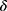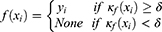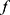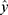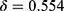Back to Journals » Nature and Science of Sleep » Volume 13
Confidence-Based Framework Using Deep Learning for Automated Sleep Stage Scoring
Authors Hong JK , Lee T , Delos Reyes RD , Hong J , Tran HH , Lee D, Jung J , Yoon IY
Received 16 August 2021
Accepted for publication 6 December 2021
Published 24 December 2021 Volume 2021:13 Pages 2239—2250
DOI https://doi.org/10.2147/NSS.S333566
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ahmed BaHammam
Jung Kyung Hong,1,2,* Taeyoung Lee,3,* Roben Deocampo Delos Reyes,4 Joonki Hong,3,4 Hai Hong Tran,4 Dongheon Lee,4 Jinhwan Jung,4 In-Young Yoon1,2
1Department of Psychiatry, Seoul National University Bundang Hospital, Seongnam, Korea; 2Seoul National University College of Medicine, Seoul, Korea; 3Korea Advanced Institute of Science and Technology, Daejeon, Korea; 4Asleep Inc., Seoul, Korea
*These authors contributed equally to this work
Correspondence: In-Young Yoon
Department of Psychiatry, Seoul National University Bundang Hospital, 82, Gumi-ro 173beon-gil, Bundang-gu, Seongnam-si, Gyeonggi-do, 463-707, Korea
Tel +82-31-787-7433
Fax +82-31-787-4058
Email [email protected]
Jinhwan Jung
R&D Division, Asleep Inc, Asleep, 15, Teheran-ro 82-gil, Gangnam-gu, Seoul, Korea
Tel +82-10-6228-7137
Email [email protected]
Study Objectives: Automated sleep stage scoring is not yet vigorously used in practice because of the black-box nature and the risk of wrong predictions. The objective of this study was to introduce a confidence-based framework to detect the possibly wrong predictions that would inform clinicians about which epochs would require a manual review and investigate the potential to improve accuracy for automated sleep stage scoring.
Methods: We used 702 polysomnography studies from a local clinical dataset (SNUBH dataset) and 2804 from an open dataset (SHHS dataset) for experiments. We adapted the state-of-the-art TinySleepNet architecture to train the classifier and modified the ConfidNet architecture to train an auxiliary confidence model. For the confidence model, we developed a novel method, Dropout Correct Rate (DCR), and the performance of it was compared with other existing methods.
Results: Confidence estimates (0.754) reflected accuracy (0.758) well in general. The best performance for differentiating correct and wrong predictions was shown when using the DCR method (AUROC: 0.812) compared to the existing approaches which largely failed to detect wrong predictions. By reviewing only 20% of epochs that received the lowest confidence values, the overall accuracy of sleep stage scoring was improved from 76% to 87%. For patients with reduced accuracy (ie, individuals with obesity or severe sleep apnea), the possible improvement range after applying confidence estimation was even greater.
Conclusion: To the best of our knowledge, this is the first study applying confidence estimation on automated sleep stage scoring. Reliable confidence estimates by the DCR method help screen out most of the wrong predictions, which would increase the reliability and interpretability of automated sleep stage scoring.
Keywords: confidence estimation, deep learning, electroencephalography, polysomnography, sleep stages, accuracy improvement
Plain Language Summary
Deploying automated sleep stage scoring in practice requires models to be both accurate and reliable. While existing works have focused on improving the accuracy, the problem of their black-box nature has hardly been solved. As the first study to introduce confidence estimation in automated sleep stage scoring, we focused on how to increase the reliability and utility of automated sleep stage scoring. Confidence estimation can serve a surveillance role for the classifier and screen out challenging epochs by showing low confidence. Therefore, only epochs with possibly wrong predictions will need manual review while accuracy is kept high. By adopting a confidence model, automated sleep stage scoring can be used in practice in a manipulable and reliable manner.
Introduction
Polysomnography (PSG) is the gold-standard procedure for analyzing and diagnosing sleep health. A PSG recording provides overnight sleep data for a patient’s sleeping at a sleep center. It mainly consists of various biosignals, including electroencephalogram (EEG), electrooculogram, electromyogram, electrocardiogram, and respiratory signals. Every 30-sec epoch of this sleep data is visually inspected by a human sleep expert and manually classified into one of five sleep stages according to the American Academy of Sleep Medicine (AASM) Scoring Manual.1 These five sleep stages are: Wake (W), Rapid eye movement (REM) sleep (R), Non-REM stage 1 (N1), Non-REM stage 2 (N2), and Non-REM stage 3 (N3). While information of sleep stages is essential for sleep diagnosis, manual annotation for sleep stages is a time-consuming and labor-intensive process. With the advancement of artificial intelligence (AI), many automated sleep stage scoring models that are trained via deep learning (DL) have been proposed to make the process more efficient.
DL-based sleep stage scoring models take advantage of huge amounts of data and recent advances in technology to automate the sleep stage scoring process. One of the earliest automated sleep stage scoring models is DeepSleepNet.2 By using convolutional and recurrent neural networks, DeepSleepNet learns spatial and temporal patterns of sleep data such that it can accurately predict sleep stages given data of raw single EEG channel as input. More recently, TinySleepNet has been proposed as a lighter version of DeepSleepNet with improved prediction capabilities and generalizability across different sleep datasets.3 Many other automated sleep stage scoring models have also been reported in the literature,4–10 with their own carefully designed model architecture and novel DL technique aimed to improve the model’s capability to classify 30-sec sleep epochs. The introduction of DL in sleep stage scoring has improved the accuracy of those automated systems by a huge margin, with reported Cohen’s kappa values of up to 0.80,10,11 which surpasses the inter-rater reliability among sleep technologists (Cohen's kappa: 0.68–0.76).12
Despite their remarkable performances for classification, DL-based sleep stage scoring models are not yet widely used in clinical environments. The lack of transparency of these models is considered as their major drawback.13 The black-box nature of DL models raises concerns regarding their reliability in practice since rationales for their predictions are not accessible. In addition, current DL-based sleep stage scoring models do not allow human experts to intervene or supervise AI-derived results. Thus, the only way to confirm their predictions is to manually review the entire PSGs. If there is a way to differentiate between reliable predictions and possibly wrong predictions, which are often mixed together within each PSG recording, clinicians would not need to review the whole PSGs. DL-based sleep stage scoring models would be more usable in clinical practice if additional information is provided with their predictions, which can specify epochs that require a manual review.
One way of doing so is by adding confidence estimation to the automated sleep stage scoring, by which values indicating how confident the model is about its predictions are given. This would allow sleep technologists and clinicians to selectively inspect those epochs screened out by the system and re-score them manually when needed. This approach may speed up the manual sleep stage scoring process while ensuring its accuracy and reliability. One of the well-known confidence estimation methods is temperature scaling14 which adopts calibrated output of neural networks as confidence to match prediction accuracy. However, the calibration does not affect the ranking of confidence estimates. In other words, wrong predictions may have the highest confidence estimate even with the calibration; thus, it is not applicable to failure prediction.15,16 A new confidence estimation model which is able to distinguish correct and wrong predictions is needed.
The objective of the study was to evaluate the utility and efficacy of confidence estimation in automatic sleep stage scoring. We proposed a novel confidence estimation model which is specified for automated sleep stage scoring to detect the wrong classification. We evaluated the performance of confidence estimation itself, compared the performance of our novel confidence model with other existing methods, and experimented with scenarios such as rejecting a fixed percentage of predictions with the lowest confidence estimates.
Methods
Datasets
Local Dataset
The dataset consisted of 3510 PSGs recorded from 1st January 2013 to 31st December 2020 at the sleep center in Seoul National University Bundang Hospital (SNUBH). For most (91.8%) subjects, the sleep study was prescribed for the purpose of clinical diagnosis, while the rest (8.2%) were for clinical trials. Exclusion criteria for the data were: (1) from patients aged <19 years or >80 years, and (2) recorded sleep time less than 240 minutes. Since the dataset was retrospectively collected from PSGs conducted in the past, additional informed consents were not available. However, the data were all anonymized. The use of this dataset in this study was approved by the Institutional Review Board (IRB) of SNUBH (IRB No. B-2011/648-102).
To increase the efficiency and save time to run numerous DL experiments, 20% of data from the SNUBH dataset were randomly selected and used for the experiments, giving a total of 702 PSGs. The mean age of examinees was 52.8±13.9 years and there were 237 (33.8%) females. The data consisted of 71 (10.1%) patients with insomnia, 347 (49.4%) patients with moderate-severe degree of sleep apnea (AHI ≥ 15), and 141 (20.1%) patients diagnosed with REM sleep behavior disorder. The mean recording time was 472.5 ± 30.1 minutes per night, ranging from 370.8 minutes to 617.6 minutes. Each PSG was visually inspected and manually annotated by sleep technologists according to AASM scoring rules and confirmed by a sleep expert.
Preprocessing
Preprocessing of the data included filtering, downsampling, and normalization. In the current study, only a single EEG signal (C3-A2) was used as input data. For filtering, a Butterworth bandpass filter with a range of 0.3 Hz to 35 Hz was applied to keep alpha (8–13 Hz), theta (4–8 Hz), delta (1–4 Hz), and sleep spindles (11–16 Hz) known to be important waveforms for sleep stage scoring according to the AASM manual. To assess the model’s performance in a realistic scenario, artifacts caused by the major body movements or by the equipment were not eliminated. Signals were then downsampled from 500 Hz to 100 Hz to reduce the computational complexity of the training of DL models. Downsampled data were then cut into 30-sec epochs. Every 11 epochs were then grouped into a sequence because when the sequence length is 10 or more, the accuracy improvement is saturated according to the previous literatures.4,5 Normalization was done using the mean and standard deviation calculated with the Welford’s algorithm, which was conducted because of the huge dataset size (600 GB). Finally, the entire dataset was divided into train, validation, and test sets with a ratio of 70:15:15 at the PSG level.
Public Dataset
To validate our framework, the Sleep Heart Health Study (SHHS) dataset from the National Heart, Lung, and Blood Institute,17,18 one of well-known open datasets, was used. We used the 125 Hz single EEG channel data from SHHS-1, and merged stages 3 and 4 according to R&K scoring rules into N3 as other existing literature.19,20 We randomly selected 50% of the data from SHHS-1, which had a total of 5793 PSG recordings, for the experiments. These data were also divided into train, valid, and test sets with a ratio of 70:15:15 at the PSG level.
Framework Architecture
Our framework was composed of a class prediction model and a confidence model (Figure 1). The classifier and the parallel confidence model both took EEG time series data as input and output sleep stages and confidence estimates, respectively. Therefore, each epoch receives a prediction of sleep stage (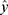 ) with a degree of certainty which is presented as a confidence value (
) with a degree of certainty which is presented as a confidence value (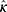 ). Through this confidence output, sleep physicians can decide which predictions of epochs are reliable and which epochs would need a manual review. If an empirical threshold for an acceptable confidence can be set, it would be even easier to screen out epochs requiring manual review by simply accepting predictions with confidence equal to or above the threshold while rejecting predictions with confidence under the threshold. Sleep physicians will only need to manually review and re-annotate epochs with low confidence which are assumed to be a small portion of full PSGs.
). Through this confidence output, sleep physicians can decide which predictions of epochs are reliable and which epochs would need a manual review. If an empirical threshold for an acceptable confidence can be set, it would be even easier to screen out epochs requiring manual review by simply accepting predictions with confidence equal to or above the threshold while rejecting predictions with confidence under the threshold. Sleep physicians will only need to manually review and re-annotate epochs with low confidence which are assumed to be a small portion of full PSGs.
 |
Figure 1 Confidence-based re-scoring framework for automated sleep stage scoring via deep learning. The confidence threshold |
Selective Classifier
In our problem, given the dataset (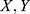 ) of PSG recordings (X) and sleep stage labels (
) of PSG recordings (X) and sleep stage labels (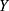 ), we defined the classifier f such that for a given time series EEG signal
), we defined the classifier f such that for a given time series EEG signal 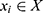 , the classifier could output a corresponding sleep stage label
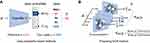
, the classifier could output a corresponding sleep stage label 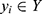 among five classes (W, N1, N2, N3, and R). The architecture of the classifier is based on TinySleepNet3 and adapted to the SNUBH dataset. As a “selective” classifier, in addition to general classification, it can selectively “accept” or “reject” the output, which is determined based on confidence in the present study. In other words, given the input xi, the selective classifier’s prediction
among five classes (W, N1, N2, N3, and R). The architecture of the classifier is based on TinySleepNet3 and adapted to the SNUBH dataset. As a “selective” classifier, in addition to general classification, it can selectively “accept” or “reject” the output, which is determined based on confidence in the present study. In other words, given the input xi, the selective classifier’s prediction  is accepted only when the confidence
is accepted only when the confidence 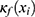 is equal to or larger than a threshold
is equal to or larger than a threshold 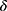 . Otherwise, the selective classifier rejects (ie, NONE) the prediction and asks for a manual review.
. Otherwise, the selective classifier rejects (ie, NONE) the prediction and asks for a manual review.
The confidence rate function  indicates how confident the classifier
indicates how confident the classifier 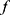 is about its prediction on a given data
is about its prediction on a given data 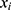 . A user-defined parameter
. A user-defined parameter 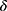 is a threshold to decide the level of confidence required to accept a prediction.
is a threshold to decide the level of confidence required to accept a prediction.
SeqConfidNet with DCR
The key component of our framework is defining the confidence rate function 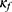 so that the confidence model can accurately estimate the accuracy of the classifier. A common way to estimate the confidence is to utilize softmax values from the last layer of the classifier. The last layer of the classifier outputs five values which are probabilities for a given epoch to be classified to each of five sleep stages. The classifier outputs the class yielding the highest probability as the final prediction, while the highest probability itself (ie, the maximum class probability) can be considered as the corresponding confidence. The Maximum Class Probability (MCP)21 can be easily obtained without an additional cost. However, it tends to be overconfident by nature. Since it is the maximum probability among the five classes, the confidence value remains high even when the prediction is incorrect.
so that the confidence model can accurately estimate the accuracy of the classifier. A common way to estimate the confidence is to utilize softmax values from the last layer of the classifier. The last layer of the classifier outputs five values which are probabilities for a given epoch to be classified to each of five sleep stages. The classifier outputs the class yielding the highest probability as the final prediction, while the highest probability itself (ie, the maximum class probability) can be considered as the corresponding confidence. The Maximum Class Probability (MCP)21 can be easily obtained without an additional cost. However, it tends to be overconfident by nature. Since it is the maximum probability among the five classes, the confidence value remains high even when the prediction is incorrect.
Because of the overconfidence of MCP, another method called True Class Probability (TCP) was introduced.15,16 TCP takes the probability of the true class as confidence (Figure 2). It is ideal in that confidence would be low for wrong predictions. However, since the information for a true class is not available in a real clinical setting where PSGs are pending for annotation, an additional neural network is required to be trained when estimating confidence that uses that information. For example, ConfidNet is a confidence model proposed to output confidence values using TCP as the confidence rate function.15 Although TCP is regarded as a better confidence estimation method than MCP, both could not reflect the actual accuracy of the classifier. Thus, using them as reference in deciding when to accept or reject predictions has limitations.
To overcome the limitation of these class probability-based methods, we propose a new confidence estimation method that uses the dropout technique to allow direct estimation of the accuracy of the sleep stage classifier. Dropout is one of the most popular DL techniques for regularization. When the dropout method is applied for a given classifier 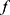 , some neurons are randomly ignored when making the prediction. As shown in Figure 2B, we can apply the dropout method to the classifier to generate multiple copies of the neural network which uses different sets of neurons for prediction. For a given sample, each of dropout-applied
, some neurons are randomly ignored when making the prediction. As shown in Figure 2B, we can apply the dropout method to the classifier to generate multiple copies of the neural network which uses different sets of neurons for prediction. For a given sample, each of dropout-applied 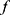 s can perform class prediction independently. The total number of correct predictions is then counted. Dropout Correct Rate (DCR) is calculated as the number of correct predictions divided by the number of dropout trials. For example, if 60 predictions are correct out of 100 dropout-applied
s can perform class prediction independently. The total number of correct predictions is then counted. Dropout Correct Rate (DCR) is calculated as the number of correct predictions divided by the number of dropout trials. For example, if 60 predictions are correct out of 100 dropout-applied 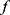 ’s predictions, the DCR is 0.6.
’s predictions, the DCR is 0.6.
In this work, we adapted the ConfidNet of Corbiere et al15,16 and designed a sequence-to-sequence confidence model (SeqConfidNet) to give a sequence of confidence values corresponding to the sequence of sleep stages predicted by the classifier. Since DCR approximates the accuracy of the prediction, the estimated confidence from SeqConfidNet with DCR is likely to reflect the actual accuracy of the class prediction. We would test not only SeqConfidNet with DCR, but also other existing confidence rate functions [MCP, temperature-scaled MCP (t-MCP), and TCP] to evaluate if SeqConfidNet with DCR could have better performance than other methods for estimating clinically meaningful confidence.
Model Architecture and Training
The model architectures of the classifier  and SeqConfidNet are both made up of four convolutional layers, one recurrent layer, and one fully connected layer. For the classifier
and SeqConfidNet are both made up of four convolutional layers, one recurrent layer, and one fully connected layer. For the classifier 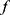 , the fully connected layer outputs five values, each showing the probability of the 30-s epoch belonging to one of five sleep stages. For SeqConfidNet, the fully connected layer only outputs one value per epoch which quantifies the confidence of classifier
, the fully connected layer outputs five values, each showing the probability of the 30-s epoch belonging to one of five sleep stages. For SeqConfidNet, the fully connected layer only outputs one value per epoch which quantifies the confidence of classifier 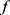 in its prediction. The detailed hyperparameters of the classifier such as filter stride, filter size, and the number of hidden layers were used with the same values as those in the original paper.3
in its prediction. The detailed hyperparameters of the classifier such as filter stride, filter size, and the number of hidden layers were used with the same values as those in the original paper.3
For model training, the classifier 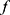 is first trained to minimize the mean squared error between the true class and the predicted class using an Adam optimizer, a learning rate of 0.0001, and a batch size of 64. Right after the classifier training, the training of the confidence model begins, with initial weights of convolutional layers set as trained weights of the classifier’s convolutional layers. To prevent the confidence model from diverging too much from the classifier, the training of SeqConfidNet freezes convolutional layers and starts with the training of recurrent and fully connected layers. After those layers are well trained, the next step is fine-tuning of SeqConfidNet where convolutional layers are trained together with recurrent and fully connected layers. Using an Adam optimizer and a batch size of 64, SeqConfidNet is trained to minimize the mean squared error between output values estimated by the confidence model and those given by the confidence rate function
is first trained to minimize the mean squared error between the true class and the predicted class using an Adam optimizer, a learning rate of 0.0001, and a batch size of 64. Right after the classifier training, the training of the confidence model begins, with initial weights of convolutional layers set as trained weights of the classifier’s convolutional layers. To prevent the confidence model from diverging too much from the classifier, the training of SeqConfidNet freezes convolutional layers and starts with the training of recurrent and fully connected layers. After those layers are well trained, the next step is fine-tuning of SeqConfidNet where convolutional layers are trained together with recurrent and fully connected layers. Using an Adam optimizer and a batch size of 64, SeqConfidNet is trained to minimize the mean squared error between output values estimated by the confidence model and those given by the confidence rate function 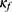 . During the training of SeqConfidNet, the learning rate was initially set at 0.0001 but reduced to 0.00003 during fine-tuning to stabilize the learning. To avoid overfitting of models, early stopping was done for training both the classifier and the SeqConfidNet.
. During the training of SeqConfidNet, the learning rate was initially set at 0.0001 but reduced to 0.00003 during fine-tuning to stabilize the learning. To avoid overfitting of models, early stopping was done for training both the classifier and the SeqConfidNet.
Evaluation Metrics
The classifier performance was evaluated by mean accuracy, Cohen’s kappa, weighted macro-F1 score, and confusion matrix. Regarding the performance of confidence estimation, five types of metrics were used as in previous works: (1) area under the receiver operating characteristic curve (AUROC),21 (2) area under the precision-recall curve (AUPR),21 (3) false positive rate (FPR) when the true positive rate (TPR) was 95% (FPR@95%TPR),15,16 (4) area under the risk-coverage curve (AURC),22,23 and (5) Excess-AURC (E-AURC).24 The AUROC is the most commonly used metric to evaluate a method’s ability of distinguishing between classes. To note, we defined a “positive class” as wrong predictions by the sleep stage classifier and the “negative class” as correct predictions because the wrong predictions were our targets to be detected here. Thus, successful rejection of wrong predictions would be a “true positive (TP)” and successful pass (non-rejection) for correct predictions would be a “true negative”. In addition to TP, false positive (FP), true negative (TN), and false negative (FN) were used to calculate precision (TP/(TP+FP)), recall=TPR (TP/(TP+FN)), FPR (FP/(FP+TN), risk (FN/(TN+FN)), and coverage (TN+FN/(TN+FN+TP+FP)), respectively. The AUPR is better in the case of two classes with greatly different base rates because it adjusts for base rates. AUROC and AUPR denote areas under the TPR-FPR curve (ROC) and the precision-recall curve while varying the confidence threshold 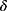 , with higher value indicating better performance. The FPR@95%TPR was the FPR at a certain threshold that gave a TPR of 95%, with a lower FPR at TPR 95% indicating a better performance. Regarding AURC, it is better to reduce the risk (missing wrong predictions) while keeping the coverage (1-rejection rate) as high as possible. E-AURC was a normalized variant of AURC by subtracting the inevitable risk. Lower values for AURC and E-AURC indicated better performances. The primary outcomes were AUROC and AUPR while FPR@95%TPR, AURC, and E-AURC were considered secondary outcomes to help evaluate the performance of confidence estimation in various aspects.
, with higher value indicating better performance. The FPR@95%TPR was the FPR at a certain threshold that gave a TPR of 95%, with a lower FPR at TPR 95% indicating a better performance. Regarding AURC, it is better to reduce the risk (missing wrong predictions) while keeping the coverage (1-rejection rate) as high as possible. E-AURC was a normalized variant of AURC by subtracting the inevitable risk. Lower values for AURC and E-AURC indicated better performances. The primary outcomes were AUROC and AUPR while FPR@95%TPR, AURC, and E-AURC were considered secondary outcomes to help evaluate the performance of confidence estimation in various aspects.
Results
Sleep Staging Performance
Classification Metrics
Figure 3 shows the confusion matrix for sleep stage classification based on local SNUBH and public SHHS sleep datasets. With the SNUBH dataset, the classifier was able to predict 85% of Wake, 59% of N1, 74% of N2, 77% of N3, and 83% of REM correctly, resulting in an average classification accuracy of 76%. The Cohen’s kappa value was 0.67 and the overall weighted F1 score was 0.76. The evaluation via the SHHS dataset showed similar results (accuracy: 82%, Cohen’s kappa: 0.75, F1 score: 0.82) (Figure 3B), confirming the efficiency and robustness of the classifier.
 |
Figure 3 Confusion matrices for sleep stage classification, comparing manual scoring vs automated scoring by the classifier using (A) the SNUBH dataset or (B) the SHHS dataset. |
Accuracy per Data Category
When categorizing the data based on specific features as displayed in Table 1, the classifier exhibited classification accuracy for certain groups. The accuracy seemed generally robust across age spans and genders except that it had a slightly lower value for the old group. However, the accuracy was reduced greatly for people with high BMI or AHI. The accuracy reduced to 68.6% for the obese group and 68.4% for people with AHI of 30 or more. When features were grouped on the level of epoch, the accuracy reduced to 64.8% for epochs with sleep apnea/hypopnea and 62.5% for epochs with respiratory arousal.
 |
Table 1 Classification Accuracy and Mean Estimated Confidence According to Clinical Features in the SNUBH Dataset |
Confidence Estimation Performance
Confidence Estimation Metrics
Results of the primary and secondary metrics for confidence estimation methods are shown in Table 2. Regarding the primary metrics, the DCR method showed the best AUROC (0.812) and the second best AUPR (0.533) following the TCP (0.538). The values of FPR@95%TPR (0.591), AURC (0.088), and E-AURC (0.057) were the best using the DCR method, followed by the values with TCP.
 |
Table 2 Performances of the Three Confidence Estimation Methods in Differentiating Wrong and Correct Predictions |
Rejection of the Least Confident Predictions
In Figure 4, the TPR is shown at each rejection rate applied. For each rejection rate, a certain percentage of predictions with the lowest confidence were rejected. A higher TPR could be interpreted as a higher rate of wrong predictions to be included in the rejection (ie, a higher detection rate for wrong predictions). By applying 20% as the rejection rate, a TPR as high as 50% was achieved using the confidence estimation by DCR. That is, almost half of the classifier’s wrong predictions could be detected, which was considered quite successful compared to the TPR of 20% at random rejection. Furthermore, if epochs with rejected predictions could be hypothesized to be manually re-scored, the classifier’s overall weighted F1 score and Cohen’s kappa value improved greatly by a maximum of 0.203 and 0.276, respectively. When the rejection rate was increased to 50%, the detection rate of wrong predictions was increased to as high as 85% and the overall accuracy of sleep staging was highly improved.
 |
Figure 4 Changes of metrics when applying different rejection rates. Rejected epochs are selected (A) randomly or (B) based on DCR. Abbreviations: TPR, true positive rate; DCR, Dropout Correct Rate. |
In addition, we compared the possible improvement of F1 score and Cohen’s kappa by re-scoring rejected epochs among the three confidence estimation methods (Figure 5). Rejecting predictions by using the DCR confidence resulted in the largest improvement of the classification accuracy among the three methods. When 40% of predictions with the lowest confidence values were replaced with original labels, DCR improved the classifier’s overall weighted F1 score by 0.015 and Cohen’s kappa value by 0.02 more than MCP did.
Confidence per Data Category
In most categories, mean estimated DCR confidence values were the closest to the classification accuracy among the confidence estimation methods (Table 1). In particular, MCP showed confidence values close to accuracy for the young, the underweight, and the normal-to-mild sleep apnea groups. However, for the older group and groups with high BMI or AHI, in which the accuracy was reduced, DCR performed the best, outputting the closest confidence values to accuracy. DCR seemed to reflect the classifier performance honestly and be less overconfident when the accuracy was reduced in specific groups.
When 20% of predictions with the lowest DCR confidence would be replaced by the correct labels, accuracy improved by as much as 10–20%. The most improvement of classification accuracy occurred for groups with low accuracies (eg, patients with old age, obesity, or severe sleep apnea) (Figure 6).
Threshold Determination
The confidence threshold 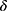 can be chosen arbitrarily by users. Here, we evaluated three systematic ways of setting the threshold. The first criterion was to set the confidence value at when TPR reached a desired value (eg, TPR = 0.95) as threshold for rejection. As shown in Table 3, this criterion resulted in a high threshold at
can be chosen arbitrarily by users. Here, we evaluated three systematic ways of setting the threshold. The first criterion was to set the confidence value at when TPR reached a desired value (eg, TPR = 0.95) as threshold for rejection. As shown in Table 3, this criterion resulted in a high threshold at 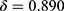 and rejections for a large portion of epochs (54.3% of correct prediction and 95.7% of wrong prediction) that required manual review. The second criterion picked the threshold as the value that kept the accuracy of remaining epochs at a desired value (eg, Accuracy = 0.85). In this case, less epochs were subjected to manual review, decreasing both FPR (21.7%) and TPR (52.3%). The third criterion was the gap maximization which set the threshold as the value that maximized the TPR while minimizing the FPR. The gap maximization criteria set the threshold at 0.724 where 29.6% of correct predictions and 82.5% of wrong predictions were rejected.
and rejections for a large portion of epochs (54.3% of correct prediction and 95.7% of wrong prediction) that required manual review. The second criterion picked the threshold as the value that kept the accuracy of remaining epochs at a desired value (eg, Accuracy = 0.85). In this case, less epochs were subjected to manual review, decreasing both FPR (21.7%) and TPR (52.3%). The third criterion was the gap maximization which set the threshold as the value that maximized the TPR while minimizing the FPR. The gap maximization criteria set the threshold at 0.724 where 29.6% of correct predictions and 82.5% of wrong predictions were rejected.
 |
Table 3 Percentages of Epochs with Correct and Wrong Predictions Chosen for Manual Review According to Each Threshold Setting Criterion |
Utility of Confidence in Sleep Staging
Examples for the utility of confidence are presented in Figure 7. The accuracy was high for the PSG of a patient with mild sleep apnea (AHI = 7.4), so was the confidence estimated by SeqConfidNet with DCR. When the threshold was used to achieve accuracy of 85% for the remaining predictions (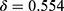 , the second selection criteria we proposed), the rejection rate was 10.4%. However, for a patient with severe sleep apnea and frequent sleep stage shifting, the accuracy of the classifier reduced to 69%. The estimated confidence also reduced greatly, resulting in 44.1% rejection rate when the same threshold was applied. Despite various accuracies for these two cases, epochs with wrong predictions were largely overlapped with a low confidence. After these rejected predictions were revised with correct sleep stages, the overall accuracy could be improved to 94–95% in both cases.
, the second selection criteria we proposed), the rejection rate was 10.4%. However, for a patient with severe sleep apnea and frequent sleep stage shifting, the accuracy of the classifier reduced to 69%. The estimated confidence also reduced greatly, resulting in 44.1% rejection rate when the same threshold was applied. Despite various accuracies for these two cases, epochs with wrong predictions were largely overlapped with a low confidence. After these rejected predictions were revised with correct sleep stages, the overall accuracy could be improved to 94–95% in both cases.
Discussion
To the best of our knowledge, this is the first study to introduce confidence estimation and selective classifier into DL-based automated sleep stage scoring, where the objective was to detect wrong predictions for sleep stages. With confidence estimation and a threshold for it, selective classifier could reject probably wrong predictions (confidence below the threshold) and only output probably correct predictions (confidence equal to or above the threshold). In addition to previously proposed confidence estimation methods (the MCP and the TCP), we proposed a novel method, DCR. We evaluated the performance of all three confidence estimation methods by their capabilities of detecting wrong predictions. DCR performed the best to differentiate correct and wrong predictions based on not only the primary metrics of AUROC but also the secondary metrics of FPT@95TPR, AURC and E-AURC. DCR also showed the greatest improvement of overall accuracy when a fixed percentage of predictions with the lowest confidence were rejected. Lastly, for groups with low accuracies, DCR confidence values were reduced concordantly with the level of accuracy, showing that the DCR method better reflected accuracy and that it was less overconfident.
In designing our classifier for automated sleep stage scoring, we adopted the TinySleepNet architecture which is light, simple, and highly robust. As expected, the performance of the classifier was robust with the local dataset (accuracy 76%) and the public dataset (accuracy 82%), which was compatible with other classifiers (accuracy 75–85%).10,11 The lower accuracy with the SNUBH dataset compared to the SHHS dataset might be explained by the fact that majority of its data were from clinical patients with various sleep disorders including sleep apnea. The classifier performed fairly well for the data without sleep respiratory events (accuracy 80%). However, the accuracy was reduced particularly for epochs with sleep respiratory events and for people with an old age, obesity, and severe sleep apnea, which was observed in other classifiers as well.6,25,26
The confidence model, the key component in our framework, provides confidence estimates for predictions of the classifier on a level of epoch. Confidence estimates can quantify how confident the model is toward its predictions. We expect a well-designed confidence function 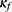 to output reliable confidence estimates, where high values (more confident) indicate trustworthy predictions and low values (less confident) refer to challenging data that require a manual review. We used SeqConfidNet with DCR as our confidence model, which was trained to output estimates of DCR. Conceptually, DCR is like virtual accuracy per prediction because it is the proportion of correctness in a number of dropout simulations for each prediction. It is worth noting that the DCR method, which outputs confidence estimates very much similar to accuracy by definition, actually shows the closest mean confidence value to accuracy (Table 1). As we intended, the mean DCR confidence value followed and reflected the accuracy well in all categories regardless of the classifier’s accuracy, while the probability-based estimation (the MCP, t-MCP, and TCP) tended to be overconfident for specific groups where accuracy reduced greatly (old, high BMI, or severe apnea groups).
to output reliable confidence estimates, where high values (more confident) indicate trustworthy predictions and low values (less confident) refer to challenging data that require a manual review. We used SeqConfidNet with DCR as our confidence model, which was trained to output estimates of DCR. Conceptually, DCR is like virtual accuracy per prediction because it is the proportion of correctness in a number of dropout simulations for each prediction. It is worth noting that the DCR method, which outputs confidence estimates very much similar to accuracy by definition, actually shows the closest mean confidence value to accuracy (Table 1). As we intended, the mean DCR confidence value followed and reflected the accuracy well in all categories regardless of the classifier’s accuracy, while the probability-based estimation (the MCP, t-MCP, and TCP) tended to be overconfident for specific groups where accuracy reduced greatly (old, high BMI, or severe apnea groups).
Regarding the ability of detecting wrong predictions, DCR showed AUROC of 0.812, AUPR of 0.930, and AURC of 0.088, which were considered as good discrimination. Metric scores of MCP or TCP were not as good as those of DCR. Detecting and re-scoring wrong predictions were intended to improve the overall accuracy. We further evaluated changes of accuracy by replacing rejected predictions with correct sleep stages. When rejection was determined by the DCR, greater improvements for both Cohen's kappa and weighted F1 score were shown compared to the rejection by the MCP or by the TCP. It might be explained by the high TPR in DCR-based rejection. That is, the DCR method can successfully detect and include more wrong predictions into rejection, leading to more potential for improvement when rejected epochs are manually re-scored.
By using DCR confidence estimates to make corrections for those predictions, the capability of DL-based automated sleep stage scoring to be used in clinical practice is maximized. PSGs can be annotated by DL-based models first and only a small portion of PSGs with possibly wrong predictions would require manual review and re-annotation. Thereby, the sleep stage scoring assisted by DL-based models can become both accurate and reliable. In addition, time complexity of DL-based models is a critical factor for practical utility, where our AI model only takes 2 seconds per PSG recording on average to output sleep stage scoring results along with confidence estimates. Therefore, for sleep physicians, the time and labor can be saved while the accuracy is kept high. Such confidence estimates can also make predictions of sleep stages transparent and manipulable.
Although our proposed framework can make DL-based sleep stage scoring models reliable, it has several limitations. First, calculating DCR confidence estimates has an additional computational overhead. Second, while the use of our confidence model is not limited for a specific classifier, the performance of confidence estimation can be highly affected by the performance of the classifier because they share the model architecture. In addition, there were limitations of the study that our experiments were conducted with only two specific datasets without external validation. Finally, the framework proposed in this study was not applied and evaluated in an actual clinical setting.
Conclusion
This study showed the potential of a confidence-based framework to improve accuracy of automated sleep stage scoring. We expect that our work may help promote the active use of DL-based automated sleep stage scoring in clinical practice and result in a reduction of workload of sleep physicians. The practical utility of the framework is needed to be validated in future works. Future study of comparing accuracy and time efficiency between full manual scoring and confidence-based framework-assisted manual scoring is needed to prove the benefit of applying our proposed model in a practical setting. Another important future research direction is to improve the generalization capability of AI models for sleep stage scoring so that AI models can perform well universally, regardless of sleep centers or recording devices.
Disclosure
The authors report no conflicts of interest in this work.
References
1. for the American Academy of Sleep Medicine; Iber C, Ancoli-Israel S, Chesson A, Quan SF. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Westchester, IL: American Academy of Sleep Medicine; 2007.
2. Supratak A, Dong H, Wu C, Guo Y. DeepSleepNet: a model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans Neural Syst Rehabil Eng. 2017;25(11):1998–2008. doi:10.1109/TNSRE.2017.2721116
3. Supratak A, Guo Y. TinySleepNet: an efficient deep learning model for sleep stage scoring based on raw single-channel EEG.
4. Phan H, Andreotti F, Cooray N, Chén OY, De Vos M. SeqSleepNet: end-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans Neural Syst Rehabil Eng. 2019;27(3):400–410. doi:10.1109/TNSRE.2019.2896659
5. Seo H, Back S, Lee S, Park D, Kim T, Lee K. Intra-and inter-epoch temporal context network (IITNet) using sub-epoch features for automatic sleep scoring on raw single-channel EEG. Biomed Signal Process Control. 2020;61:102037. doi:10.1016/j.bspc.2020.102037
6. Sun H, Ganglberger W, Panneerselvam E, et al. Sleep staging from electrocardiography and respiration with deep learning. Sleep. 2020;43(7). doi:10.1093/sleep/zsz306
7. Jia Z, Cai X, Zheng G, Wang J, Lin Y. SleepPrintNet: a multivariate multimodal neural network based on physiological time-series for automatic sleep staging. IEEE Trans Artif Intell. 2021;1:248–257.
8. Kim HJ, Lee M, Lee SW. End-to-end automatic sleep stage classification using spectral-temporal sleep features.
9. Phan H, Chén OY, Koch P, Mertins A, De Vos M. Fusion of end-to-end deep learning models for sequence-to-sequence sleep staging.
10. Biswal S, Sun H, Goparaju B, Westover MB, Sun J, Bianchi MT. Expert-level sleep scoring with deep neural networks. J Am Med Inform Assoc. 2018;25(12):1643–1650.
11. Fiorillo L, Puiatti A, Papandrea M, et al. Automated sleep scoring: a review of the latest approaches. Sleep Med Rev. 2019;48:101204. doi:10.1016/j.smrv.2019.07.007
12. Danker-Hopfe H, Anderer P, Zeitlhofer J, et al. Interrater reliability for sleep scoring according to the Rechtschaffen & Kales and the new AASM standard. J Sleep Res. 2009;18(1):74–84. doi:10.1111/j.1365-2869.2008.00700.x
13. Goldstein CA, Berry RB, Kent DT, et al. Artificial intelligence in sleep medicine: background and implications for clinicians. J Clin Sleep Med. 2020;16(4):609–618. doi:10.5664/jcsm.8388
14. Guo C, Pleiss G, Sun Y, Weinberger KQ On calibration of modern neural networks. International Conference on Machine Learning; 2017;1321–1330. PMLR
15. Corbière C, Thome N, Bar-Hen A, Cord M, Pèrez P. Addressing failure prediction by learning model confidence. arXiv preprint arXiv:1910.04851; 2019.
16. Corbière C, Thome N, Saporta A, Vu T-H, Cord M, Pèrez P. Confidence estimation via auxiliary models. IEEE Trans Pattern Anal Mach Intell. 2020;2020:54.
17. Quan SF, Howard BV, Iber C, et al. The sleep heart health study: design, rationale, and methods. Sleep. 1997;20(12):1077–1085.
18. Zhang G-Q, Cui L, Mueller R, et al. The national sleep research resource: towards a sleep data commons. J Am Med Inform Assoc. 2018;25(10):1351–1358. doi:10.1093/jamia/ocy064
19. Li Q, Li Q, Liu C, Shashikumar SP, Nemati S, Clifford GD. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol Meas. 2018;39(12):124005. doi:10.1088/1361-6579/aaf339
20. Yan R, Li F, Zhou DD, Ristaniemi T, Cong F. Automatic sleep scoring: a deep learning architecture for multi-modality time series. J Neurosci Methods. 2021;348:108971. doi:10.1016/j.jneumeth.2020.108971
21. Hendrycks D, Gimpel K. A baseline for detecting misclassified and out-of-distribution examples in neural networks. ICLR; 2017.
22. El-Yaniv R, Wiener Y. On the foundations of noise-free selective classification. J Mach Learn Res. 2010a;11(May):1605–1641.
23. Jiang H, Kim B, Guan MY, Gupta MR. To trust or not to trust a classifier. NeurIPS; 2018.
24. Geifman Y, Uziel G, El-Yaniv R. Bias-reduced uncertainty estimation for deep neural classifiers.
25. Korkalainen H, Aakko J, Nikkonen S, et al. Accurate deep learning-based sleep staging in a clinical population with suspected obstructive sleep apnea. IEEE J Biomed Health Info. 2019;24(7):2073–2081. doi:10.1109/JBHI.2019.2951346
26. Zhang X, Xu M, Li Y, et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020;24:581–590. doi:10.1007/s11325-019-02008-w
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.