved in any aspect of this study. All other authors have no conflicts of interest to declare.
References
1. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020;(October). doi:10.1038/s41579-020-00459-7
2. Ni W, Yang X, Yang D, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):1–10. doi:10.1186/s13054-020-03120-0
3. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi:10.1016/j.cytogfr.2020.05.003
4. Felsenstein S, Herbert JA, Mcnamara PS, Hedrich CM. COVID-19: immunology and treatment options. Clin Immunol J. 2020;215:108448. doi:10.1016/j.clim.2020.108448
5. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi:10.1016/S0140-6736(20)31022-9
6. Geleris J, Sun Y, Platt J, et al. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N Engl J Med. 2020;382:2411–2418. doi:10.1056/nejmoa2012410
7. Boulware DR, Pullen MF, Bangdiwala AS, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N Engl J Med. 2020;1–9. Doi:10.1056/nejmoa2016638.
8. Kumar G, Kumar D, Singh NP. Therapeutic approach against 2019-nCoV by inhibition of ACE-2 receptor. Drug Res (Stuttg). 2020. doi:10.1055/a-1275-0228
9. Raza A, Muhammad F, Bashir S, et al. Antiviral and immune boosting activities of different medicinal plants against Newcastle disease virus in poultry. Worlds Poult Sci J. 2015;71(3):523–532. doi:10.1017/S0043933915002147
10. Ganguly B, Umapathi V, Rastogi SK. Nitric oxide induced by Indian ginseng root extract inhibits Infectious Bursal Disease virus in chicken embryo fibroblasts in vitro. J Anim Sci Technol. 2018;60(1):4–8. doi:10.1186/s40781-017-0156-2
11. Williams-Orlando C. Human immunodeficiency virus and herbal medicine. Altern Complement Ther. 2017;23(2):51–59. doi:10.1089/act.2017.29099.cwo
12. Cai Z, Zhang G, Tang B, Liu Y, Fu X, Zhang X. Promising anti-influenza properties of active constituent of withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: computational study. Cell Biochem Biophys. 2015;72(3):727–739. doi:10.1007/s12013-015-0524-9
13. Bale S, Venkatesh P, Sunkoju M, Godugu C. An adaptogen: withaferin a ameliorates in vitro and in vivo pulmonary fibrosis by modulating the interplay of fibrotic, matricelluar proteins, and cytokines. Front Pharmacol. 2018;9(MAR):1–16. doi:10.3389/fphar.2018.00248
14. Kumar V, Dhanjal JK, Kaul SC, Wadhwa R, Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J Biomol Struct Dyn. 2020. doi:10.1080/07391102.2020.1772108
15. Kumar V, Dhanjal JK, Bhargava P, et al. Withanone and withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J Biomol Struct Dyn. 2020. doi:10.1080/07391102.2020.1775704
16. Balkrishna A, Pokhrel S, Singh H, et al. Withanone from Withania somnifera attenuates SARS-CoV-2 RBD and host ACE2 interactions to rescue spike protein induced pathologies in humanized zebrafish model. Drug Des Dev Ther. 2020.
17. Sinha K, Mishra N, Singh J, Khanuja S. Tinospora cordifolia (Guduchi), a reservoir plant for therapeutic applications: a review. Indian J Tradit Knowl. 2004;3(3):257–270.
18. Pruthvish R, Gopinatha SM. Antiviral prospective of Tinospora cordifolia on HSV-1. Int J Curr Microbiol Appl Sci. 2018;7(1):3617–3624. doi:10.20546/ijcmas.2018.701.425
19. Kalikar M, Thawani V, Varadpande U, Sontakke S, Singh R, Khiyani R. Immunomodulatory effect of Tinospora cordifolia extract in human immuno-deficiency virus positive patients. Indian J Pharmacol. 2008;40(3):107–110. doi:10.4103/0253-7613.42302
20. Akram M, Tahir IM, Shah SMA, et al. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother Res. 2018;32(5):811–822. doi:10.1002/ptr.6024
21. Singh S, Agrawal SS. Anti-asthmatic and anti-inflammatory activity of Ocimum sanctum. Int J Pharmacogn. 1991;29(4):306–310. doi:10.3109/13880209109082904
22. Vinaya BLK, Kamdod MA, Swamy M, Swamy M. Bronchodilator activity of Ocimum sanctum Linn. (tulsi) in mild and moderate asthmatic patients in comparison with salbutamol: a single-blind cross-over study. Int J Basic Clin Pharmacol. 2017;6(3):511. doi:10.18203/2319-2003.ijbcp20170543
23. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi:10.1016/j.cell.2020.02.052
24. Whitt MA. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169(2):365–374. doi:10.1007/978-3-030-57317-1_5
25. Rentsch MB, Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6(10). doi:10.1371/journal.pone.0025858
26. Balkrishna A, Pokhrel S, Varshney A. Tinospora cordifolia (Giloy) may curb COVID-19 contagion: tinocordiside disrupts the electrostatic interactions between ACE2 and RBD. Comb Chem High Throughput Screen. 2020;23(1):1–12. doi:10.2174/1386207323666201110152615
27. Foster KA, Oster CG, Mayer MM, Avery ML, Audus KL. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp Cell Res. 1998;243:359–366. doi:10.1006/excr.1998.4172
28. Gubernatorova EO, Gorshkova EA, Polinova AI, Drutskaya MS. IL-6: relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020;53:13–24. doi:10.1016/j.cytogfr.2020.05.009
29. Yang J, Hooper WC, Phillips DJ, Talkington DF. Regulation of proinflammatory cytokines in human lung epithelial cells infected with Mycoplasma pneumoniae. Infect Immun. 2002;70(7):3649–3655. doi:10.1128/IAI.70.7.3649-3655.2002
30. Wang J, Wang Q, Han T, et al. Soluble interleukin-6 receptor is elevated during influenza A virus infection and mediates the IL-6 and IL-32 inflammatory cytokine burst. Cell Mol Immunol. 2015;12:633–644. doi:10.1038/cmi.2014.80
31. Crestani B, Cornillet P, Dehoux M, Rolland C, Guenounou M, Aubier M. Alveolar type II epithelial cells produce interleukin-6 in vitro and in vivo: regulation by alveolar macrophage secretory products. J Clin Invest. 1994;94(2):731–740. doi:10.1172/JCI117392
32. Balkrishna A, Solleti SK, Verma S, Varshney A. Application of humanized zebrafish model in the suppression of SARS-CoV-2 spike protein induced pathology by tri-herbal medicine coronil via cytokine modulation. Molecules. 2020;25(21):5091. doi:10.3390/molecules25215091
33. Wang W, Ye L, Ye L, et al. Up-regulation of IL-6 and TNF-alfa induced by SARS-coronavirus spike protein in murine macrophages via NF-kB pathway. Virus Res. 2007;128:1–8. doi:10.1016/j.virusres.2007.02.007
34. Dosch SF, Mahajan SD, Collins AR. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi:10.1016/j.virusres.2009.01.005
35. Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID-19. Immunity. 2020;53:19–25. doi:10.1016/j.immuni.2020.06.017
36. Buszko M, Park J, Verthelyi D, Sen R, Young HA, Rosenberg AS. The dynamic changes in cytokine responses in COVID-19: a snapshot of the current state of knowledge. Nat Immunol. 2020;21(10):1146–1151. doi:10.1038/s41590-020-0779-1
37. Yang Y, Islam MS, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16(10):1708–1717. doi:10.7150/ijbs.45538
38. Rastogi S, Narayan D, Harsh R. COVID-19 pandemic: a pragmatic plan for ayurveda intervention. J Ayurveda Integr Med. 2020. doi:10.1016/j.jaim.2020.04.002
39. Shree P, Mishra P, Selvaraj C, et al. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants–Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)–a molecular docking study. J Biomol Struct Dyn. 2020;1–14. Doi:10.1080/07391102.2020.1810778.
40. Tripathi MK, Singh P, Sharma S, Singh TP, Ethayathulla AS, Kaur P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J Biomol Struct Dyn. 2020;1–14. doi:10.1080/07391102.2020.1790425
41. Sagar V, Kumar AH. Efficacy of natural compounds from tinospora cordifolia against SARS-CoV-2 protease, surface glycoprotein and RNA polymerase. Biol Eng Med Sci Rep. 2020;6(1):6–8. doi:10.5530/bems.6.1.2
42. Chowdhury P. In silico investigation of phytoconstituents from Indian medicinal herb “Tinospora cordifolia (giloy)” against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J Biomol Struct Dyn. 2020;1–18. doi:10.1080/07391102.2020.1803968
43. Carino A, Moraca F, Fiorillo B, et al. Hijacking SARS-CoV-2/ACE2 receptor interaction by natural and semi-synthetic steroidal agents acting on functional pockets on the receptor binding domain. Front Chem. 2020;8:1–26. doi:10.3389/fchem.2020.572885
44. Pavlova NI, Savinova OV, Nikolaeva SN, Boreko EI, Flekhter OB. Antiviral activity of betulin, betulinic and betulonic acids against some enveloped and non-enveloped viruses. Fitoterapia. 2003;74(5):489–492. doi:10.1016/S0367-326X(03)00123-0
45. Lai W, Huang L, Ho P, Li Z, Montefiori D, Chen CH. Betulinic acid derivatives that target gp120 and inhibit multiple genetic subtypes of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2008;52(1):128–136. doi:10.1128/AAC.00737-07
46. Hong EH, Song JH, Kang KB, Sung SH, Ko HJ, Yang H. Anti-influenza activity of betulinic acid from Zizyphus Jujuba on influenza A/PR/8 virus. Biomol Ther. 2015;23(4):345–349. doi:10.4062/biomolther.2015.019
47. Tohmé MJ, Giménez MC, Peralta A, Colombo MI, Delgui LR. Ursolic acid: a novel antiviral compound inhibiting rotavirus infection in vitro. Int J Antimicrob Agents. 2019;54(5):601–609. doi:10.1016/j.ijantimicag.2019.07.015
48. Lin WY, Yu YJ, Jinn TR. Evaluation of the virucidal effects of rosmarinic acid against enterovirus 71 infection via in vitro and in vivo study. Virol J. 2019;16(1):1–9. doi:10.1186/s12985-019-1203-z
49. Swarup V, Ghosh J, Ghosh S, Saxena A, Basu A. Antiviral and anti-inflammatory effects of rosmarinic acid in an experimental murine model of Japanese encephalitis. Antimicrob Agents Chemother. 2007;51(9):3367–3370. doi:10.1128/AAC.00041-07
50. Yan Y, Shen X, Cao Y, Zhang J, Wang Y. Discovery of anti-2019-nCoV agents from 38 Chinese patent drugs toward respiratory diseases via docking screening. Preprints. 2020. doi:10.20944/preprints202002.0254.v2
51. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi:10.1016/j.cell.2020.06.043
52. Zhang L, Jackson CB, Mou H, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv Prepr Serv Biol. 2020. doi:10.1101/2020.06.12.148726
53. Padhi AK, Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol Transl Sci. 2020;17–20. doi:10.1021/acsptsci.0c00113
54. Ou J, Zhou Z, Dai R, et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. bioRxiv. 2020;1–30. Doi:10.1101/2020.03.15.991844.
55. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi:10.1073/pnas.2002589117
56. Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101(9):925–940. doi:10.1099/jgv.0.001453
57. Harcourt J, Tamin A, Lu X, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020;26(6):1266–1273. doi:10.3201/EID2606.200516
58. Butterweck H, Weber A. A potential mechanism for triggering cytokine storm in Covid-19 patients. COJ Biomed Sci Res. 2020;1(2):
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.