Back to Journals » Infection and Drug Resistance » Volume 14
Construction of a Risk Prediction Model for Subsequent Bloodstream Infection in Intestinal Carriers of Carbapenem-Resistant Enterobacteriaceae: A Retrospective Study in Hematology Department and Intensive Care Unit
Authors Wang Y, Lin Q, Chen Z, Hou H, Shen N, Wang Z , Wang F, Sun Z
Received 23 October 2020
Accepted for publication 20 January 2021
Published 2 March 2021 Volume 2021:14 Pages 815—824
DOI https://doi.org/10.2147/IDR.S286401
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yue Wang,1 Qun Lin,1 Zhongju Chen,1 Hongyan Hou,1 Na Shen,1 Zhen Wang,2 Feng Wang,1 Ziyong Sun1
1Department of Laboratory Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, People’s Republic of China; 2Department of Pharmacy, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei Province, People’s Republic of China
Correspondence: Ziyong Sun
Department of Laboratory Medicine, Tongji Hospital, No. 1095 Jiefang Avenue, Wuhan, Hubei Province, 430030, People’s Republic of China
Email [email protected]
Background: To establish a risk prediction model for carbapenem-resistant Enterobacteriaceae (CRE) bloodstream infection (BSI) in intestinal carriers.
Methods: CRE screenings were performed every two weeks in hematology department and intensive care unit (ICU). Patients with positive CRE rectal swab screening were identified using electronic medical records from 15 May 2018 to 31 December 2019. Intestinal carriers who developed CRE BSI were compared with those who did not develop CRE infection. A 1:1 matched case-control study was conducted. The control group was selected by stratified random sampling based on the department to ensure that all the departments were represented. Univariate logistic analysis, multivariate logistic analysis and stepwise regression analysis were carried on a variety of patient factors and microbial factors.
Results: A total of 42 cases were included. Multivariate analysis showed that gastrointestinal injury (OR 86.819, 95% CI 2.584– 2916.592, P=0.013), tigecycline exposure (OR 14.991, 95% CI 1.816– 123.737, P=0.012) and carbapenem resistance score (OR 11.236, 95% CI 1.811– 69.700, P=0.009) were independent risk factors for CRE BSI in intestinal carriers (P< 0.050). They were included in the Logistic regression model to predict BSI. According to receiver operating characteristic (ROC) curve analysis, the cut-off value of the model was 0.722, and the sensitivity, specificity and area under the curve (AUC) were 90.5%, 85.7% and 0.921, respectively.
Conclusion: The risk prediction model based on gastrointestinal injury, tigecycline exposure and carbapenem resistance score of colonizing strain can effectively predict CRE BSI in patients with CRE colonization. Early CRE screening and detection for inpatients in key departments may promote early warning and reduce the risk of nosocomial infection of CRE.
Keywords: carbapenem-resistant Enterobacteriaceae, colonization, bloodstream infection, risk factor, risk prediction model
Background
Carbapenem-resistant Enterobacteriaceae (CRE) has attracted widespread attention due to its rapid growth, treatment difficulty, high mortality and high economic burden.1–3 The course of CRE bloodstream infection (BSI) in immunocompromised patients is usually abrupt and fatal.4 A cohort study of the impact of CRE infections on mortality of patients presenting with sepsis showed that patients with CRE infections had significantly higher 30-day mortality: 63.8% versus 33.4% (P<0.010).5 Therefore, the study of CRE BSI has important clinical significance.
It is known that CRE colonization was an independent risk factor for CRE infection.6 Could we effectively prevent CRE infection by CRE de-colonization? It might take a lot of effort for little return. In fact, the majority of CRE carriers would not suffer from CRE infection. A retrospective study showed that only 16.5% (299/1806) of 1806 patients with CRE colonization subsequently developed CRE infection.7 Excessive de-colonization might lead to the waste of medical resources and the abuse of antibiotics.
The World Health Organization (WHO) recommended that surveillance cultures for asymptomatic CRE colonization should be performed with the guidance of local epidemiology and risk assessment.8 Populations that should be considered for such surveillance include patients with previous CRE colonization, patient contacts of CRE colonized or infected patients and patients with a history of recent hospitalization in endemic CRE settings.8 Since 15th May 2018, our institute had carried out regular CRE screening for all inpatients in intensive care unit (ICU) and hematology department, two high-risk departments of CRE infection. It provided a very suitable observation population for this study. Standardized screening and isolation made the research results more valuable for clinical reference.
It is well known that the occurrence of infection depends on the interaction between pathogen and host. We aimed to establish a risk prediction model for CRE BSI in CRE carriers simultaneously based on the pathogenic features of colonizing CRE strains and host risk factors, in order to early identify high-risk inpatients and to prevent CRE BSI.
Methods
Study Design and Setting
CRE infection control project was carried out in hematology department (215 beds) and ICU (52 beds) since 15 May 2018 in a general teaching hospital in Wuhan, China. At the beginning of the project, all inpatients were screened and grouped. All the newly admitted patients were assessed according to the guidance from the European Centre for Disease Prevention and Control.9 The patients who met the requirements were screened for CRE and reexamined regularly (Figure 1). Data from 15 May 2018 to 31 December 2019 were collected. The cases with positive rectal swab CRE screening results in hematology department and ICU were enrolled.
 |
Figure 1 CRE screening, enrollment and follow-up. *The control group were selected by stratified random sampling based on the department to ensure that all the departments were represented. |
CRE intestinal colonization was defined as a positive result of CRE rectal swab screening without invasive infection. CRE BSI was defined as an infection in which CRE strains were isolated from one or more blood cultures and had clinical infection symptoms. These symptoms included fever (≥38°C) or low temperature (<36°C), shivering, increased (count >10.0×109/L, especially when “nuclear shift to the left”) or decreased (count <3.0×109/L) leukocyte count, skin and mucosa hemorrhage, coma, multiple organ failure, decreased blood pressure, increased C-reactive protein. The length of admission in the case group and the control group was more than 48 hours. Cases of CRE BSI occurred before admission or within 48 hours after admission were excluded to ensure that the cases are nosocomial infection cases, since the study focused on risk factors of nosocomial infection. Patients with CRE already, patients with spontaneous or drug-induced de-implantation, and patients with inconsistent positive results were excluded. The uninfected patients were reexamined for CRE screening at least once before discharge. Cases without reexamination were also excluded. Patients with CRE BSI subsequent to CRE intestinal colonization were included in the case group. Patients without secondary CRE infection were included in the control group. Patients in the control group were finally 1:1 matched with case group using stratified random sampling based on the department. Random sampling was carried out by using a random number generator program.
Microbiology
The rectal swabs were inoculated directly to a chromogenic agar plate containing carbapenem as selective agent (CHROMagar, France) for CRE screening. All the isolated bacteria were identified by MALDI-TOF mass spectrometer (Bruker Daltonics, USA). The sensitivity of meropenem and imipenem was detected by Kirby-Bauer method. The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) M100-ED30 breakpoints.10 NG-Test CARBA 5 (NG Biotech, France) was used to detect five common carbapenemase types: KPC, NDM, IMP, VIM and OXA.
Variables and Definitions
The data were collected retrospectively, through electronic medical record. All variables potentially related to BSI were collected: general information (gender, age, department), underlying conditions (hypertension, diabetes, solid organ tumor, hematological malignancy, impaired immune function, gastrointestinal injury), invasive procedures and devices (solid organ transplantation, hematopoietic stem cell transplantation, surgery, mechanical ventilation, central venous catheter, urinary catheter, gastric tube, drainage tube), antibiotic exposure and duration, length of stay from CRE screening to outcome (occurrence of CRE BSI or discharge), strain factors (colonizing bacteria, carbapenem resistance score, and carbapenemase producing).
Impaired immune function included receiving radiotherapy and chemotherapy, agranulocytosis, long-term or massive hormone therapy, and HIV infection. Gastrointestinal injury included gastrointestinal bleeding or perforation, ostomy or excision of stomach and intestine, gastroenteritis, cholecystitis and pancreatitis. Antibiotic exposure and duration were considered from CRE screening to CRE BSI onset for the case group, or to hospital discharge for the control group.
The inhibition zone diameters of meropenem and imipenem were discontinuous numerical variables, so we used carbapenem resistance score as a categorical variable to represent the resistance of carbapenems. When the inhibition zone diameters of meropenem and imipenem were both >6mm, it was recorded as “1”; when either the inhibition zone diameter of meropenem or imipenem was equal to 6mm, it was recorded as “2”; when the diameter of meropenem and imipenem were both equal to 6mm, it was recorded as “3”.
Statistical Analysis
Carriers who developed CRE BSI were compared with those who did not develop CRE infection. Pearson chi-square test was used for binary data. Nonparametric Mann–Whitney rank sum test was used for ordered categorical data. T-test was used for measurement data. The variables with P<0.200 in univariate analysis were included in multivariate analysis. Then, stepwise regression analysis was carried out to determine the parameters in the risk prediction model and establish the logistic regression model. The P value, OR value and 95% confidence interval of each factor were calculated. P < 0.050 was considered statistically significant. The receiver operating characteristic (ROC) curve was used to evaluate the predictive ability of the model.
Results
Patient Cohort
During the study period, 12,754 rectal swabs (7456 inpatients) were screened. Eight hundred and sixty patients being admitted to hematology department and ICU with positive CRE rectal swab screening were identified. After exclusion criteria were applied, a total of 73 patients met the standard. Twenty-one patients developed CRE BSI were included in the case group, matched with 21 patients who did not have CRE infection at a ratio of 1:1 (Figure 1).
Univariate Analysis for Variables
Patient Factors
Demographic and clinical factors of case and control groups are summarized in Table 1. There was no significant difference in gender, age, invasive procedures and devices, or length of stay from CRE screening to outcome (occurrence of CRE BSI or discharge). The median time from positive screening to CRE BSI was 13 days (range from 1 to 202).
 |
Table 1 Univariate Analysis of Risk Factors for Subsequent BSI in CRE Intestinal Carriers |
In addition, there was no significant difference in most underlying conditions such as hypertension, diabetes, solid organ tumor, hematological malignancy and impaired immune function (P>0.050), except gastrointestinal injury. Gastrointestinal injury was more frequently identified in case group than in control group (P=0.014). Tigecycline exposure was more prevalent in case group than in control group (P=0.011), whereas the other antibiotic exposure and duration had no significant difference.
Microbial Factors
Microbial factors are shown in Table 1. Among 42 colonized bacteria, 31 (73.8%) were K. pneumoniae, 9 (21.4%) were E. coli, 2 (4.8%) were Citrobacter spp. K. pneumoniae were more common isolates in the case group than in the control group, 85.7% versus 61.9% (P=0.079), while E. coli were more detected in the control group than in the case group, 33.3% versus 9.5% (P=0.060). But the difference was not statistically significant (P>0.050).
Carbapenem resistance score was “3” in 23 cases (54.8%), “2” in 5 cases (11.9%), “1” in 14 cases (33.3%). A score of “3” was significantly more common in the case group than in the control group (P=0.001), whereas a score of “1” was more prevalent in the control group than in the case group (P=0.009). All 23 isolates with a score of “3” were K. pneumoniae. K. pneumoniae with a carbapenem resistance score of “3” accounted for 94.4% (17/18) of all K. pneumoniae in the case group and 46.2% (6/13) in the control group, meanwhile K. pneumoniae with a carbapenem resistance score of “1” accounted for 5.6% (1/18) and 53.8% (7/13) in the control group. There were significant differences (P<0.050).
KPC-type carbapenemase was detected in 54.8% (23/42) isolates, which were all K. pneumoniae. NDM-type carbapenemase was detected in 23.8% (10/42) isolates, which E. coli accounted for 70.0% (7/10). Besides, KPC-, NDM-, IMP-, VIM- or OXA-type carbapenemase was not detected in the other 9 colonized bacteria. Carbapenemase-producing was more commonly detected in the case group than in the control group (P=0.008), especially KPC (P=0.001). All 23 K. pneumoniae isolates with a score of “3” were identified producing KPC.
Development and Validation of CRE BSI Risk Prediction Model
Variables with P<0.200 in univariate analysis were then included in multivariate binary logistic regression analysis. Gastrointestinal injury (OR 86.819, 95% CI 2.584–2916.592, P=0.013), tigecycline exposure (OR 14.991, 95% CI 1.816–123.737, P=0.012) and carbapenem resistance score (OR 11.236, 95% CI 1.811–69.700, P=0.009) were independent risk factors for CRE BSI in intestinal carriers (P<0.05). They were chosen as valuable markers for risk prediction model of CRE BSI (Table 2). The logistic regression model was established as follows:
 |
Table 2 Multivariate Binary Logistic Regression Analysis of Risk Factors for Subsequent BSI in CRE Intestinal Carriers |
PV = 1/(1+e^-(−8.488+4.464×gastrointestinal injury+2.707×tigecycline exposure+2.419×carbapenem resistance score))
Abbreviation: PV, predictive value; e, natural logarithm.
Variable: carbapenem resistance score, it was given 1, 2 or 3 according to the inhibition zone diameter; gastrointestinal jury and tigecycline exposure, “Yes” and “No” were given 1 and 0, respectively.
By ROC curve analysis (Figure 2), area under the curve (AUC) was 0.921. The cut-off value of the model was 0.722. The sensitivity and specificity of the model were 90.5% and 85.7%, respectively.
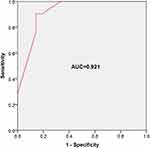 |
Figure 2 ROC curve analysis was performed to evaluate the predictive ability of risk prediction model. AUC=0.921, cut-off value = 0.722, sensitivity = 90.5%, specificity = 85.7%. |
Discussion
Of the 7456 patients screened for CRE, 860 were founded CRE colonization, with the colonization rate of 11.5%. Among all CRE colonizers, only 2.4% (21/860) had subsequent CRE BSI, of which 57.1% (12/21) had adverse outcomes. Actually, CRE screening by rectal swabs could not reflect the CRE colonization of the intestinal flora, because the selected positions were not the whole intestinal flora. Whether and when CRE colonizers need to be de-colonization is a clinical unsolved puzzle. The potential risk factors of infection in different sites are different. Due to the critical consequences of CRE BSI, our study focused on the identification of high-risk patients with CRE bacteremia to provide a theoretical basis for de-colonization.
Establishing a risk prediction model of CRE BSI in ICU and hematology department is considered to be difficult, since the underlying conditions and treatment methods of patients in these two departments are obviously different. To eliminate this interference factor, control group were selected by stratified random sampling based on the department, and matched with the case group. Finally, a total of 42 cases (24 from ICU and 18 from hematology department, respectively) were successfully enrolled. Amazingly, univariate Analysis showed that most demographic and clinical factors of case and control groups such as gender, age, hypertension, diabetes, solid organ tumor, hematological malignancy, impaired immune function, invasive procedures and devices, antibiotic exposure and duration did not have significant difference (P>0.050), except gastrointestinal injury (P=0.014) and tigecycline exposure (P=0.011). After analyzing both patient factors and microbial factors, gastrointestinal injury (OR 86.819, 95% CI 2.584–2916.592, P=0.013), tigecycline exposure (OR 14.991, 95% CI 1.816–123.737, P=0.012) and carbapenem resistance score (OR 11.236, 95% CI 1.811–69.700, P=0.009) were chosen as valuable markers for risk prediction model of CRE BSI.
Patients with hematologic malignancies and hematopoietic stem cell transplant recipients were reported at high risk of developing invasive infections due to enteric bacteria because of chemotherapy-induced neutropenia and gastrointestinal mucositis.11 In fact, gastrointestinal mucositis is a complex inflammatory reaction of the mucous membranes, a side effect of both chemotherapy and radiotherapy.12 Its severity is difficult to assess. Interestingly, gastrointestinal injury with definite diagnosis including gastrointestinal bleeding or perforation, ostomy or excision of stomach and intestine, gastroenteritis, cholecystitis and pancreatitis was found to be an independent risk factor and could be included in the model for risk assessment of CRE BSI in CRE carriers.
Tigecycline has a large volume of distribution and high concentration in gallbladder, colon and pulmonary tissue. In contrast, the serum concentrations of tigecycline are relatively low.13 After a single 100mg dose of tigecycline, serum concentrations of tigecycline rapidly declined from a mean value of 1.94mg/L to 0.31mg/L between 3min and 1h after the end of the infusion. The subsequent concentrations in serum slowly declined to mean values of 0.22ng/mL and 0.07mg/L at 4h and 24h after the start of the tigecycline infusion.14 Tigecycline is not approved for the treatment of BSI because its serum concentrations are generally deemed not adequate. Therefore, conventional dose of tigecycline (100mg initially, followed by 50mg q12h) cannot prevent CRE BSI. In addition, tigecycline has a wide antibacterial spectrum, which is active against a wide range of Gram-positive and -negative aerobic and anaerobic bacteria.13 In most large-scale monitoring studies, the sensitivity of tigecycline in Enterobacteriaceae was kept at a high level of >90.0%.15–17 CRE isolates also showed high susceptibility to tigecycline (89.7%).18 It should be noted that tigecycline resistance rate of K. pneumoniae in hematopoietic stem cell transplant patients could reach 16.0%.19 Taken together, we inferred that tigecycline exposure increased the risk of CRE BSI might be due to intestinal flora disorder. It needs to be verified by intestinal flora analysis rather than CRE screening. Moreover, it suggested us that more attention should be paid to avoid tigecycline exposure in the treatment of CRE carriers.
A longitudinal large-scale CRE study in China showed that the majority of clinical CRE isolates were Klebsiella pneumoniae (1201/1801, 66.7%), followed by Escherichia coli (282/1801, 15.7%).18 From 2015 to 2016, 532 non-repetitive clinical CRE isolates from 14 hospitals in Hubei Province collected by us. K. pneumoniae accounted for 81.6% (434/532), and E. coli accounted for 8.3% (44/532), which showed similar results to previous report. Most isolates showed high-level carbapenems resistance. The minimum inhibitory concentrations (MIC) of meropenem in 92.3% (491/532) CRE strains and MIC of imipenem in 91.4% (486/532) CRE stains were greater than or equal to 32μg/mL. The carbapenem inhibition zone diameters of these strains were usually 6 mm. In contrast, the MIC value distribution of CRE strains isolated from rectal swabs were significantly different. According to the analysis of 192 strains of colonized bacteria, it was found that the MIC value distribution of meropenem was bimodal, with a minor peak at 4μg/mL and a major peak at 64μg/mL. Thus, a hypothesis was proposed that strains isolated from rectal swabs with high resistance to carbapenems might relate with the occurrence of infection. The hypothesis was verified by this study. It was also noticed that all these isolates were KPC-producing K. pneumoniae. In fact, the outbreak of carbapenem-resistant hypervirulent K. pneumoniae strain carrying blaKPC-2 gene has been reported in China.20 This strain posed a substantial threat to human health due to its simultaneously hypervirulent, multidrug-resistant, and highly transmissible. Our results suggested that there might be an epidemic of this strain. Further verification is needed and control measures should be taken.
There were few studies on the correlation between bacterial characteristics of colonized CRE and the subsequent occurrence of infection currently. Giannella et al established a prediction model of carbapenem-resistant K. pneumoniae (CR-KP) BSI following CR-KP colonization, based on whether there were ICU admission, radiotherapy and chemotherapy, abdominal invasive operation and multi-site CR-KP colonization.21 The sensitivity and specificity were 93.0% and 42.0%, respectively. In our model, carbapenem resistance score of colonizing CRE was chosen as a valuable marker along with gastrointestinal injury and tigecycline exposure. The specificity of the model was as high as 85.7%, and the sensitivity was 90.5%. The carbapenem resistance score was calculated according to whether the disk diameters of meropenem and imipenem were 6 mm or not. Comparing with MIC, the advantage was that it could be evaluated at the same time as the CRE screening. It would greatly simplify the detection and evaluation process, and would be helpful for the promotion and use of the model. In fact, the model could be expected to be used in clinic, because the data of three indicators are easy to obtain. Gastrointestinal injury and tigecycline exposure history could be captured through electronic medical record. Early warning of CRE carriers in high-risk of CRE BSI through electronic information technology would help clinicians make a decision of using drugs in time for de-colonization or preemptive treatment. It might reduce the occurrence or death risk of CRE BSI. At the same time, the application of this model might reduce the abuse of antibiotics for low-risk patients, and might reduce the occurrence of drug resistance.
However, some limitations of this study should be noted. The number of cases was small because of the low incidence rate of CRE BSI. There might be a selection bias in sampling. Besides, the model is currently only suitable for hematology department and ICU. It could not be extended to other areas due to different epidemiological distribution of CRE strains. Further studies are still needed.
Conclusions
To summarize, gastrointestinal injury, tigecycline exposure and carbapenem resistance score of colonizing bacteria could effectively predict the risk of subsequent BSI in CRE carriers. Our findings suggested that carbapenem susceptibility test of colonized bacteria in critical patients could identify patients at high risk of CRE infection and prevent CRE BSI as early as possible. The application of this risk prediction model might reduce the incidence and mortality of CRE infection. It might also avoid unnecessary use of antibiotics in low-risk groups to reduce the selective pressure of antibiotics, and further reduce the occurrence of CRE.
Abbreviations
CRE, carbapenem-resistant Enterobacteriaceae; BSI, bloodstream infection; ICU, intensive care unit; ROC, receiver operating characteristic; AUC, area under the curve; MICs, minimum inhibitory concentrations; PV, predictive value; CR-KP, carbapenem-resistant Klebsiella pneumoniae.
Data Sharing Statement
All data generated or analysed during this study are included in this published article.
Ethics Approval and Consent to Participate
The study was approved by the ethical committee of Huazhong University of Science and Technology, Wuhan, China ([2018]107). This study was conducted in accordance with the Declaration of Helsinki. The data are anonymous, and the requirement for informed consent was therefore waived.
Consent for Publication
Written informed consent for publication was obtained from all participants.
Acknowledgments
This study was supported by the National Mega Project on Major Infectious Disease Prevention (2017ZX10103005-007) and Natural Science Foundation of Hubei Province (2019CFB666).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests.
References
1. Centers for Disease Control and Prevention, USA. Antibiotic resistance threats in the United States, 2019; 2019. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html.
2. Peri AM, Potoski BA, Harris PNA, Paterson DL, Righi E. Antimicrobial treatment challenges in the era of carbapenem resistance. Diagn Microbiol Infect Dis. 2019;94:413–425. doi:10.1016/j.diagmicrobio.2019.01.020
3. Bartsch SM, McKinnell JA, Mueller LE, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect. 2017;23:
4. Jaiswal SR, Gupta S, Kumar RS, et al. Gut colonization with carbapenem-resistant Enterobacteriaceae adversely impacts the outcome in patients with hematological malignancies: results of a prospective surveillance study. Mediterr J Hematol Infect Dis. 2018;10:e2018025. doi:10.4084/mjhid.2018.025
5. Sabino S, Soares S, Ramos F, Moretti M, Zavascki AP, Rigatto MH. A cohort study of the impact of carbapenem-resistant Enterobacteriaceae infections on mortality of patients presenting with sepsis. mSphere. 2019;4:e00052–19. doi:10.1128/mSphere.00052-19
6. McConville TH, Sullivan SB, Gomez-Simmonds A, Whittier S, Uhlemann AC. Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One. 2017;12:e0186195. doi:10.1371/journal.pone.0186195
7. Tischendorf J, de Avila RA, Safdar N. Risk of infection following colonization with carbapenem-resistant Enterobactericeae: a systematic review. Am J Infect Control. 2016;44:539–543. doi:10.1016/j.ajic.2015.12.005
8. World Health Organization. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities; 2017. Available from: https://www.who.int/infection-prevention/publications/guidelines-cre/en/.
9. Magiorakos AP, Burns K, Rodríguez Baño J, et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob Resist Infect Control. 2017;6:113. doi:10.1186/s13756-017-0259-z
10. Clinical and Laboratory Standards Institute, USA. Performance standards for antimicrobial susceptibility testing, M100 30th edition; 2020. Available from: https://clsi.org/standards/products/microbiology/documents/m100/.
11. Pouch SM, Satlin MJ. Carbapenem-resistant Enterobacteriaceae in special populations: solid organ transplant recipients, stem cell transplant recipients, and patients with hematologic malignancies. Virulence. 2017;8:391–402. doi:10.1080/21505594.2016.1213472
12. Kuiken NSS, Rings EHHM, Blijlevens NMA, Tissing WJE. Biomarkers and non-invasive tests for gastrointestinal mucositis. Support Care Cancer. 2017;25:2933–2941. doi:10.1007/s00520-017-3752-2
13. Frampton JE, Curran MP. Tigecycline. Drugs. 2005;65:2623–2637. doi:10.2165/00003495-200565180-00008
14. Rodvold KA, Gotfried MH, Cwik M, Korth-Bradley JM, Dukart G, Ellis-Grosse EJ. Serum, tissue and body fluid concentrations of tigecycline after a single 100mg dose. J Antimicrob Chemother. 2006;58:1221–1229. doi:10.1093/jac/dkl403
15. Sader HS, Flamm RK, Jones RN. Tigecycline activity tested against antimicrobial resistant surveillance subsets of clinical bacteria collected worldwide (2011). Diagn Microbiol Infect Dis. 2013;76:217–221. doi:10.1016/j.diagmicrobio.2013.02.009
16. Decousser JW, Woerther PL, Soussy CJ, Fines-Guyon M, Dowzicky MJ. The tigecycline evaluation and surveillance trial; assessment of the activity of tigecycline and other selected antibiotics against gram-positive and gram-negative pathogens from France collected between 2004 and 2016. Antimicrob Resist Infect Control. 2018;7:68. doi:10.1186/s13756-018-0360-y
17. Liu XJ, Lyu Y, Li Y, Xue F, Liu J. Trends in antimicrobial resistance against Enterobacteriaceae strains isolated from blood: a 10-year epidemiological study in mainland China (2004–2014). Chin Med J (Engl). 2017;130:2050–2055. doi:10.4103/0366-6999.213407
18. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
19. Averbuch D, Tridello G, Hoek J, et al. Antimicrobial resistance in gram-negative rods causing bacteremia in hematopoietic stem cell transplant recipients: intercontinental prospective study of the infectious diseases working party of the European bone marrow transplantation group. Clin Infect Dis. 2017;65:1819–1828. doi:10.1093/cid/cix646
20. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
21. Giannella M, Trecarichi EM, De Rosa FG, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infection among rectal carriers: a prospective observational multicentre study. Clin Microbiol Infect. 2014;20:1357–1362. doi:10.1111/1469-0691.12747
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
