Back to Journals » Cancer Management and Research » Volume 12
Construction and Characterization of KRAS Immune Lipid Magnetic Balls for Colorectal Cancer Circulating Tumor Cells
Authors Feng C , Wang J, Yang X, Zang X, Zhou H , Zhang E , Li H, Liu B, Chen S, Wang Y, Wang P, Gao L , Liu J
Received 17 April 2020
Accepted for publication 11 August 2020
Published 13 October 2020 Volume 2020:12 Pages 10067—10075
DOI https://doi.org/10.2147/CMAR.S258565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Chengbao Feng,1 Jingjing Wang,1 Xue Yang,2 Xuefang Zang,1 Haikun Zhou,1 Erying Zhang,1 Haifei Li,1 Bo Liu,1 Shumin Chen,1 Ying Wang,1 Peng Wang,1 Lixue Gao,1 Jinyan Liu1
1Department of Medical Oncology, No. 2 Hospital of Baoding, Baoding City, People’s Republic of China; 2Baoding Children’s Hospital, Baoding City, People’s Republic of China
Correspondence: Jinyan Liu Email [email protected]
Objective: The purpose of this study was to prepare and characterize a lipid magnetic ball modified with KRAS antibodies on the surface and to isolate circulating tumor cells of colorectal cancer with KRAS mutations.
Methods: The microemulsion method was used to form lipid bilayers to encapsulate Fe3O4 nanoparticles with superparamagnetism to form lipid magnetic balls, and KRAS antibodies were formed on the surface to form KRAS immune lipid magnetic balls.
Results: Compared with traditional EpCAM antibody-modified lipid magnetic balls, it can effectively improve the capture ability of colorectal cancer circulating tumor cells with KRAS mutation, the capture rate reaches 92.9%, and the capture results are consistent with clinical diagnosis and pathology.
Conclusion: Our results showed that KRAS antibody-modified lipid magnetic balls can be used in the diagnosis and treatment of KRAS colorectal cancer.
Keywords: colorectal cancer, circulating tumor cells, KRAS mutation, lipid magnetic balls
Introduction
The incidence of colorectal cancer (CRC) is second only to lung cancer and breast cancer worldwide, accounting for about 10% of the total cancer worldwide.1,2 Colorectal cancer is occult and the main cause of death is recurrence and metastasis. The overall 5-year survival rate is 50% −60%.3,4 Circulating tumor cell (CTC) refers to tumor cells that fall off from the primary or metastatic foci into the peripheral blood circulation. CTC carries the genetic and phenotypic information of tumor tissues, which can guide patients with CRC to take drugs and detect resistance.5,6 At present, the detection and research of CTC have been verified in colorectal cancer, non-small cell lung cancer, breast cancer and so on.7–9
KRAS gene (Kirsten rat sarcoma viral oncogene) participates in the regulation of signal pathways such as MAPK, PI3K and Ral-GEFs.10–12 They are commonly found in digestive malignant tumors such as colorectal cancer, pancreatic cancer, and bile duct cancer,13–15 and among non-digestive system malignancies such as endometrial cancer, breast cancer, lung cancer,16–18 they are closely related to the occurrence, development, invasion and metastasis of these tumors. The occurrence of KRAS mutation is a negative predictor of CRC patients’ drugs targeting EGFR (epidermal growth factor receptor).19,20 Yang et al’s research shows that KRAS mutations can be detected in CTCs of CRC patients, and 30% of patients without mutations have secondary KRAS mutations during the course of targeted therapy.21
Magnetic activated cell sorting (MACS) method is currently the most widely used and mature CTC capture method. Most of them are lipid magnetic balls modified with epithelial cell adhesion molecule (EpCAM) antibodies to isolate CTCs, but they are not ideal for CTC capture through epithelial–mesenchymal transition.22,23 This study uses KRAS antibody-modified lipid magnetic balls to perform CTC detection on CRC patients, which can screen patients with KRAS mutations and better guide medications for CRC patients. Compared with traditional EpCAM lipid magnetic balls, more advantages in CRC.
Materials and Methods
Sample Collection
Peripheral blood was collected from 55 patients with pathologically diagnosed colorectal cancer in the Second Hospital of Baoding, Hebei Province. Of these, tissue samples of 15 patients were collected at the same time. The enrolled patients did not receive any adjuvant therapy. This study was approved by the Ethics Committee of the Second Hospital of Baoding, Hebei Province, and informed consent of each research participant was obtained, and an informed consent was signed in accordance with the “Helsinki Declaration”. LoVo human colorectal cancer KRAS mutant cell line was purchased from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Materials and Instruments
F-12K broth, fetal calf serum, trypsin were purchased from Gibco, 1-ethyl-3-(3-dimethylammoniumpropyl) carbonate (EDC), N-hydroxysuccinimide (NHS), Cholesterol, and other commonly used chemical reagents were purchased from Sinopharm, EpCAM lipid magnetic balls, Fe3O4, carboxymethyl chitosan hexadecyl quaternary ammonium salt (HQCMC), 1.2-dioleylphosphatidylcholine (DOPC), dimethyl octadecyl epoxypropyl ammonium chloride (GHDC) were purchased from Huzhou Liyuan Medical Laboratory Co., Ltd., KRAS gene upstream and downstream primers and PCR kit (B639289-0100) were purchased from Shanghai Sangon Biotech Co., Ltd., KRAS antibody (ab180772) was purchased from Abcam, BI-90Plus laser particle size analyzer/Zeta potentiometer was purchased from Brook-Haven Company, XL-30 environmental scanning electron microscope was purchased from PHILIPS, Netherlands, OLYMPUS B×61 fluorescence microscope was purchased from Olympus, Japan, and the MPMS-XL-7 vibration sample magnetometer was purchased from American Quantum Design Corporation.
Preparation of KRAS Lipid Magnetic Balls and EpCAM Lipid Magnetic Balls
KRAS-Lipid magnetic ball (K-LMB) and EpCAM-Lipid magnetic ball (Ep-LMB) were prepared according to reference.24–26 Parallel experiments were carried out to select the appropriate antibody concentration and reagent concentration. Dissolve cholesterol, DOPC, GHDC, HQCMC, and Fe3O4 in dichloromethane, and add a 0.1 mol/L PBS solution at the same time. Mix the mixture by vigorous stirring and heat it to 25°C to completely emulsify it, forming the Lipid magnetic ball (LMB). Mechanical agitation is required during this period to avoid agglomeration or aggregation of iron oxide nanoparticles and nanocolloids. Take 0.6mg of KRAS antibody and dissolve it in 10mL of isopropanol. Add coupling agents EDC and NHS and mix with the nanomagnetic balls. Stir at 24°C for 24 hours to obtain KRAS antibody-modified lipid magnetic balls. The prepared lipid magnetic spheres were separated from the remaining solution and stored by magnetic separation.
K-LMB Characterization
Take 10μL K-LMB sample and dilute it with 1mL distilled water, then use BI-90Plus laser particle size analyzer/Zeta potentiometer to measure particle size and potential. To observe K-LMB with atomic force microscope (AFM) and aggregation occurred or not. Dilute 10μL K-LMB sample with 1mL distilled water, take 50 μL and coat it on the mica tablet, use non-contact tap mode (frequency = 325 Hz), scan at 1.0 Hz and observe after drying. Dilute 10 μL K-LMB and LMB samples in 1 mL of distilled water, measure the absorbance at 280 nm with an ultraviolet spectrophotometer. The magnetic properties of K-LMB were measured using a vibrating sample magnetometer.
K-LMB’s Ability to Capture LoVo Colorectal Cancer Cells
The cultured LoVo colorectal cancer cells were added to the 7.5mL PBS solution at a gradient of 10, 20, 50, 100, 200 cells/mL after counting, and divided into K-LMB group and Ep-LMB group, each group has triplicate samples, the prepared K-LMB was tested for its ability to capture LoVo colorectal cancer cells. Add 20 μL of K-LMB to a 7.5 mL sample, and incubate at room temperature for 15 min, mix once every 5 min. Insert the centrifuge tube into a magnetic separator for 10 min, discard the supernatant, and add PBS solution for washing twice. Add 10 μL of FITC CK19 monoclonal antibody (CK19-FITC), 20 μL DAPI staining solution, 10 μL PE-labeled CD45 antibody (CD45-PE), mix and stain for 15 min in the dark. After the staining, wash twice with deionized water to wash off unbound antibody and DAPI. Magnetically separate for 5 min. Finally, 15 μL of deionized water was added to the centrifuge tube to resuspend the CTC, evenly coat the poly-L-lysine-treated anti-off slides. After the liquid drops are dried, observe and count under a fluorescence microscope.
K-LMB and Ep-LMB Capture CTC Tests for CRC Patients
First, the peripheral blood of 55 patients with CRC and the tissues of 15 patients with CRC were tested for KRAS gene, and the cases with KRAS mutation were counted. Then, K-LMB and Ep-LMB were used to perform CTC tests on peripheral blood samples of 35 patients, and the isolated CTCs were subjected to immunofluorescence identification and KRAS gene detection.
Detection of KRAS Mutations in CRC Patient Tissues and CTCs
The KRAS mutations in the CRC tissues and CTCs were detected by PCR. The upstream primers of KRAS were 5´-GGGGAGGGCTTTCTTTGTGTA-3´, and the downstream primers were 5´-GTCCTGAGCCTGTTTTGTGTC-3´. The reaction parameters were: pre-denaturation at 95°C for 6min; denaturation at 95°C for 10s, annealing at 55°C for 10s, and extension at 72°C for 30s, a total of 50 cycles. The experiment was repeated independently at least 3 times, and 3 replicates were set for each sample.
Statistical Analysis
All the data in this study were analyzed by SPSS 20.0 statistical analysis software. The comparison between the two groups was analyzed by independent sample t test. Statistical standards of α = 0.05 and P <0.05 were used to judge that the difference was statistically significant. All data are the result of 3 independent repeated experiments.
Results
We synthesized the required KRAS lipid magnetic balls and performed characterization and verification of the effect of separating CTC. The technical route is shown in Figure 1.
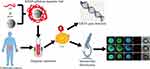 |
Figure 1 K-LMB preparation and CTC detection technology route. |
K-LMB Characterization
A series of characterization tests were performed on K-LMB. Figure 2 shows the particle size and potential distribution, AFM image, and UV absorption spectrum of K-LMB. The synthesized lipid magnetic nanoparticles are spherical, with an average particle size of 198.3 ± 4.2 nm, and the distribution is between 110.1 and 356.2 nm. The particle size distribution is narrow and relatively uniform. The magnetic ball has a charge of +29.7 mV. The AFM image shows that the nanoparticles are spherical with a size of about 250 nm, and the shape is more regular. There is no agglomeration and the results are consistent with the particle size detection results. The ultraviolet absorption spectrum showed that the lipid magnetic sphere had no absorption peak at 280 nm, and K-LMB showed a broad absorption peak at 280 nm, which had protein characteristics, indicating that the surface of the lipid magnetic sphere was connected by an EDS-NHS coupling reaction. KRAS antibody with good specificity. Figure 3 shows the results of the magnetic performance test of K-LMB. The hysteresis loop of K-LMB passed the origin, and the coercive force and remanence are close to zero. Under the action of an external magnetic field, K-LMB dispersed in the solution can be collected, and when the magnetic field disappears, K-LMB can be re-dispersed into the solution after shaking, which indicates that the prepared lipid magnetic balls are super-cis magnetic particle.
 |
Figure 2 (A) The particle size distribution of K-LMB; (B) the potential distribution of K-LMB; (C) the atomic force image of K-LMB; (D) the ultraviolet absorption spectrum of K-LMB and LMB. |
 |
Figure 3 (A) The magnetic response of K-LMB under an external magnetic field; (B) the hysteresis loop of K-LMB. |
Capability of K-LMB to Capture LoVo Colorectal Cancer Cells
Using the prepared K-LMB to capture the KRAS mutation of LoVo colorectal cancer cells, the experiment was performed to simulate the separation effect of CTC. The results are shown in Figure 4. Compared with Ep-LMB, the recovery of LoVo colorectal cancer cells by K-LMB is more stable at various concentrations. The capture efficiency of K-LMB is 90%, so it can be used for CTC capture of CRC clinical samples with KRAS mutations.
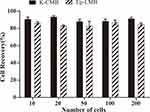 |
Figure 4 Capability of K-LMB and Ep-LMB to capture KRAS mutated LoVo colorectal cancer cells. |
K-LMB Positive Rate of CTC in CRC Patients
K-LMB and Ep-LMB were used to perform CTC detection of the blood of 55 CRC patients. The immunofluorescence identification of CTC captured by K-LMB is shown in Figure 5. CTC of CRC patients has obvious cell morphology under white light, CK19-FITC green fluorescence is strongly positive, DAPI blue fluorescence is strongly positive, the two fluorescence overlaps after superposition, and CD45 staining does not show fluorescence. The captured cell is determined to be colorectal Cancer CTC.
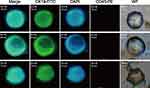 |
Figure 5 CTC immunofluorescence identification results of CRC patients captured by K-LMB. |
The CTC isolation rate of 55 CRC patients enrolled was 100% (n = 55) using Ep-LMB and 54.5% (n = 30) using K-LMB. The counting results are shown in Figure 6.
 |
Figure 6 CTC test results of 55 CRC patients using K-LMB and Ep-LMB, respectively. |
The Effect of K-LMB on CTC Isolation in Patients with KRAS Mutant CRC and Its Relationship with Clinicopathological Characteristics
KRAS mutations were analyzed in the blood of 55 patients with CRC and in tissues of 15 patients with CRC. Among them, 26 patients had KRAS mutations in the blood and 5 patients had KRAS mutations in the tissues, and 16 patients had KRAS mutations detected in both tissues and blood. KRAS mutation detection was performed on 30 K-LMB isolated CTCs, and the results showed that the KRAS mutation rate was 100% (n = 30). In general, K-LMB detected 92.9% of CTCs with KRAS mutation in CRC patients. In order to clarify the correlation between KRAS mutations and other clinicopathological characteristics in CRC, the clinical and pathological characteristics of 55 patients with CRC were statistically analyzed, and important clinical parameters such as age, tumor size, lymph node metastasis, distant metastasis and TNM staging were selected for correlation, correlation analysis is shown in Table 1. The results showed that KRAS mutation was not related to other clinicopathological features (P <0.05).
 |
Table 1 Relationship Between KRAS Mutations and Clinical Characteristics in Blood from 55 CRC Patients and Tissues from 15 CRC Patients |
Discussion
Liquid biopsy has become a hot spot in current cancer treatment, with more and more new technologies and methods having benefited patients with colorectal cancer. Immune nanomagnetic balls have been widely used and studied in colorectal cancer due to their unique advantages.27 Compared with the traditional EpCAM lipid magnetic spheres, the lipid magnetic spheres with KRAS antibodies prepared by our research can screen patients with KRAS mutations in CRC, and the positive detection rate is high. The prepared KRAS lipid magnetic balls have uniform specifications, stable properties and high specificity. They are ideal magnetic balls for CTC separation of CRC patients.
The membrane-like structure of the lipid magnetic ball minimizes cell damage during CTC separation. The use of KRAS antibody can better personalize the EGFR-targeted therapy of CRC patients before and after drug resistance and improve detection Specificity. There have been studies on the preparation of EGFR antibody-modified lipid magnetic balls for CTC detection of CRC and detection of KRAS mutations, with a KRAS mutation rate of 71.4% (5/7).28 The number of samples included in this study is too small and not representative. We used KRAS antibody-modified lipid magnetic balls to be more targeted.
At present, there have been studies on CTC screening for KRAS mutations in CRC. Taieb et al analyzed 3934 MSS stage III CRC patients found that the patients with KRAS mutations had a much shorter postoperative recurrence time, survival time after relapse, and overall survival time, and the overall survival risk with KRAS mutations is 1.62 compared with wild-type patients, indicating that KRAS mutations are independent predictors of postoperative recurrence, post-relapse survival, and overall survival deterioration in colorectal cancer.29 In addition, some research tested the tumor tissue and CTC for KRAS mutations at the same time. The test results show that the consistency rate of the primary tumor tissue and CTC mutations can reach 70%.30 The expression of EpCAM on CTC is very variable. The epithelial characteristics CTC undergoes interstitial transformation, making EpCAM magnetic balls unsuitable for other tumors.31 Our research can directly screen CTCs with KRAS mutations in patients with CRC, which can guide patients’ medication before treatment, determine prognosis, and more accurately detect whether patients are suitable for targeted therapy.
In summary, the KRAS antibody-modified lipid magnetic balls prepared in this study are more suitable for CTC detection in CRC than EpCAM lipid magnetic balls, and it has been confirmed that KRAS mutations can be screened by CTC in the CRC in the past to achieve the possibility of early diagnosis and prognostic evaluation in patients with CRC.
Acknowledgment
The detection analysis and magnetic beads preparation in this project are mainly assisted by Huzhou Lieyuan Medical Laboratory Co., Ltd.
Disclosure
The authors report no conflict of interests in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637–1649. doi:10.1136/gutjnl-2014-309086
3. Tzeng CW, Alola TA. Colorectal liver metastases. J Gastrointestinal Surg. 2013;17(1):195–201. doi:10.1007/s11605-012-2022-3
4. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
5. Batth IS, Mitra A, Rood S, et al. CTC analysis: an update on technological progress. Translational Res. 2019;212:14–25. doi:10.1016/j.trsl.2019.07.003
6. Hao YX, Fu Q, Guo YY, et al. Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: a meta-analysis. Onco Targets and Therapy. 2017;10:945–953. doi:10.2147/OTT.S123954
7. Buim M, Fanelli M, Souza VS, et al. Detection of KRAS mutations in circulating tumor cells from patients with metastatic colorectal cancer. Cancer Biol Ther. 2015;16(9):1289–1295. doi:10.1080/15384047.2015.1070991
8. Liu DG, Xue L, Li J, et al. Epithelial-mesenchymal transition and GALC expression of circulating tumor cells indicate metastasis and poor prognosis in non-small cell lung cancer. Cancer Biomark. 2018;22(3):417–426. doi:10.3233/CBM-170995
9. Rugo HS, Cortes J, Awada A, et al. Change in Topoisomerase 1-Positive Circulating Tumor Cells Affects Overall Survival in Patients with Advanced Breast Cancer after Treatment with Etirinotecan Pegolr. Clin Cancer Res. 2018;24(14):3348–3357. doi:10.1158/1078-0432.CCR-17-3059
10. Bahrami A, Hassanian SM, ShahidSales S, et al. Targeting RAS signaling pathway as a potential therapeutic target in the treatment of colorectal cancer. J Cell Physiol. 2018;233(3):2058–2066. doi:10.1002/jcp.25890
11. Neuzillet C, Tijeras-raballand A, Mestier L, et al. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141(2):160–171. doi:10.1016/j.pharmthera.2013.10.001
12. Dienstmann R, Elez E, Argiles G, et al. Analysis of mutant allele fractions in driver genes in colorectal cancer-biological and clinical insights. Mol Oncol. 2017;11(9):1263–1272. doi:10.1002/1878-0261.12099
13. Prieur A, Cappellini M, Habif G, et al. Targeting the Wnt Pathway and Cancer Stem Cells with Anti-progastrin Humanized Antibodies as a Potential Treatment for K-RAS-Mutated Colorectal Cancer. Clin Cancer Res. 2017;23(17):5267–5280. doi:10.1158/1078-0432.CCR-17-0533
14. Karmakar S, Kaushik G, Nimmakayala R, et al. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. 2019;54:63–71. doi:10.1016/j.semcancer.2017.11.020
15. Cavalloni G, Peraldoneia C, Sassi F, et al. Establishment of a patient-derived intrahepatic cholangiocarcinoma xenograft model with KRAS mutation. Bmc Cancer. 2016;16(1):90. doi:10.1186/s12885-016-2136-1
16. Koo KH, Jeong WJ, Cho YH, et al. K-Ras stabilization by estrogen via PKCdelta is involved in endometrial tumorigenesis. Oncotarget. 2015;6(25):21328–21340. doi:10.18632/oncotarget.4049
17. Liu R, Li J, Lai Y, et al. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int J Biol Macromol. 2015;81:491–497. doi:10.1016/j.ijbiomac.2015.08.016
18. Wang L, Zhao Y, Xiong Y, et al. K-ras mutation promotes ionizing radiation-induced invasion and migration of lung cancer in part via the Cathepsin L/CUX1 pathway. Exp Cell Res. 2018;362(2):424–435. doi:10.1016/j.yexcr.2017.12.006
19. Bokemeyer C, Köhne CH, Ciardiello F, et al. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. Eur J Cancer. 2015;51(10):1243–1252. doi:10.1016/j.ejca.2015.04.007
20. Van Cutsem E, Lenz HJ, Köhne CH, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol. 2015;33(7):692–700. doi:10.1200/JCO.2014.59.4812
21. Yang MJ, Chiu HH, Wang HM, et al. Enhancing detection of circulating tumor cells with activating KRAS oncogene in patients with colorectal cancer by weighted chemiluminescent membrane array method. Ann Surg Oncol. 2010;17(2):624–633. doi:10.1245/s10434-009-0831-8
22. Hyun KA, Koo GB, Han H, et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget. 2016;7(17):24677–24687. doi:10.18632/oncotarget.8250
23. Wu F, Zhu J, Mao Y, et al. Associations between the Epithelial-Mesenchymal Transition Phenotypes of Circulating Tumor Cells and the Clinicopathological Features of Patients with Colorectal Cancer. Dis Markers. 2017;2017:9474532. doi:10.1155/2017/9474532
24. Liang XF, Shi BZ, Wang K, et al. Development of self-assembling peptide nanovesicle with bilayers for enhanced EGFR-targeted drug and gene delivery. Biomaterials. 2016;82:194–207. doi:10.1016/j.biomaterials.2015.12.015
25. Ding J, Wang K, Tang WJ, et al. Construction of Epidermal Growth Factor Receptor Peptide Magnetic Nanovesicles with Lipid Bilayers for Enhanced Capture of Liver Cancer Circulating Tumor Cells. Anal Chem. 2016;88(18):8997–9003. doi:10.1021/acs.analchem.6b01443
26. Liang XF, Li XY, Chang J, et al. Properties and Evaluation of Quaternized Chitosan/Lipid Cation Polymeric Liposomes for Cancer-Targeted Gene Delivery. Langmuir. 2013;29(27):8683–8693. doi:10.1021/la401166v
27. Kondo Y, Hayashi K, Kawakami K, et al. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017;17(1):311. doi:10.1186/s12885-017-3305-6
28. Kuai JH, Wang Q, Zhang AJ, et al. Epidermal growth factor receptor-targeted immune magnetic liposomes capture circulating colorectal tumor cells efficiently. World J Gastroenterol. 2018;24(3):351–359. doi:10.3748/wjg.v24.i3.351
29. Taieb J, Le Malicot K, Shi Q, et al. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J Natl Cancer Inst. 2017;109.
30. Musella V, Pietrantonio F, Di Buduo E, et al. Circulating tumor cells as a longitudinal biomarker in patients with advanced chemorefractory, RAS-BRAF wild-type colorectal cancer receiving cetuximab or panitumumab. International Journal of Cancer. 2015;137(6):1467–1474. doi:10.1002/ijc.29493
31. Nicolazzo C, Gradilone A, Carpino G, et al. Molecular Characterization of Circulating Tumor Cells to Study Cancer Immunoevasion. Methods Mol Biol. 2019;1884:247–258.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
