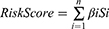Back to Journals » International Journal of General Medicine » Volume 14
Construction of a Novel Lung Adenocarcinoma Immune-Related lncRNA Pair Signature
Authors Qi X , Chen G, Chen Z, Li J, Chen W, Lin J, Lin L
Received 17 June 2021
Accepted for publication 29 July 2021
Published 11 August 2021 Volume 2021:14 Pages 4279—4289
DOI https://doi.org/10.2147/IJGM.S325240
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xiangjun Qi,1 Guoming Chen,1 Zhuangzhong Chen,2 Jing Li,1,3 Wenmin Chen,1 Jietao Lin,2 Lizhu Lin2,4
1The First Clinical Medical College of Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China; 2Department of Oncology, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China; 3Department of Oncology, The First Affiliated Hospital of Hunan University of Chinese Medicine, Changsha, People’s Republic of China; 4Cancer Project Team of China Center for Evidence Based Traditional Chinese Medicine, Guangzhou, People’s Republic of China
Correspondence: Jietao Lin; Lizhu Lin
The First Affiliated Hospital of Guangzhou University of Chinese Medicine, No. 16 Jichang Road, Baiyun District, Guangzhou, 510405, People’s Republic of China
Tel +86 20-36596356
Email [email protected]; [email protected]
Background: A growing number of studies have demonstrated that immune-related long noncoding ribonucleic acids (irlncRNAs) are potential prognostic factors for lung adenocarcinoma. Two-gene combination patterns could improve the sensitivity of prognostic models, providing us a novel signature construction concept that we applied to lung adenocarcinoma.
Methods: Gene expression and clinical data were downloaded from the Lung Adenocarcinoma project of The Cancer Genome Atlas (TCGA) database. We applied a co-expression analysis with immune genes obtained from the ImmPort database to recognize irlncRNA. The matrix of irlncRNA pairs was established by a cyclic comparison of each lncRNA pair expression level. Univariate and multivariate Cox regressions and Lasso penalized regression analysis were applied to construct the risk model. Patients with lung adenocarcinoma were divided into high- and low-risk groups, according to the Akaike Information Criterion (AIC) values of the receiver operating characteristic (ROC) curve. Then, we evaluated our signature under various clinical settings: clinical-pathological characteristics, tumor-infiltrating immune cells, checkpoint-related biomarkers, targeted therapy, and chemotherapy.
Results: Based on the 239 differently expressed irlncRNAs, we constructed an 11-irlncRNA pair signature. The area under the curve (AUC) of the ROC curve for the signature to predict the 4-year survival rate was 0.819, and the cut-off point was recognized as 1.003. Subsequent analysis showed that our signature can effectively distinguish unfavorable survival outcomes, prognostic clinic-pathological characteristics, and specify tumor infiltration status. Highly expressed immune checkpoint-related genes, as well as higher chemosensitivity, were correlated to the low-risk group.
Conclusion: We constructed a novel lung adenocarcinoma irlncRNA signature with promising prognostic value using the TCGA database, based on paired irlncRNAs and not relying on lncRNAs special expression levels.
Keywords: lung adenocarcinoma, immune-related lncRNA, immune infiltrate, survival analysis, prognosis
Introduction
Lung cancer is a major cause of cancer-related deaths worldwide. The 5-year survival rate for lung cancer patients is below 20% in most countries, accounting for approximately 2.2 million new cases and 1.8 million deaths in 2020.1 Lung adenocarcinoma (LUAD) is the most common historical subtype of lung cancer - accounting for more than 40% of the cases.2 Immunotherapy, especially Immune Checkpoint Inhibitors (ICIs), has been regarded as a promising therapy for LUAD patients, who do not harbor a known mutation, such as in the epidermal growth factor receptor gene. For example, Pembrolizumab and Nivolumab - anti-programmed cell death-1 (PD-1) antibodies - are approved for advanced non-small cell lung cancer treatment.3,4 Also, another anti-programmed cell death ligand-1 (PD-L1) antibody, Atezolizumab is approved.5 Additionally, clinical trials such as CHIO3, to evaluate the response of operable stage III A/B non-small cell lung cancer patients to Durvalumab, are in progress.
Long non-coding RNAs (lncRNAs) are a type of RNA with a length greater than 200 nucleotides. Although they are not translated into proteins, lncRNAs play an essential role in gene transcription and expression. Studies have suggested that lncRNAs are associated with many tumor-related biological processes. For example, overexpression of LINC00472 was proved to inhibit Epithelial-Mesenchymal Transition (EMT) processes of A549 and PC-9 cells via YBX1 modulation, a known cancer-promoting gene.6 Additionally, a growing number of studies showed that lncRNAs are related to many aspects of cancer-immune activities including immune infiltrating cells with an identified role in immunotherapy responses.7 For example, in patients with non-small cell lung cancer, a higher expression level of PD-1 may be accompanied by CD8+TILs depletion.8 Therefore, a novel understanding of the relationship between lncRNA and tumor-infiltrating immune cells deserves to be explored.
Several immune-related lncRNAs (irlncRNAs) LUAD signatures have been established and demonstrate potential diagnostic and predictive value. The Areas Under Curve (AUC) values of Receiver Operating Characteristic (ROC) curve calculated for the LUAD dataset of The Cancer Genome Atlas database (TCGA, https://tcga-data.nci.nih.gov/tcga/) ranged from 0.686 to 0.783 in the established signature. All signature were constructed based on lncRNAs’ expression levels,9–14 and two research analyzed the relationship between infiltration immune cell subtypes and risk score.13,15
A two-gene combination strategy to seek cancer-related biomarkers from gene expression big data was identified as more sensitive in multiple cancers.16 Hong et al17 constructed a novel irlncRNA signature for hepatocellular carcinoma with paired irlncRNAs, and the AUC of the 5-year ROC curve in their model was 0.904, which showed the combination strategy potential to construct prognostic signatures. However, few studies have applied this strategy to LUAD.
Materials and Methods
Identification of Immune-Related lncRNA
The transcriptome profiling (RNAseq) fragments per kilobase million (FPKM) data from the LUAD project were downloaded from the TCGA database. The expression profile data of each sample was combined in an RNAseq expression matrix. Gene Transfer Format (GTF) files, downloaded from Ensemble database (http://asia.ensembl.org) for annotation, were used to isolate lncRNAs from the RNAseq expression matrix. The list of immune genes was obtained from the ImmPort database (http://www.immport.org) and an immune genes-related expression matrix was also extracted from the RNAseq expression matrix. Immune-related lncRNAs (irlncRNAs) were screened by a correlation between immune genes and lncRNAs. Those with correlation coefficients > 0.4 and p < 0.001 were recognized as irlncRNAs. We identified differential expression of irlncRNAs (DEirlncRNAs) using the R limma package and set the log Fold Change (FC) > 1.5 along with a False Discovery Rate (FDR) < 0.05 to screen irlncRNAs. A DEirlncRNAs heatmap was drawn with the R pheatmap package.
Construction of the DEirlncRNA Pairs Matrix
The construction of the DEirlncRNA pairs matrix was performed based on each lncRNAs pair expression level. If the expression level of lncRNA A was higher than that of lncRNA B, it was recorded in the matrix as 1, otherwise, as 0. In the 0-or-1 matrix constructed, pairs without a certain rank were not appropriate for predicting patients’ survival outcomes. Finally, when the number of lncRNA pairs with an expression of 0, or 1, accounted for less than 20% of the total pairs, they were considered an invalid match.
Clinical Data Acquisition
Clinical data of LUAD patients were downloaded from the TCGA-LUAD project. Patients without documented survival, or survival status, were excluded.
Establishment of DEirlncRNAs RiskScore Model
First, we conducted a univariate Cox regression analysis with a p-value of 0.05 to screen DEirlncRNA pairs candidates with prognostic value. Then, we performed a Least Absolute Shrinkage and Selection Operator (LASSO) regression with 10-fold cross-validation. To improve model stability, the LASSO procedure was repeated 1000 times with a random stimulation, and pairs with frequency superior to 100 times were reserved for stepwise Cox proportional hazard regression analysis. A multivariate Cox regression analysis was also conducted. The ROC curve analysis for 1-, 2-, 3-, 4- and 5-years was plotted and the AUC values were calculated. RiskScore calculation for each patient was conducted as follows:
The “βi” and “Si” represent each prognostic DEirlncRNA pair coefficient and expression status, respectively. Patients were divided into high- and low-risk groups according to the optimal Akaike Information Criterion (AIC) values, where the maximum inflection point of the 4-year ROC curve was regarded as the risk cut-off point. The R survival, glmnet, survivalROC, and survminer packages were utilized for these operations.
Constructed Risk Model Clinical Prognostic Signatures
To valid the cut-off point and clarify the survival differences between high- and low-risk groups, we carried out a Kaplan–Meier analysis, and the survival curve was plotted. ROC curves for 4-year as well as for clinical characteristics such as the TNM stage, age, and gender were visualized. Then, we evaluated the association between the risk model and clinical characteristics with a χ2 or a Wilcoxon signed-rank test. To better explore if the risk model can be regarded as an independent prognostic predictor, we used univariate and multivariate Cox regression analyses. The following R packages were applied in these operations: survival, glmnet, pbapply, survivalROC, survminer, pHeatmap, and ggupbr.
Tumor-Infiltrating Immune Cells Estimation
We calculated infiltration values of immune cell subtypes for TCGA-LUAD dataset samples based on 7 algorithms: TIMER,18,19 QUANTISEQ,20,21 EPIC,22 CIBERSORT,23 XCELL,24,25 MCPcounter,26 and CIBERSORT-ABS.27 A Wilcoxon signed-rank test was used to explore infiltration value differences between high- and low-risk groups, and a p < 0.05 was considered to be significant. The relationship of infiltration immune cell subtypes and RiskScore value was assessed by a Spearman correlation analysis. The following R packages were applied in these operations: limma, ggplot2, ggtext, scales, and ggpubr.
Immunosuppressive Molecules Expression
We compared the gene expression levels of CTLA4, PDCD1 that were related to ICIs between high- and low-risk groups in our risk model. We used limma and ggpubr R packages in that case.
Chemotherapeutic and Targeted Therapy Response Prediction
Chemotherapeutic and targeted therapy is commonly used in LUAD, so we decided to use the pRRophetic R package to calculate the half-maximal Inhibitory Concentration (IC50) of anticancer drugs (gefitinib, erlotinib docetaxel, cisplatin, vinorelbine, and paclitaxel) for each TCGA-LUAD dataset sample. Gene expression and drug sensitivity data were downloaded from the Genomics of Drug Sensitivity in Cancer website (GDSC, https://www.cancerrxgene.org). Additionally, we compared the IC50 difference between high- and low-risk groups in our risk model by a Wilcoxon signed-rank test.
Results
Verification of DEirlncRNAs in TCGA-LUAD Database Samples
The research roadmap of our study is represented in Figure 1. A total of 594 transcriptome profiling data - 535 cancer and 59 normal samples - were obtained from the TCGA-LUAD project. A summary list containing 2483 immune genes was downloaded from the ImmPort database. After annotation of the transcriptome profiling data with GTF files, an expression matrix containing 14,087 lncRNAs was extracted. Then, the correlation analysis was conducted between lncRNAs and immune genes, 1201 irlncRNAs were identified according to the pre-defined filtering conditions (Supplementary Table S1), 293 of which were DEirlncRNAs with a log FC > 1.5 and FDR < 0.05, including 249 upregulated and 44 downregulated. The DEirlncRNAs volcano map and heatmap are shown in Figure 2.
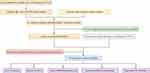 |
Figure 1 Study’s roadmap. |
 |
Figure 2 Identification of DEirlncRNAs: (A) heatmap and (B) volcano map of 293 DEirlncRNAs. |
RiskScore Model Establishment and Validation
A total of 31,914 DEirlncRNA pairs were obtained by comparing the expression level of each DEirlncRNA pair. After univariate Cox and LASSO regressions, eleven pairs retained prognostic significance were included in the risk model (Figure 3), among which 7 DEirlncRNAs had hazard ratio (HR) values greater than 1, and the remaining 4 including LINC02036|AL365181.2, AC026355|LINC01547, AC104958.2|AC031985.3 and AC007991.2|AL365181.2 were less than 1. In the multivariate Cox regression model, seven DEirlncRNAs pairs were further confirmed to regarded as independent prognostic factors. Then, the ROC curve analysis for 1-, 2-, 3-, 4- and 5-years was plotted and the AUC values were calculated (Figure 4A and B). All AUC values were over 0.7 and the 4-year ROC curve presented the maximum (0.819). Additionally, we compared the 4-year ROC curve with common clinical characteristics ROC curve (Figure 4D). It is apparent that the prognostic value of our RiskScore model is superior to the clinical characteristics, including age, gender, sex, tumor stage and TNM stage, which all have AUC values below 0.7. According to the AIC, we identified 1.003 as the cut-off point (Figure 4C), and patients in TCGA-LUAD with a risk score higher than 1.003 were classified as the high-risk group and those above 1.003 as the low-risk group.
Constructed Model Effectiveness Validation
Figure 5A and B shows the distribution of risk scores and survival status of patients in the high- and low-risk groups and suggested that patients in the low-risk have a higher survival rate. A Kaplan–Meier analysis verified that patients in the low-risk group exhibited a better prognosis than those in the high-risk one (p < 0.001) (Figure 5C). The relationships between clinical characteristics (TNM stage, tumor stage, age, and gender) and risk score are presented in Figure 6. TNM and tumor stage were significantly related to the risk score. Patients with advanced lung adenocarcinoma had significantly higher risk score compared to early stage. To further explore the prognostic value of the constructed model, a univariate and multivariate Cox regression analysis were conducted and the results showed that tumor stage (p < 0.001, HR = 1.577, 95% CI [1.348–1.845]), T stage (p < 0.001, HR = 1.579, 95% CI [1.296–1.923]), M stage (p = 0.037, HR = 1.843, 95% CI [1.038–3.272]), N stage (p < 0.001, HR = 1.706, 95% CI [1.405–2.072]) and riskScore (p < 0.001, HR = 1.363, 95% CI [1.291–1.438]) can be regarded as prognostic factors, while the riskScore (p < 0.001, HR = 1.334, 95% CI [1.257–1.417]) was the one that had an independent prognostic role.
 |
Figure 5 Survival characteristics of patients with lung adenocarcinoma: (A) risk scores distribution. (B) Survival outcomes distribution. (C) Survival curve in low- and high-risk groups. |
Comparison with Other Existing Prognostic Signatures for Lung Adenocarcinoma
We have compared our risk model with other existing immune-related lncRNAs prognostic model for lung adenocarcinoma based on TCGA dataset. In a signature consisting of 7 immune-related lncRNAs, the ROC curve for 5-year overall survival (OS) corresponded to an AUC value of 0.702.12 Time-ROC curve analysis of a 10 immune-related lncRNAs signature in the TCGA dataset in 5-year was 0.783.9 The AUC value of ROC curve for a 4 immune-related lncRNAs prognostic signature, consisted of ITGB1-DT, ABALON, TMPO-AS1 and VIM-AS1, plotted by Zhang et al was 0.75614 and The 5-year AUC value in another 4 immune-related lncRNAs prognostic signature including HSPC078, DRAIC, AP004608.1 and MIR223HG was 0.63.13 The 1-year AUC value in a 5 immune-related lncRNAs signature based on TCGA was 0.686.11 The AUC of the 6 immune-related lncRNA signature of Miao et al was 0.761 in training cohort and 0.723 in testing cohort.15 In our constructed signature, the highest AUC value was 0.819, and the AUC value of the 5-year ROC curve was 0.791. Comparing with other existing prognostic models, our signature has superior predictive sensitivity and specificity.
Constructed Model Immune Landscape
We presented the differences in tumor-infiltrating immune cells between high- and low-risk groups, based on 7 algorithms, with a boxplot (Supplementary Figure S1). All differences were significant and we could infer that B cells, T cells CD8, and T cell CD4 memory resting were tested for high expression in the low-risk group (Supplementary Table S2). Additionally, a Spearman correlation analysis was further conducted (Figure 7A) and the detailed results are listed in Supplementary Table S3. B cell, CD8+ T cell, Macrophage M2, and T CD4+ T cell [except for T cell CD4+ (non-regulatory)], T cell CD4+ memory activated, T cell CD4+ Th2 were negatively correlated to risk scores, while macrophage M0 and cancer-associated fibroblast were positively correlated. Tow ICIs-related genes were tested in the constructed model (Figure 7B and C) and the results suggested that a higher CTLA4 expression was associated with the low-risk group, while there was not a statistical difference in PDCD1.
Constructed Model Effectiveness for Drug Response Prediction
We evaluated the difference in IC50 between high- and low-risk groups for 7 drugs: 2 EGFR-TKIs and 5 chemotherapeutic agents (Figure 7D). The results showed that the high-risk group is significant correlated with lower IC50.
Discussion
LncRNA possesses many biological functions and has gained increasing attention for its value as a prognostic factor in malignant tumors. Although several prognostic irlncRNA signatures of lung adenocarcinoma based on the TCGA-LUAD dataset have been developed, they are all dependent on specific lncRNA expression levels of transcripts.9–14 Therefore, this is the first study to apply a two-gene combination strategy to establish lung adenocarcinoma signatures.
First, we obtained DEirlncRNAs by a differential co-expression strategy using an immune gene list from the ImmPort database, while previous studies retrieved them from the Molecular Signature Database or the Gene Set Enrichment Analysis databases.9,10,12,14 Additionally, Wang et al obtained DEirlncRNAs by differential expression analysis between high- and low-ESTIMATE score groups.13 Second, the DEirlncRNAs pairs matrix was arranged based on a cyclic comparison of each lncRNA pair expression level. We constructed the matrix to minimize sample errors derived from expression variations. Third, univariate and LASSO penalized regression analysis was performed to establish the risk signature and a ROC curve was plotted, since its commonly used in bioinformatics research. Fourth, the novel signature prognostic value was evaluated in different situations: various patients’ clinical characteristics, tumor-infiltrating cells, drug response, and ICI-related genes.
A total of 11 irlncRNA pairs were included in our signature, of which LINC02036|AL365181.2, AC026355.2|LIN01547, AC104958.2|AC125807.2, and AC007991.2|AL365181.2 were regarded as risk protective factors. They were identified for the first time, except AC007991.2, which has been verified as a protective autophagy-related lncRNA (HR: 0.609 p = 0.004) in bladder cancer.28 The remaining 7 pairs were considered as risk-related factors. LINC01116, DRAIC, AC022784-1, and LINC01843 were explored in former irlncRNA LUAD signatures.11–13 Previous studies have implied that overexpressed LINC01116 may promote tumor proliferation, metastasis, and drug resistance in LUAD.29–31 DRAIC was recognized to have a role in migration and invasion regulation, as well as a predictor in various cancers.32 The ROC curve indicated that the signature was stable and reliable - AUC superior to most existing signatures. We demonstrated that our signature was significantly related to patients’ LUAD prognosis, and proved to be an independent prognostic factor by univariate and multivariate Cox regression analysis. These findings suggested that our signature could potentially help clinicians in the LUAD patients’ treatment.
Tumor-infiltrating immune cells have been identified as a pivotal component of the tumor immune microenvironment and are involved in the response to immunotherapy. Immune scores are commonly applied to guide immunotherapy and evaluate prognosis. As mentioned in the previous reference, lung cancer patients in the high-immune score group had significantly longer disease-free and overall survival than those in the low-immune score group.33 Due to the complexity and defects of each immune infiltrate algorithm, we performed a correlation analysis between risk scores and immune scores using 7 algorithms. Prior studies have investigated the correlation between TIMER estimated infiltration value and risk scores of a 4-irlncRNA signature including HSPC078, DRAIC, AP004608.1, and MIR223HG in LUAD patients. The results showed that CD4+ T cell, CD8+ T cells, neutrophils, macrophages, and myeloid dendritic cells were significantly negatively associated with risk scores.13 In the immune landscape that we constructed, B cells (B cell memory and B cell plasma) were significantly negatively correlated to the risk score. T cells - T cell CD4+ central memory, T cell CD4+ effector memory, T cell CD4+ memory resting, T cell CD8+, T cell follicular helper, T cell NK, and T cell regulatory (Tregs) - were significantly negatively correlated to risk score, except for T cell CD4+ (non-regulatory), T cell CD4+ memory activated, and T cell CD4+ Th2. These findings suggested that the 7 algorithm combination that we performed presented a more abundant immune landscape compared to individual algorithms. We assessed CTLA4 and PDCD1 expression levels and the results showed that the signature was significantly related to CTLA4. Although CTLA4 and PDCD1 expression have an irreplaceable guiding role in current immunotherapy, this enriches the immune landscape of our signature. Our model suggested that the low-risk group was associated with sensitivity to chemotherapeutics (docetaxel, cisplatin, vinorelbine, and paclitaxel), and targeting-therapy (gefitinib and erlotinib). This indicated that our signature may help clinicians with the LUAD’s treatment.
Finally, we constructed a novel irlncRNA LUAD signature using the TCGA database, based on paired irlncRNAs, and that does not rely on lncRNAs’ specific expression levels. Further analysis confirmed that the prognostic model could be used as an independent prognostic factor for LUAD patients and presented an abundant immune landscape.
Ethics Statement
TCGA is an open-source database in which the patients involved have received ethical approval. Users can download the relevant data for research and publication for free. Therefore, there are no ethical issues.
Acknowledgments
The authors would like to express their appreciation to TCGA database for providing their platforms and China Center for Evidence Based Traditional Chinese Medicine, China Academy of Chinese Medical Sciences for funding.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Administration of Traditional Chinese Medicine Project (2019XZZX-ZL001), Traditional Chinese Medicine Bureau of Guangdong Province Project (20201095) and Pilot project of Integrated traditional Chinese and western medicine clinical collaboration for major and difficult diseases (lung cancer).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Zheng M. Classification and pathology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):447–468. doi:10.1016/j.soc.2016.02.003
3. Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, Phase 3 trial. Lancet. 2019;393(10183):1819–1830. doi:10.1016/S0140-6736(18)32409-7
4. Wu YL, Lu S, Cheng Y, et al. Nivolumab versus docetaxel in a predominantly Chinese patient population with previously treated advanced NSCLC: checkMate 078 randomized Phase III Clinical Trial. J Thorac Oncol. 2019;14(5):867–875. doi:10.1016/j.jtho.2019.01.006
5. West H, McCleod M, Hussein M, et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20(7):924–937. doi:10.1016/S1470-2045(19)30167-6
6. Deng X, Xiong W, Jiang X, et al. LncRNA LINC00472 regulates cell stiffness and inhibits the migration and invasion of lung adenocarcinoma by binding to YBX1. Cell Death Dis. 2020;11(11):945. doi:10.1038/s41419-020-03147-9
7. Yu WD, Wang H, He QF, Xu Y, Wang XC. Long noncoding RNAs in cancer-immunity cycle. J Cell Physiol. 2018;233(9):6518–6523. doi:10.1002/jcp.26568
8. Thommen DS, Koelzer VH, Herzig P, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24(7):994–1004. doi:10.1038/s41591-018-0057-z
9. Li Y, Shen R, Wang A, et al. Construction of a prognostic immune-related LncRNA risk model for lung adenocarcinoma. Front Cell Dev Biol. 2021;9:648806. doi:10.3389/fcell.2021.648806
10. Jin D, Song Y, Chen Y, Zhang P. Identification of a seven-lncRNA immune risk signature and construction of a predictive nomogram for lung adenocarcinoma. Biomed Res Int. 2020;2020:7929132. doi:10.1155/2020/7929132
11. Mu L, Ding K, Tu R, Yang W. Identification of 4 immune cells and a 5-lncRNA risk signature with prognosis for early-stage lung adenocarcinoma. J Transl Med. 2021;19(1):127. doi:10.1186/s12967-021-02800-x
12. Li JP, Li R, Liu X, et al. A seven immune-related lncRNAs model to increase the predicted value of lung adenocarcinoma. Front Oncol. 2020;10.
13. Wang J, Yin X, Zhang YQ, Ji X. Identification and validation of a novel immune-related four-lncRNA signature for lung adenocarcinoma. Front Genet. 2021;12:111.
14. Zhang B, Wang R, Li K, et al. An immune-related lncRNA expression profile to improve prognosis prediction for lung adenocarcinoma: from bioinformatics to clinical word. Front Oncol. 2021;11:671341. doi:10.3389/fonc.2021.671341
15. Miao H, Chen D, Li R, et al. Identification of an immune-related six-long noncoding RNA signature as a novel prognosis biomarker for adenocarcinoma of lung. Biosci Rep. 2021;41(1):BSR20202444. doi:10.1042/BSR20202444
16. Lv Y, Lin SY, Hu FF, et al. Landscape of cancer diagnostic biomarkers from specifically expressed genes. Brief Bioinform. 2020;21(6):2175–2184. doi:10.1093/bib/bbz131
17. Hong W, Liang L, Gu Y, et al. Immune-related lncRNA to construct novel signature and predict the immune landscape of human hepatocellular carcinoma. Mol Ther Nucleic Acids. 2020;22:937–947. doi:10.1016/j.omtn.2020.10.002
18. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
19. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–w514. doi:10.1093/nar/gkaa407
20. Finotello F, Mayer C, Plattner C, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11(1):34. doi:10.1186/s13073-019-0638-6
21. Plattner C, Finotello F, Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol. 2020;636:261–285.
22. Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6:e26476. doi:10.7554/eLife.26476
23. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
24. Aran D. Cell-type enrichment analysis of bulk transcriptomes using xCell. Methods Mol Biol. 2020;2120:263–276.
25. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi:10.1186/s13059-017-1349-1
26. Dienstmann R, Villacampa G, Sveen A, et al. Relative contribution of clinicopathological variables, genomic markers, transcriptomic subtyping and microenvironment features for outcome prediction in stage II/III colorectal cancer. Ann Oncol. 2019;30(10):1622–1629. doi:10.1093/annonc/mdz287
27. Tamminga M, Hiltermann TJN, Schuuring E, Timens W, Fehrmann RS, Groen HJ. Immune microenvironment composition in non-small cell lung cancer and its association with survival. Clin Transl Immunol. 2020;9(6):e1142. doi:10.1002/cti2.1142
28. Wan J, Guo C, Fang H, Xu Z, Hu Y, Luo Y. Autophagy-related long non-coding RNA is a prognostic indicator for bladder cancer. Front Oncol. 2021;11:647236. doi:10.3389/fonc.2021.647236
29. Zeng L, Lyu X, Yuan J, et al. Erratum: long non-coding RNA LINC01116 is overexpressed in lung adenocarcinoma and promotes tumor proliferation and metastasis. Am J Transl Res. 2021;13(4):3919–3920.
30. Wang J, Gao J, Chen Q, et al. LncRNA LINC01116 contributes to cisplatin resistance in lung adenocarcinoma. Onco Targets Ther. 2020;13:9333–9348. doi:10.2147/OTT.S244879
31. Shang B, Li Z, Li M, et al. Silencing LINC01116 suppresses the development of lung adenocarcinoma via the AKT signaling pathway. Thorac Cancer. 2021;12(14):2093–2103. doi:10.1111/1759-7714.14042
32. Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive nexus. Mol Cancer Res. 2015;13(5):828–838. doi:10.1158/1541-7786.MCR-15-0016-T
33. Han K, Qian K, Zhao T, Liu XS, Zhang Y. Prediction of prognosis of patients with lung cancer in combination with the immune score. Biosci Rep. 2021;41(5). doi:10.1042/BSR20203431
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.