Back to Journals » OncoTargets and Therapy » Volume 11
Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma
Authors Cao S, Chen G, Yan L, Li L, Huang X
Received 18 June 2018
Accepted for publication 14 August 2018
Published 24 October 2018 Volume 2018:11 Pages 7385—7394
DOI https://doi.org/10.2147/OTT.S177524
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Sai Cao,1,* Guohao Chen,2,* Liumei Yan,3,* Libo Li,2 Xianying Huang4
1Department of Thoracic Surgery, Nanfang Hospital, Southern Medical University, Guangdong 510515, China; 2Department of General Surgery, Dongguan People’s Hospital, Dongguan 523000, China; 3Department of Gastroenterology, Wuzhou Red Cross Hospital, Wuzhou 543000, China; 4Department of Vascular Surgery, Nanfang Hospital, Southern Medical University, Guangdong 510515, China
*These authors contributed equally to this work
Purpose: Accumulating evidence demonstrates that circRNAs regulate diverse cellular processes and cancer progression. However, it remains unclear whether circRNAs play any functional role in esophageal squamous cell carcinoma (ESCC).
Materials and methods: The significance of circRNA_100876 in ESCC was analyzed by studying circRNA_100876 expression in ESCC tissues and the association between circRNA_100876 expression and clinicopathologic parameters. The biological effects of circRNA_100876 knockdown by lentivirus-mediated siRNAs on cell proliferation, cell cycle, apoptosis, and migration were investigated in vitro and in vivo.
Results: CircRNA_100876 expression was upregulated (P<0.05) and was negatively correlated with survival outcome (P<0.05) in ESCC. Inhibition of proliferation, migration, invasion, and epithelial–mesenchymal transition progression was confirmed after circRNA_100876 depletion.
Conclusion: Dysregulation of circRNA_100876 expression leads to poor prognosis in ESCC by accelerating cell proliferation and metastasis.
Keywords: circRNA_100876, esophageal squamous cell carcinoma, metastasis, epithelial–mesenchymal transition
Introduction
Esophageal cancer is one of the most common malignancies worldwide.1,2 Although a combination of radical esophageal cancer resection and chemotherapy has been widely used, the 5-year overall survival rate is still relatively low owing to the difficulty in early diagnosis of esophageal cancer.3,4 Consequently, the search for effective molecular markers for screening, early diagnosis, and prognosis for the high-risk groups of esophageal cancer is a clinical hotspot. In recent years, molecular studies have helped elucidate many abnormal biological events that occur during the development of esophageal cancer and thus aided in screening a series of specific therapeutic targets and biological markers, which could contribute to better understanding of the underlying biology.5–7
CircRNA is a novel endogenous RNA, which is highly expressed in the eukaryotic transcriptome. In contrast to lncRNAs and miRNAs, circRNAs have a covalently closed continuous loop, instead of 5′–3′ polarity and polyadenylated tails.8,9 Owing to this special stable structure, circRNAs can participate in various biological functions with increased biological stability. CircRNAs have a high degree of tissue-specific expression, indicating that their roles might differ for various sites and species. The multiple functions of circRNAs have gradually been elucidated, for example, circRNAs can act as miRNA sponges, regulate splicing and transcription, bind to proteins, and transport specific RNAs.8,10 In recent years, emerging evidence has shown that circRNAs may also play a key role in tumor development.11,12
CircRNA_100876 is a circRNA oriented from chr11:71668272–71671937.13 This circRNA (encoded by RNF121) can be regulated as a ceRNA. CircRNA_100876 regulates the expression of MMP13, which is usually correlated with cancer occurrence and development.13–15 To date, circRNA_100876 has been found to be highly expressed in lung cancer, with its high expression being negatively correlated with the prognosis of non-small-cell lung cancer,16 suggesting that circRNA_100876 promotes metastatic potential.
The expression pattern and biological function of circRNA_100876 in esophageal squamous cell carcinoma (ESCC) remain unknown. Therefore, we analyzed circRNA_100876 expression in ESCC and tried to clarify the relationship between clinicopathologic parameters and circRNA_100876 expression in ESCC, including the prognostic value of circRNA_100876. More importantly, we knocked down circRNA_100876 expression in esophageal cancer cells, observed its effects on proliferation and metastasis, and analyzed the possible mechanisms underlying these effects.
Materials and methods
Tissue samples
Human ESCC and paired adjacent noncancerous tissues were obtained from 74 patients who had undergone radical excision without receiving any other treatment before surgery and been pathologically diagnosed with ESCC between 2011 and 2014. All specimens were immediately frozen in liquid nitrogen after excision and stored at −80°C. The Ethics Committee of Nanfang Hospital, Southern Medical University approved this research. The informed consent form regarding specimen use for scientific research was signed by each patient.
Cell culture
The ESCC cell lines, Eca-109 and TE-1, were obtained from the National Infrastructure of Cell Line Resource (Shanghai, China), the KYSE-410 and KYSE-150 cell lines were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany), and the immortalized esophageal epithelial cell line, HEEC, was obtained from ScienCell Research Laboratory (Carlsbad, CA, USA). KYSE-410, KYSE-150, and HEEC cells were cultured in Roswell Park Memorial Institute 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific); ECA-109 and TE-1 cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS. The cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
RNA interference
To establish cell lines with stable knockdown, lentiviruses expressing siRNAs against circRNA_100876 (siRNA-1 and siRNA-2 cells) and the negative control (NC cells) sequence (Genechem, Shanghai, China) were constructed. TE-1 and KYSE-150 cells were transfected with a mixture of lentiviruses (multiplicity of infection, 100) and 5 μg/mL polybrene. After 72 hours, the transfected cells were used for further analysis. The siRNAs targeting circRNA_100876 and NC sequences are presented in Table 1.
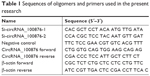  | Table 1 Sequences of oligomers and primers used in the present research |
Total RNA extraction and real-time quantitative PCR (RT-qPCR) assays
Total RNA was extracted from the frozen tissues and cell lines with TRIzol reagent (Thermo Fisher Scientific) and 500 ng total RNA was reverse transcribed to cDNA with the PrimeScript RT Master Mix (TaKaRa, Dalian, China) as per the respective manufacturer’s instruction. Then, the relative circRNA_100876 expression, normalized to expression of the endogenous control β-actin, was determined by RT-qPCR assays using the SYBR Premix Ex Taq II Kit (TaKaRa) on the StepOnePlus system (Thermo Fisher Scientific). Fold changes were calculated using the 2−ΔΔCt method.17 The primers used for RT-qPCR assay are presented in Table 1.
Cell viability and proliferation assays
Cell viability and proliferation were analyzed using Cell Counting Kit-8 (CCK-8), colony-formation, and 5-ethynyl-2´-deoxyuridine (EdU)-incorporation assays.
For the CCK-8 assay, 3×103 cells per well were seeded into 96-well plates. Their viability was measured at 0, 24, 48, 72, and 96 hours after adherence by using CCK-8 (Solarbio, Beijing, China) as per the manufacturer’s protocol; absorbance was recorded at 450 nm using a microplate reader.
For the colony-formation assay, cells (200/well) were seeded into six-well plates. After incubation for another 2 weeks, colonies with diameter greater than 1 mm were counted after staining with 0.5% crystal violet.
For the EdU-incorporation assay, 3×103 cells for each well were seeded into 96-well plates. The Cell-Light EdU imaging kit (RiboBio, Guangzhou, China) was used in accordance with the manufacturer’s instructions. Images were captured under an inverted fluorescence microscope (Olympus, Tokyo, Japan).
Animal study
The anti-growth effect of circRNA_100876 silencing on ESCC cell line was determined in vivo after receiving approval from the Ethics Committee of Nanfang Hospital, Southern Medical University. Specific-pathogen-free–grade male nude mice (age 3–4 weeks) were bought from the Experimental Animal Center of Guangdong Province, Guangzhou, China. The transfected TE-1 cells were cultured, harvested, and washed with PBS buffer, and then subcutaneously injected into the flanks of the mice (1×107/200 μL for each nude mouse). All mice were raised and maintained in specific-pathogen-free–grade environment in accordance with the National Institutes of Health USA (2011) Guide for the Care and Use of Laboratory Animals.18 The tumor volumes were measured and calculated every 4 days using the following formula: volume = (length × width × width)/2.
Flow cytometric analyses
Cell cycle distribution and apoptotic cell proportion were analyzed on a FACSCanto II flow cytometer (BD Biosciences) and detected using a cell cycle detection kit (Keygen, Nanjing, China) and the Annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (Keygen), respectively, as per the manufacturers’ instructions.
Transwell migration and invasion assays
The cell migration and invasion abilities were evaluated by Transwell assays performed using Transwell chambers (0.8 μm; Corning Incorporated, Corning, NY, USA) with or without Matrigel coating (Corning Incorporated). The complete medium was added to the lower chambers, and 5×104 cells suspended in serum-free medium were then added into the upper compartments. After 24-hour incubation, nonmigrating and noninvading cells were removed from the upper surface of the membranes, while the cells remaining on the other side were fixed and stained with 0.5% crystal violet and digitally imaged.
Western blotting assay
Total proteins were extracted from cell lines by using RIPA (Beyotime, Shanghai, China) buffer supplemented with the protease inhibitor phenylmethanesulfonyl fluoride (1:100; Beyotime). After detecting the protein concentrations by using the bicinchoninic acid (Beyotime) method, equivalent total protein was separated by 10% SDS-PAGE (Fdbio Science, Hangzhou, China) and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were then blocked with TBST solution containing 5% skimmed milk (Solarbio) for 1 hour at room temperature and incubated with primary antibodies at 4°C overnight. The primary antibodies used (all from Proteintech, Rosemont, IL, USA) were as follows: anti-cyclin D1 rabbit polyclonal antibody (1:500), anti-cyclin B1 rabbit polyclonal antibody (1:1,000), anti-E-cadherin rabbit polyclonal antibody (1:1,000), anti-N-cadherin rabbit polyclonal antibody (1:1,000), anti-vimentin rabbit polyclonal antibody (1:2,000), anti-snail rabbit polyclonal antibody (1:500), and anti-β-actin rabbit polyclonal antibody (1:5,000). Subsequently, the membranes were incubated with HRP-labeled goat anti-rabbit secondary antibody (1:10,000; Proteintech) for 1 hour at room temperature; the protein bands were visualized with an enhanced chemiluminescence substrate kit (Fdbio Science) on FluorChem E system (ProteinSimple, San Jose CA, USA). The densitometric analysis of the protein bands was conducted using AlphaView SA version 3.4.0.0 (ProteinSimple).
Statistical analyses
Results have been presented as mean ± standard error of the mean. All statistical analyses were performed using the Pearson chi-squared test, two-tailed Student’s t-test, or ANOVA, as appropriate, with either GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) or IBM SPSS Statistics 20.0 (IBM Corporation, Armonk, NY, USA). Survival data were evaluated using the Kaplan–Meier method and the log-rank test. P<0.05 was considered statistically significant.
Results
CircRNA_100876 was upregulated in ESCC and predicted poor prognosis
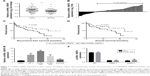
To determine the potential role of circRNA_100876 in ESCC, the relative circRNA_100876 expression in ESCC tissues, the correlation between circRNA_100876 expression and clinicopathologic characteristics, and postoperative survival times were analyzed. RT-qPCR showed that circRNA_100876 expression was significantly higher in ESCC tissues than in adjacent normal tissues (P<0.001; Figure 1A); in ESCC tissues, it was upregulated in 71.6% of the samples (53/74; Figure 1B). Using the medium expression value of circRNA_100876 as the cut-off point for the Pearson chi-squared test and Kaplan–Meier’s plot, it was found that circRNA_100876 expression was strongly correlated with tumor invasion depth (P=0.017), lymph node metastasis (P=0.027), and vascular invasion (P=0.036; Table 2). Patients with higher circRNA_100876 expression had shorter recurrence-free survival (median, 41 vs 23 months; P=0.029; Figure 1C) and overall survival time (median, 46 vs 30 months; P=0.021; Figure 1D) than those with lower circRNA_100876 expression, indicating that circRNA_100876 expression was linked to the clinical progression of ESCC and might represent a promising prognostic biomarker.
CircRNA_100876 was knocked down in ESCC cells
To gain insight into the biological significance of circRNA_100876 in ESCC development and progression, loss-of-function experiments were performed. Endogenous circRNA_100876 expression levels were studied in a panel of ESCC cell lines. CircRNA_100876 expression was upregulated in the ESCC cell lines, especially in TE-1 and KYSE-150 cells, compared with the human immortal esophageal epithelial cell line HEEC (Figure 1E). Subsequently, TE-1 and KYSE-150 cells with stably silenced circRNA_100876 expression were established, and validated with RT-qPCR (Figure 1F).
CircRNA_100876 knockdown suppressed cell proliferation in vitro and in vivo
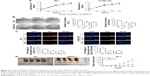
In vitro analysis with CCK-8 and colony-formation assays showed that circRNA_100876 knockdown markedly reduced the proliferative capacity of ESCC cells (Figure 2A and B). A similar tendency was noted in the EdU-incorporation assay: the proportion of EdU-positive cells decreased in circRNA_100876-silenced ESCC cells (Figure 2C). Besides, the anti-growth effect of TE-1 cells transfected with siRNA-1 or NC sequence was compared using in vivo tumorigenicity assay. The average volume of xenograft tumors grown from circRNA_100876-knockdown TE-1 cells was 75.27±7.51 mm3 at day 16, while that of the mice inoculated with NC sequence-infected TE-1 cells was 472.20±142.71 mm3, which was significantly larger than that of circRNA_100876 silencing group (P<0.05; Figure 2D). What is more, the difference in tumor size between the two groups rose over time and peaked at day 28, the endpoint (444.88±99.68 vs 1,561.32±349.22 mm3, P<0.05; Figure 2D). These results showed that circRNA_100876 silencing can restrict ESCC cell growth both in vitro and in vivo.
CircRNA_100876 knockdown led to G2/M phase arrest and apoptosis in vitro
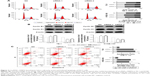
To determine the initial mechanism by which circRNA_100876 modulated ESCC cell proliferation, the alterations in the cell cycle and apoptosis were also analyzed after circRNA_100876 knockdown. The cell populations with circRNA_100876 knockdown showed increased percentages of G2/M phase cells and decreased percentages of G1 phase cells; there was no obvious change in S phase cells (Figure 3A). Consistent with the aforementioned data, the expression of the G2/M phase checkpoint protein cyclin B1 was upregulated, while the G1 phase checkpoint protein cyclin D1 decreased when circRNA_100876 was depleted (Figure 3B). Flow cytometric analysis based on the Annexin V/propidium iodide method showed that the apoptotic cell count (Annexin V positive) was significantly higher in circRNA_100876 knockdown groups than in the control group (Figure 3C). Hence, circRNA_100876 depletion restricted ESCC progression, partially by eliminating cell cycle arrest and inhibiting apoptosis.
CircRNA_100876 knockdown suppressed migration, invasion, and epithelial–mesenchymal transition (EMT) in vitro
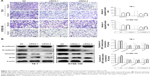
CircRNA_100876 promotes extracellular matrix degradation by competing with MMP13 for miR-136.13 Given that relative circRNA_100876 expression was significantly correlated with invasion depth, lymph node metastasis, and vascular invasion, we hypothesized that circRNA_100876 might participate in regulating ESCC metastasis. The metastatic potential of ESCC cells with circRNA_100876 knockdown was markedly inhibited, in accordance with the decrease in migrating and invading cells shown by Transwell assays (Figure 4A). The EMT process was also restrained after circRNA_100876 knockdown, which was characterized by upregulation of epithelial-phenotype E-cadherin and downregulation of mesenchymal-phenotype N-cadherin, vimentin, and the EMT-associated transcription factor Snail (Figure 4B). Therefore, we concluded that circRNA_100876 promoted cell migration, invasion, and EMT progression in ESCC, thus facilitating cancer metastasis.
Discussion
CircRNAs have been implicated in cancer biology.11,19 They are associated with the occurrence and development of several human cancer types, wherein they may function as cancer promoters or suppressors.20–26 Altered circ_100876 expression has been identified in lung squamous cell carcinoma.16 However, little is known about its role in ESCC. To our knowledge, the current study is the first to analyze the expression pattern and biological value of circ_100876 in ESCC. Circ_100876 expression levels in ESCC tissues were markedly higher than those in the corresponding noncancerous tissues. Moreover, there was a significant correlation between elevated circ_100876 expression and the presence of lymph node metastasis, advanced tumor staging, and poor prognosis of ESCC patients. Our findings indicate that circRNA_100876 should be considered as a crucial molecular marker for ESCC patients.
To elucidate the function of circRNA_100876 in ESCC occurrence and development, in vitro and in vivo assays were performed using circRNA_100876-targeting siRNAs to knock down circRNA_100876 in the ESCC cell lines. CircRNA_100876 knockdown could suppress proliferation, induce apoptosis of ESCC cells, and arrest cell cycle in the G2/M phase. CircRNA_100876 depletion reduced tumor growth in nude mice. These results validated the hypothesis that circRNA_100876 may function as an oncogene and serves an important function in ESCC cell proliferation. In addition, circRNA_100876 knockdown remarkably weakened the migration and invasion capabilities of ESCC cells, indicating that circRNA_100876 might enhance metastasis in ESCC. These findings coupled with our discovery of the positive relationship between tissue circRNA_100876 expression and lymph node metastasis suggested that elevated circRNA_100876 expression in ESCC tissues might enhance distant metastasis and thus result in unfavorable outcomes in ESCC patients.
EMT, an important hallmark of cancer progression, involves transformation of epithelial cells to a mesenchymal cell phenotype via a pathophysiological mechanism.27–29 The relationship between EMT and circRNAs in cancer metastasis has attracted considerable attention recently; most research has focused on the expression of the EMT-associated regulatory factor, including the epithelial marker E-cadherin and the mesenchymal markers N-cadherin and vimentin.30,31 In the current study, we found that circRNA_100876 depletion led to upregulation of the protein levels of E-cadherin and downregulation of N-cadherin and vimentin in ESCC cells, suggesting that absence of circRNA_100876 might reverse EMT process. Therefore, we believe that circRNA_100876 might promote metastasis by activating EMT in ESCC.
Conclusion
Taken together, circRNA_100876 is found to be upregulated in ESCC tissues and may contribute to ESCC initiation and progression. Knockdown of circRNA_100876 suppressed proliferation, migration, and invasion of ESCC cells through modulating EMT process. Our analysis of circRNA_100876 provides new insights into disease mechanisms; circRNA_100876 should be considered as a potential biomarker and therapeutic target for ESCC. Our study revealed important findings, but is still limited by lack of comprehensive analysis of the functional role of circRNA_100876 as an endogenous sponge. The clinical significance of our data is limited because only tissue samples were used from humans; it would increase greatly if circRNA_100876 could be successively detected in the circulatory system. CircRNAs exhibit strong regulatory effects in cancer; they combine with miRNAs to regulate their function.32 Additional studies to elucidate the functional mechanisms of circRNA_100876 in ESCC and other cancers are essential.
Acknowledgment
We thank the Editage Company for the language revision service.
Disclosure
The authors report no conflicts of interest in this work.
References
Zhang N, Wen D, Shan B, et al. Clustering and geographic variation of upper gastrointestinal cancers in a high-risk region of esophageal cancer in northern China. Asian Pac J Cancer Prev. 2011;12(1):193–198. | ||
Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol. 2017;27(3):215–221. | ||
Yamanouchi S, Sako Y, Suemitsu S, et al. Early esophageal cancer with epidermization diagnosed and treated with endoscopic resection. Clin J Gastroenterol. 2018;11(1):29–33. | ||
Guo LW, Huang HY, Shi JF, et al; Health Economic Evaluation Working Group, Cancer Screening Program in Urban China (CanSPUC). Medical expenditure for esophageal cancer in China: a 10-year multicenter retrospective survey (2002–2011). Chin J Cancer. 2017;36(1):73. | ||
Uemura N, Kondo T. Current status of proteomics of esophageal carcinoma. Expert Rev Proteomics. 2016:1029–1040. | ||
Wen Y, Hu X. Expression of esophageal carcinoma related gene 4 (ECRG4) and its clinical significance in prognosis of esophageal carcinoma. Int J Clin Exp Pathol. 2015;8(11):14772–14778. | ||
Liu X, Wang Z, Zhang G, et al. High TRAF6 expression is associated with esophageal carcinoma recurrence and prompts cancer cell invasion. Oncol Res. 2017;25(4):485–493. | ||
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. | ||
Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. | ||
Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. | ||
Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL, Zl J. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. | ||
Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. | ||
Liu Q, Zhang X, Hu X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572. | ||
Hsu CP, Shen GH, Ko JL. Matrix metalloproteinase-13 expression is associated with bone marrow microinvolvement and prognosis in non-small cell lung cancer. Lung Cancer. 2006;52(3):349–357. | ||
Yu X, Wei F, Yu J, et al. Matrix metalloproteinase 13: a potential intermediate between low expression of microRNA-125b and increasing metastatic potential of non-small cell lung cancer. Cancer Genet. 2015;208(3):76–84. | ||
Yao JT, Zhao SH, Liu QP, et al. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. | ||
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. | ||
Nrccftuotgft C, Uol A. Guide for the Care and Use of Laboratory Animals. Publication No. 85-23(rev). 2011;327(3):963–965. | ||
Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. | ||
Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging. 2017;9(6):1585–1594. | ||
Chen L, Zhang S, Wu J, et al. CircRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. | ||
Nair AA, Niu N, Tang X, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7(49):80967–80979. | ||
Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. | ||
Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. | ||
Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. | ||
Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. | ||
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. | ||
Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial–mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172(7):973–981. | ||
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. | ||
Zhang X, Zhou H, Jing W, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers. 2018;2018:3073467–3073469. | ||
Yang L, Han B, Zhang Y, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial–mesenchymal transition. Autophagy. 2018;14(3):404–418. | ||
Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.