Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 12
Concepts Driving Pharmacogenomics Implementation Into Everyday Healthcare
Authors Giri J , Moyer AM , Bielinski SJ , Caraballo PJ
Received 5 July 2019
Accepted for publication 7 October 2019
Published 30 October 2019 Volume 2019:12 Pages 305—318
DOI https://doi.org/10.2147/PGPM.S193185
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Jyothsna Giri,1 Ann M Moyer,2 Suzette J Bielinski,3 Pedro J Caraballo4,5
1Center for Individualized Medicine, Mayo Clinic, Rochester, MN, USA; 2Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA; 3Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA; 4Department of Internal Medicine, Mayo Clinic, Rochester, MN, USA; 5Center for the Science of Health Care Delivery, Mayo Clinic, Rochester, MN, USA
Correspondence: Pedro J Caraballo
Center for the Science of Health Care Delivery, Mayo Clinic, 200 First Street SW, Rochester, MN 55904, USA
Tel +1 507 284 8586
Fax +1 507 284 5370
Email [email protected]
Abstract: Pharmacogenomics (PGx) is often promoted as the domain of precision medicine with the greatest potential to readily impact everyday healthcare. Rapid advances in PGx knowledge derived from extensive basic and clinical research along with decreasing costs of laboratory testing have led to an increased interest in PGx and expectations of imminent clinical translation with substantial clinical impact. However, the implementation of PGx into clinical workflows is neither simple nor straightforward, and comprehensive processes and multidisciplinary collaboration are required. Several national and international institutions have pioneered models for implementing clinical PGx, and these initial models have led to a better understanding of unresolved challenges. In this review, we have categorized and explored the most relevant of these challenges to highlight potential gaps and present possible solutions. We describe the ongoing need for basic and clinical research to drive further developments in evidence-based medicine. Integration into daily clinical workflows introduces new challenges requiring innovative solutions; specifically those related to the electronic health record and embedded clinical decision support. We describe advances in PGx testing and result reporting and describe the critical need for increased standardization in these areas across laboratories. We also explore the complexity of the PGx knowledge required for clinical practice and the need for educational strategies to ensure adequate understanding among members of current and future healthcare teams. Finally, we evaluate knowledge obtained from previous implementation efforts and discuss how to best apply these learnings to future projects. Despite these challenges, the future of precision medicine appears promising due to the rapidity of recent advances in the field and current multidisciplinary efforts to effectively translate PGx to everyday clinical practice.
Keywords: precision medicine, pharmacogenomics, clinical implementation, clinical decision support, delivery of health care, medication therapy management
Introduction
Pharmacogenomics (PGx), the highlight of the National Institutes of Health’s Precision Medicine Initiative (PMI), has the potential to immediately impact the care of patients in a clinically meaningful fashion. The PMI aims to understand how patient-specific factors, including genetics, can help clinicians determine the best approach to prevent or treat disease in an individual patient. With over 90% of individuals having actionable genetic findings that could be used by clinicians to inform on the choice of medication prescribed, this area of genomic medicine could have a significant positive impact on healthcare, potentially affecting every patient.1–3
Over the past few decades, significant research has led to the identification of a large number of PGx variants with demonstrated clinical utility. These variants have been incorporated into US Food and Drug Administration (FDA) labeling for medications and into consensus guidelines to facilitate their clinical use.4–7 Ideally, selecting therapies based in part on the genetic profile of individual patients could make it possible to alleviate or avoid adverse drug reactions, maximize drug efficacy, improve the overall patient experience, and reduce health care costs. However, the incorporation of PGx into clinical practice has been slow, and challenges have been identified by several programs and institutions that have explored different options for implementing PGx into clinical workflows.5,6,8 However, these efforts have also revealed some useful strategies to improve future implementation projects.
The aim of this review is to describe the most relevant concepts driving PGx implementation (Figure 1), grouping them into discrete categories to better understand potential challenges and opportunities (Table 1).
 |
Table 1 Pharmacogenomics Implementation Challenges And Opportunities |
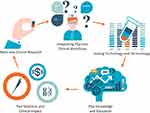 |
Figure 1 Concepts driving pharmacogenomics implementation into everyday healthcare. Abbreviation: PGx, Pharmacogenomics. |
Pharmacogenomics Research
Basic Research
Identification and functional characterization of the genetic variation that influences medication efficacy or predisposes to adverse reactions is an active area of research. These findings are the foundation upon which clinical studies that generate evidence supporting the use of PGx in clinical care are built.4
Historically, most studies have utilized a candidate gene approach, where variants in genes encoding drug metabolizing enzymes, transporters, and drug targets were evaluated for relationships with drug response.9 Later, tools for performing genome-wide association studies became more accessible, allowing researchers to explore variants across the genome and to discover variants in genes not previously known to be related to the safety or efficacy of a medication. After identification of gene(s) of therapeutic interest via either candidate gene studies or genome-wide approaches, research to date has been focused primarily on individual variants within the coding regions of these genes and, in particular, amino acid changes within these regions. However, there are known examples of splicing variants and promoter variants that alter transcription and are associated with medication response as well.10–12 Future studies exploring non-coding regions in greater depth may lead to identification of additional sources of genetic variation and strengthen associations between specific genes and medication response.
Augmented human intelligence (AHI) is emerging as a strategy that may identify additional genes and pathways that influence pharmacotherapy. Many medications are metabolized through complex pathways, yet studies have traditionally been limited by current knowledge of genes encoding components of these pathways and have been difficult to perform due to the large sample sizes required and inability to account for the potentially additive nature of small changes across many components within a complex pathway. While the use of genome-wide association study (GWAS) has allowed for an unbiased approach to the identification of additional novel variants/genes that could be incorporated into studies, this technique does not allow for identification of smaller signals with an additive effect. AHI approaches coupled with large datasets may allow for the evaluation of multiple variables simultaneously, including both genetic and non-genetic, to identify novel genes/variants and patterns where multiple variables may act in an additive manner. As such, AHI may significantly drive the field of pharmacogenomics forward.
As genes and variants are identified, experimental evidence is necessary to determine the functional consequence and relevance of each variant in order to translate that information to the clinic. However, functional genomic studies are labor-intensive making it impractical to study every identified variant with traditional techniques.9,13–16 Computational methods to predict the effect of changes in amino acids have been developed, but are not yet sufficiently reliable for clinical use.17 High-throughput methodologies are being developed and will be necessary to generate the data required for clinical translation.18–21
Clinical Research
After the initial identification of a potentially important variant of a gene, subsequent clinical studies including large sample sizes and patients of diverse ethnic backgrounds are necessary to replicate the findings. Due to the expense of sequencing, historically a targeted genotyping approach has been used. Many historic studies focused on relatively small, Caucasian (European-descent) populations with limited inclusion of ethnic minorities. Therefore, common variants that may have a significant impact on medication response or toxicity in other populations were not included in the design of subsequent studies. This may lead to conflicting results in studies evaluating the relationship between genetic variation and drug response, particularly when studies include subjects from different ethnic backgrounds.22 Even among well-studied populations, rare variants have not been the focus of many studies. However, with decreasing costs of sequencing allowing for studies with larger numbers of participants common variants in non-Caucasian populations and rare variants in all populations are now being identified.23–26
Even with robust research findings, clinical translation of basic research findings remains a challenge. For example, despite the growing body of research characterizing PGx variants with clinical value (Table 2) and leading to curation of over 650 medications in PharmGKB, there are currently only approximately 132 medications that have annotated clinical guidelines.4,27 These clinical annotations are based on works published by professional societies such as the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Royal Dutch Association for the Advancement of Pharmacy - Pharmacogenetics Working Group (DPWG), the Canadian Pharmacogenomics Network for Drug Safety (CPNDS). Establishing the clinical relevance of PGx variants is challenging due to the multitude of other confounding factors that can impact medication response including age, sex, lifestyle, diet, drug-drug interactions, the microbiome, and epigenetic changes influencing gene expression. Incorporation of these other factors into study design, or at a minimum reporting information on subgroups for use in subsequent meta-analyses, may clarify findings and facilitate translation into clinical use.
 |
Table 2 Commonly Tested Pharmacogenes And Their Association With Published Guidelines For Clinical Implementation |
While randomized trials are typically considered the gold standard for confirming the clinical relevance of a variant, it is not feasible to perform a randomized trial for each variant that may impact medication response, especially considering that, as of this writing, PharmGKB included 21,693 variant annotations. More recently, pragmatic and observational studies have been conducted to assess the impact of individual PGx variants on clinical outcomes.28 Pragmatic studies reflect the utility of testing for a variant in a real-world setting and may be more generalizable than randomized controlled studies. Large scale studies involving many patients have historically been rare but are becoming more widely available.3,29
Ultimately, PGx is predicted to evolve from purely interrogating genetic variants to guide therapeutic decisions to a “multi-omic” approach where other information, including metabolomics and proteomics, will also be incorporated into clinical decision making.9 Recent advancements in computational techniques including artificial intelligence may facilitate the integration of these large datasets. This may improve our understanding of the mechanism of action of medications and how specific variants and/or genes lead to differences in efficacy or toxicity, as well as potentially allow for novel relationships between medications and pathways to be discovered.
Research Opportunities And Recommendations
Ongoing research is essential to continue to advance the field of PGx and to generate the necessary data to allow for its widespread clinical implementation. Increased funding is needed to allow for studies that include a wider breadth of variants and medications. While targeted genotyping approaches are generally less expensive, using sequencing-based approaches will identify both common and rare variants that can influence PGx phenotypes. Furthermore, inclusion of diverse populations will lead to identification of important variants in non-Caucasian populations. These variants can then be studied further to uncover and explain differences in medication response among populations. Advanced computational platforms and artificial intelligence tools are emerging and they will be crucial to advance PGx research. These tools may support more sophisticated in silico assessment of variants, perhaps obviating the need for costly and time-consuming functional genomic studies of each variant. In addition they may allow for combinations of variants in several genes in the same pathway to be studied together.30,31 Although these tools currently may have limitations, they show promise.
Integrating Pharmacogenomics Into Clinical Workflows
All clinical workflows are now highly integrated with the electronic health record (EHR). This shift has increased the availability of tools that enable better delivery of data and knowledge to clinicians during clinical encounters. However, PGx test results and the knowledge needed to interpret such data for direct patient care are complex and still poorly standardized requiring the use of clinical decision support (CDS) tools integrated in the EHR and clinical workflows.32 Yet, EHR vendors have not developed the necessary applications that can achieve this integration seamlessly. Significant challenges have been identified including the format of the PGx test results, lack of standards, interfaces with the laboratories, storage of the PGx data in EHR and integration with CDS.33
In the United States, many laboratories have adopted a reporting style that includes medication recommendations in addition to genotypes and phenotypes. However, the format provided by many laboratories often does not include discrete fields and therefore does not facilitate direct migration into the EHR. PGx test results are often only provided in PDF format and must then be scanned into the EHR. These formatting issues, combined with the lack of a unique repository within the EHR for storing these data, present a barrier to their clinical use, as the awareness of available PGx test results may be limited solely to the provider who ordered the test, and therefore not drawn upon for future patient encounters with other clinicians. Similarly, in a PDF format, medication recommendations are not updated as new knowledge emerges and the test results are not compatible with automated CDS functionality, which is necessary for providing point-of-care guidance regarding clinically actionable test results. Similarly, the lack of a standardized process for reporting test results from different laboratories makes their translation, interpretation and integration into workflows difficult. The optimal process for receiving PGx data on individual patients involves an interface that directly delivers structured PGx results into the EHR for reliable, automated entry into discrete fields. This can be costly to develop and requires significant information technology (IT) resources. These structured data are essential for the implementation of CDS which is an essential component for a successful implementation.34 To circumvent the use of PDF reports and to allow for CDS functionality, some clinics have taken to manual entry of test results. However, this is not ideal due to the time and resources required and the obvious potential for data entry errors. Further, clinicians require training and corresponding educational resources on any system that is implemented, and the institution must secure resources to support the ongoing maintenance and upkeep of PGx data and related processes within the EHR either by an internal team or an external vendor. Lastly, while patients are enthusiastic about clinicians using their PGx test results to dictate clinical care, as of now, test results are not readily transferable between institutions within the US due to limitations in EHR interoperability.
A somewhat unique approach to delivering PGx information that may overcome some of the challenges of EHR interoperability leverages a paper-based, scannable quick response (QR) code.35 The Ubiquitous Pharmacogenomics (UPGx) Consortium is promoting and evaluating the “Safety-Code card”, with a QR code to access patient’s PGx test results online. This unique approach is geared towards health care systems without access to results and tools in the EHR and represents a step toward to portability of PGx results.36 The potential downside to this approach is that the patient must remember to share their “Safety-Code card” with their provider who in turn must know how to use it.
To work toward overcoming the aforementioned challenges related to data migration and integration, a new era of PGx implementation initiatives is creating a broader understanding of techniques that can be used to incorporate genomic medicine into clinical workflows and is paving the way for more aligned generation and sharing of knowledge.37 The Electronic Medical Records and Genomics (eMERGE) Network,38 Vanderbilt Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) program,39 Mayo Clinic RIGHT study40,41 in the US, and the U-PGx Preemptive Pharmacogenomic Testing for Preventing Adverse Drug Reactions (PREPARE) study in Europe are examples of academic programs conducting preemptive sequence-based PGx testing on a research basis. These programs are also evaluating clinical return of results to patients and their EHR using scannable QR code and/or CDS tools for both preemptive and reactive PGx testing to promote the use of PGx information at the point-of-care.33 The results of these and other ongoing efforts continue to build on each other, leading to better and more optimal methods for implementing PGx.
Clinical Workflow Opportunities And Recommendations
To effectively integrate PGx into clinical workflows requires institutional-level support and investment of resources. If a CDS approach is selected, as is often the case in the US, identifying appropriate IT resources and creating teams in-house who can build-out and support customized CDS capabilities for PGx-based alerts to meet the needs of the local practice is ideal. However, due to the resources required to build these systems, purchasing ready-made CDS tools may be a better option for some institutions. Careful research into the functionality and compatibility of available products is recommended to ensure integration with existing systems and to avoid the need to change products in the future. Another opportunity that gained popularity due to the work of U-PGx is the Safety-Code card that resolves the issue of portability of results across institutions as well circumvents challenges with integration into the electronic health record. The utilization of a QR code with PGx test information that can be accessed using a mobile phone is a strategy that perhaps the hospitals in the US can implement based on the demonstrated success by UPGx.36
Research and clinical implementation efforts that include placing PGx test results in the EHR are necessary to evaluate health outcomes and potential cost-savings associated with PGx-driven prescribing.42 The evidence from these evaluations is needed to encourage reimbursement from private health insurance and inclusion of PGx testing in government health programs. Selecting a laboratory partner with the capability to automatically populate results in discrete fields within the EHR and that effectively communicates in advance any changes to testing that may require updates to CDS systems is highly recommended. Working with a single laboratory, or just a small number of laboratories if necessary, is also recommended due to the current variability in available tests and result formats. Efforts to increase standardization are currently underway within the PGx clinical laboratory community and, if successful, represent an opportunity to further simplify and increase PGx adoption.
Pharmacogenomics Testing And Terminology
Testing Technology
Advances in genetic testing technology have allowed for clinical PGx laboratories to transition from performing expensive tests focused on a single gene to more affordable multiplexed panel tests that include many genes. This is an important advancement in that it allows for the possibility of preemptive testing, particularly in cases where the patient does not yet need medication but would like to have PGx data available for consultation during future clinic visits.
While the majority of clinically available tests today utilize a targeted genotyping-based approach, where only pre-defined genetic variants are included and reported, as the cost of sequencing continues to decline, clinical testing will likely transition to sequencing-based approaches. This has several implications. First, this implies that future improvements in testing technology are expected; indicating that patients who desire to undergo preemptive testing with today’s targeted genotyping-based tests should not expect to be able to use their test results indefinitely and may require additional testing in the future. Second, it is important for providers to recognize that only the variants included in the test design can be detected and reported. At present, most laboratories include the most common variants identified in the Caucasian population in a given gene, but there is significant variability in whether and which additional variants are included.43,44 Conventionally, when no genetic variants are detected, a result of *1/*1 and “normal metabolizer” is reported. Therefore, if a test does not include a variant that is common in a particular population, more false negative (*1/*1) results will be reported, whereas a test that is designed to include that common variant will detect and report it.
While techniques have been developed for the targeted genotyping of complex loci, such as CYP2D6 and the HLA region, new tools will be required to handle highly homologous and highly polymorphic regions as laboratories transition to sequencing. Currently, most clinical grade high-throughput sequencing utilizes short-read next-generation sequencing (NGS) technology. In this case, algorithms and/or special techniques may be a useful solution to interrogate these complex loci.45–47 Alternatively, several platforms are available that can produce long reads of several kilobases, which have been successfully used for these loci.Historically, PGx testing has been performed by individual laboratories with little concern for standardization, in part because testing was initially limited to a small number of laboratories with expertise in this area and in part because testing was not widespread, and data portability issues had not been raised. Now that PGx is becoming more mainstream, there is a desire to standardize testing to allow for consistency in implementation and triggering of CDS alerts across clinics as well as consistency in the generation and utilization of guidelines for clinical care, regardless of where the genetic testing was performed. These standards will also help improve data portability and the transfer of patient data from one clinic to the next to allow for a more consistent and seamless patient care experience. Underscoring the need for standardization, Bousman and Dunlop performed a study examining the degree of concordance in test result interpretation among four commercial PGx panels for psychiatry in a cohort of five patients.48 The tests evaluated each provided medication recommendations in addition to genotype and phenotype results. The level of agreement among these four panels with respect to medication recommendations ranged from 55% for antipsychotics to 84% for mood stabilizers, leading the investigators to conclude that these tests cannot be considered equivalent or interchangeable at this time.
Ongoing Standardization Efforts
Several organizations and institutions have already made significant progress in standardization, developing the tools and recommendations necessary to ensure high quality, standardized testing. The Pharmacogenomics Working Group of the Association for Molecular Pathology (AMP) Clinical Practice Committee along with the College of American Pathologists (CAP) recently published recommendations for clinical cytochrome P450 (CYP) 2C19 and CYP2C9 genotyping allele selection in an effort to standardize the tests performed.28,49 In these publications, two “tiers” of alleles were recommended. A minimum panel of alleles that should be included in tests (tier 1) was selected because: 1) they were present at an appreciable minor allele frequency in a population, 2) they had a known functional impact on enzyme function and, 3) there were available reference materials related to the allele. An expanded set of alleles (tier 2) included those that did not meet all three of the above criteria. This work should aid both the clinical laboratories designing tests, as well as assist providers in selecting the most appropriate tests for their patients. Guidance for other genes is also needed and being created.
In addition to the AMP/CAP standardization efforts previously mentioned, the Pharmacogene Variation Consortium (PharmVar)50 has recently been organized by PGx experts and clinicians to continue the cataloguing of allelic variation of PGx genes and haplotype structure, an undertaking historically managed by the Karolinska Institute.51 This includes curating the star allele nomenclature for CYP genes and other PGx genes, which is now used almost exclusively to relay patient genotypes on PGx test reports. Furthermore, the Genetic Testing Reference Materials Coordination Program (GeT-RM), a Center for Disease Control (CDC) project, has been tasked with establishing a community process for creating reference materials, quality control measures, and proficiency testing that can be used by PGx laboratories developing tests and as control data.28,52,53 Laboratories are also now able to subscribe to proficiency testing programs through the College of American Pathologists and other vendors to ensure accuracy in test reporting, which may subsequently result in enhanced consistency across testing platforms.54
Challenges In Standardization
While many of these standardization efforts currently underway will serve to harmonize and align clinical PGx testing and outputs to some degree, challenges still exist that may prevent PGx testing from ever becoming fully standardized to the same degree as other analytical tests. First, when laboratories transition from targeted genotyping to sequencing, many rare or private variants will be identified adding additional challenges (Figure 2). A previous study estimated that 30-40% of functional variability in pharmacogenes may be due to rare variants.23 The current star allele nomenclature system would require a new specific name to identify that allele. This would be possible in the research setting, where reporting is not as time-sensitive, but is not ideal in a clinical setting, where reporting a patient result cannot wait for assignment of a new allele name. While the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) criteria for variant classification are widely used in clinical hereditary genetics practice, they are not designed for use with PGx.55 While in silico prediction tools have improved, they are still not considered sufficient for clinical classification of variants on their own; therefore, in the absence of functional studies for rare variants, many may be classified as variants of uncertain significance and lead to ambiguity in phenotype prediction. In addition, professional judgement is required for variant interpretation, so despite all attempts at standardization, differences in clinical opinion may still exist. Lastly, as PGx research continues, there will also be new genes identified that impact medication response. Developing a new test for clinical use takes time, and differences exist among laboratories with regard to the amount of time required to implement a new test following initial gene discovery. Additionally, the same challenges in standardization of nomenclature and interpretation will likely be present every time a new gene is implemented.
Laboratory Testing Opportunities And Recommendations
Clinical PGx testing technologies will continue to advance, and the PGx community will need to continue to work toward aligning testing practices and nomenclature across laboratories to ensure high quality testing and consistent interpretation of results. To this end, efforts to better standardize testing of well-established PGx genes would likely have the greatest yield and could positively influence how future genes and variants are incorporated into testing panels. Due to their wide reach and accessibility, repositories and programs like PharmVar and GeT-RM should be leveraged to further align the field.
At an institutional level, close collaboration and frequent communication between members of an institution’s multidisciplinary PGx team will be required to maintain consistency across testing and reporting procedures. The active engagement of a laboratory director experienced in collaborating with multidisciplinary PGx teams is also highly beneficial. While support of an internal laboratory directory may not always be available for smaller programs that do not perform in-house testing, these sites can still collaborate with a reputable PGx testing laboratory that has the expertise needed to assist clients in PGx implementation. Lastly, where possible, clinics may benefit from investing in novel platforms and support tools that can interact directly with the EHR to deliver and interpret PGx results. This would further enhance consistency within individual clinics and would help remove some of the nuance in testing and reporting that arises due to human intervention in PGx workflows.
Test Selection And Clinical Impact
Test Utilization And Selection
One ongoing challenge related to PGx testing in the clinic stems from the lack of universal acceptance of its utility on the part of clinicians and third-party payers, despite considerable evidence in the form of clinical studies and published PGx guidelines. In addition, inclusion of pharmacogenomic information in the Summary of Product Characteristics (SmPC) or package inserts is often not consistent among countries, may lag behind the literature and not align with published guidelines, and the recommendations included in published guidelines may not be harmonized among different organizations. For example, despite FDA-approved labeling warning that CYP2C19 poor metabolizers have a diminished effect of clopidogrel when used during percutaneous coronary interventions in the setting of acute coronary syndrome, the use of PGx testing for CYP2C19 in clinical practice in the United States is still limited. The American Heart Association guidelines do not recommend routine CYP2C19 testing prior to initiating therapy with clopidogrel. The lack of sufficient prospective trials demonstrating patient benefit related to a reduction in undesirable outcomes has been quoted as a barrier to recommend routine testing.56 In contrast to the American Heart Association, theDPWG and CPIC agree that the use of clopidogrel is contraindicated for CYP2C19 poor metabolizers. However, the DPWG includes dipyridamole as an additional alternative to prasugrel and ticagrelor in the case of a percutaneous coronary intervention when there are no other contraindications, whereas CPIC does not classify recommendations by specific indication and lists only ticagrelor and prasugrel as alternatives.4,57
With the growing number of PGx tests on the market in the US, once a clinician has decided a patient would benefit from testing the next hurdle is knowing which test to choose. It can be difficult for providers, especially those who are not familiar with PGx testing, to select the most appropriate test for their patients. Providers often do not have a good understanding of the differences that exist between laboratories and the tests offered and their potential limitations.32 In fact, most clinicians report the same unfamiliarity with PGx testing and available testing options as they do with genomic technologies in general. In a survey of 300 clinicians in the United States, of whom 60% were primary care physicians, 80% indicated that they had not ordered a PGx test in the past year. When asked why they had not, the most common answer, reported by nearly 70% of physicians polled, was “not knowing what test to order”.58
Clinicians who care for patients of ethnic minorities must be careful to select a test that includes the alleles relevant in that population. Different alleles may be more common in non-Caucasian populations but are not necessarily included in today’s targeted genotyping-based clinical tests. If an inappropriate test is used with a limited allele selection, important variants may be missed, resulting in an incorrect phenotype prediction. As such, misclassification of PGx phenotypes may disproportionately affect ethnic minority patients. The potential clinical significance of using currently available PGx panels in predicting response to drugs and toxicity for individual ethnic minority patients is unknown, particularly for those groups that have not been included in research studies. Additional research on the prevalence of variants across different populations and incorporation of these variants into standardized testing protocols are necessary to make PGx-based medicine available and useful to all patients.
Pharmacogenomics Testing Reimbursement
Reimbursement of PGx testing is not yet common and poses one of the greatest barriers to large scale clinical implementation, one that may not be overcome until significantly more evidence of clinical utility is available. A handful of studies have demonstrated significant cost savings when PGx testing is used prior to the administration of medications to treat conditions such as depression. However, building robust studies that can demonstrate the economic value and utility of PGx testing is challenging and therefore there are limited economic data available.59 In addition, based on the principles of genetics and allelic distribution, payers must consider the prevalence of any given allele within a population and the PGx phenotype that corresponds with that allele. Consequently, payers may be hesitant to reimburse testing given the odds that a patient has an allele that is predicted to affect either safety or efficacy. More recently in the United States, the United Health Group, one of the major third-party payers has reported coverage of psychiatric PGx panel testing based on some pre-requisite criteria.60 In Europe specifically the U-PGx project was funded by the European Commission to conduct preemptive PGx testing to make this information available for all European citizens in this consortia.36 Statistically speaking, the number of patients with intermediate phenotypes belonging to any given health plan is likely to be much higher than an extreme PGx phenotype (ultra-rapid or poor). However, because the majority of research to date has focused on the more extreme phenotypes, and not intermediate phenotypes, significant evidence in support of testing to ameliorate more severe outcomes is more likely to exist and can be used to support reimbursement.
The decreased cost of PGx testing has led to broader patient acceptance of out-of-pocket payment. The combination of more affordable testing and increased patient awareness has made PGx testing a consumer-driven initiative, thereby challenging providers to integrate it into their clinical practice. At the same time, several national agencies like CPIC,5 PharmGKB4 are working on strategies to reach out to payers for PGx reimbursement while recognizing existent barriers.61
Consumer-Initiated Testing
Due to the recent influx in direct-to-consumer and consumer-initiated PGx testing, patients now have the ability to select their own genetic tests and access their genetic information without necessarily involving a healthcare provider in the process. However, the clinical utility of such tools is yet to be determined. For example, while recently approving one such direct-to-consumer test for commercial use, the FDA also indicated that the test results must be confirmed clinically prior to use.62 Shortly thereafter, the FDA also issued a safety warning to providers and consumers alike about PGx tests that are making health-related claims that are not supported by evidence. As discussed previously, while evidence is mounting, most studies showing the efficacy of clinical PGx testing are limited to either non-randomized open-label trials or a few small randomized trials, and the relevance and utility of these studies is still unclear; similarly, there is still a dearth of studies examining the efficacy of direct-to-consumer PGx testing as well.63,64
Test Selection Opportunities And Recommendations
Due to rapid advances in testing technologies, the number of available options for PGx testing in the clinic is only expected to increase in the future. However, because of the known gaps in clinician education and knowledge regarding PGx testing, it is not surprising that determining which test to order for which patient is a significant barrier to the more widespread adoption of PGx-based medicine. This further underscores the need for more targeted clinician education, not just on the foundational science of PGx and its translation and clinical application, but the very practical knowledge of understanding the menu of tests currently available, the panel of genes assayed by each, and potential limitations of currently available testing options. It is also important to implement clinical tools into workflows that are able to provide information to clinicians on appropriate testing options at the point of care. Similarly, the increase in direct-to-consumer marketing of PGx testing has generated a lot of interest in the patient community. This interest and excitement should be leveraged and seen as an opportunity to also educate patients on how PGx may positively impact their care.
Even as the number of PGx-trained clinicians increases, reimbursement may still present a barrier to more widespread adoption of testing in the clinic. However, several opportunities present themselves. The demand by third-party payers for economic evidence to support PGx testing will potentially lead to the design and conduct of large cohort studies that will not only provide data on economic outcomes but can also be leveraged to gain additional clinical insights into the utility of testing for certain alleles in larger populations than may have been studied previously. Additionally, as integration of PGx in the clinic is expected to improve individual patient outcomes, the opportunity to thoroughly examine the utility of currently-supported healthcare models may present itself, which will likely lead to further reductions in healthcare costs while increasing the overall quality of care.
Pharmacogenomics Knowledge And Education
One of the most significant barriers to implementing PGx into clinical workflows is related to the knowledge of clinicians regarding all steps of the process, from the potential impact of PGx-based medicine on patient outcomes to knowing which test to prescribe to understanding how to use the results of testing once obtained. For example, in one study, 65% of primary care physicians agreed or strongly agreed that PGx is or will be a useful tool to predict the likelihood of a therapy’s effectiveness or to reduce adverse events, and over half believed that they should be able to provide information to a patient about available PGx tests.65 Yet, only 13% of these prescribers felt sufficiently informed to use PGx test results to guide their prescribing. Similarly, assessments of PGx knowledge in inpatient and outpatient pharmacists following targeted PGx education also revealed that overall retention of information is minimal.66 In addition, it is still unknown what specific skill sets are needed by each care team member in order to discuss a patient’s PGx testing results.67 These reported knowledge and confidence gaps create an imperative to ensure healthcare teams are adequately prepared to support a healthcare model where prescribing is based on the results of PGx testing.
The challenge with PGx education is two-fold. The first challenge is related to educating currently practicing members of the healthcare team for whom the rate of adoption and level of comfort with ordering and interpreting PGx test results remains low.68–70 Practicing healthcare providers face several barriers to acquiring PGx knowledge, which include lack of dedicated time, complicated jargon, and frequently changing evidence and guidelines. The second challenge is integrating PGx education into standard curricula across medical disciplines in order to prepare future practitioners. Already, the pharmacy education curriculum has included PGx as one of its required core competencies.71 However, this is not yet the case across all medical disciplines. Further, traditional didactic teaching methods may not be sufficient to adequately inform students about PGx. At institutions that have included PGx as part of their medical education programs, learners have reported difficulties translating the information into clinical practice.72
The use of traditional education approaches for PGx learning is also complicated by the continually evolving nature of PGx. Rapidly expanding PGx knowledge derived from basic and clinical research is published in primary literature sources and is slowly translated into tertiary literature such as textbooks, which therefore become quickly outdated. Changing the traditional didactic approach to PGx education into a more adaptable, evolving, opportune and easily accessible format is paramount to the successful delivery of PGx education to both students and practicing clinicians. Not surprisingly, readily available, online educational resources that provide information on interpreting test results and providing medication recommendations have been identified as preferred resources.58
Limited standardization across testing laboratories with regard to terminology, the format used to report results, and the specific genes and alleles tested in a given panel continues to make PGx education a challenge. National and international organizations like the CPIC,5 the Dutch Pharmacogenomics Working Group (DPWG),73 and the Canadian Pharmacogenomics Network for Drug Safety,74 are addressing this concern by creating recommendations and guidelines, which increase standardization and provide an excellent resource to supplement educational efforts. PharmGKB, facilitates identification of and access to relevant publications, and assigns a level of evidence for the associations between genes and medications that can serve as a quick reference to clinicians prior to reading the primary literature.27 The PharmGKB website also contains additional helpful tools, such as haplotype tables and IT resources.50,75
Another positive step toward educating clinicians is the FDA’s approach of including data on PGx biomarkers directly in the labeling of over 250 currently-approved medications, increasing the accessibility of this information. However, most clinicians have had limited experience navigating product labeling and not all biomarkers included in the labeling are directly actionable in the clinic. This further highlights the need for a stronger educational foundation in PGx for clinicians prior to encountering it in their daily practice.
Education Opportunities And Recommendations
The current challenges related to PGx education present an opportunity to better prepare healthcare teams of the future. To this end, integration of PGx education into medical, pharmacy, and nursing training programs should be strongly encouraged to ensure each of these future providers has a foundation in basic PGx knowledge and understands how to apply this information to clinical decision making. In addition, increasing the number of pharmacy residency programs focused on PGx, as well as more widely integrating PGx pharmacists into healthcare teams, could help ensure an adequate clinical workforce with expertise in PGx to facilitate its widespread adoption in the future. A good example for leveraging pharmacists to initiate PGx into clinical practice has been show by Bank et al Netherlands has a nation-wide fully integrated CDS along with guidelines from DPWG and a pharmacist lead approach to PGx has shown to be feasible and had an 18% rate of adoption in primary care. 76
While addressing the knowledge gaps for current practitioners with busy clinical schedules is more challenging, several online PGx certificate programs are currently available for those who seek them out. Due to time constraints and competing priorities, integration of PGx into traditional continuing medical education offerings as well as short online educational modules may make this education more accessible and practical. While adoption of PGx guidelines by traditional medical societies has been slow, other national organizations are working to gain acceptance of PGx-based medicine and to disseminate existing guidelines through the typical channels where providers receive information. Finally, large implementation studies may lead to increased provider exposure to PGx, and with this exposure, an increased experience and level of comfort in using PGx information in daily clinical practice is anticipated.
Final Remarks
For PGx to have the greatest impact on patient outcomes, it must be fully integrated into clinical practice and not merely a tool used as needed by certain clinicians and only when conditions allow. Developing infrastructure tools and systems to support the storage, standardization, display, and translation of test results in a consumable format and over time is key but still not sufficient without adequate education and continued training of current and future clinicians. While these systems can highlight, important PGx information in real-time, provider intervention and clinical judgement will still be required to customize the interpretation of the test results to individual patient scenarios. All the challenges presented here are highly interrelated and will not be overcome by individual institutions alone. PGx testing laboratories, national health organizations, medical associations, and academic institutions must collaborate to address these highlighted barriers and, in doing so, promote a more widespread and aligned integration of PGx into clinical practice.
Acknowledgments
The authors would like to recognize Lucy Hodge and Stacy A Johnson for their help with manuscript preparation and the Center for Individualized Medicine and the Center for the Science of Health Care Delivery at Mayo Clinic for their support. This work was supported in part by NIH grant U01HG006379. Dr. Caraballo is additionally funded by grants NSF 1602198 and LM 11972.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bush WS, Crosslin DR, Owusu-Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160–169. doi:10.1002/cpt.350
2. Ji Y, Skierka JM, Blommel JH, et al. Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next-generation dna sequencing and a customized cyp2d6 genotyping cascade. J Mol Diagn. 2016;18(3):438–445. doi:10.1016/j.jmoldx.2016.01.003
3. Van Driest SL, Shi Y, Bowton EA, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95(4):423–431. doi:10.1038/clpt.2013.229
4. PharmGKB.org. PharmGKB dosing guidelines. 2017; Available from: https://www.pharmgkb.org/view/dosing-guidelines.do.
5. Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166:56–67. doi:10.1002/ajmg.c.31390
6. Etienne-Grimaldi M-C, Boyer J-C, Thomas F, et al. UGT1A1 genotype and irinotecan therapy: general review and implementation in routine practice. Fundam Clin Pharmacol. 2015;29(3):219–237. doi:10.1111/fcp.12117
7. US Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labeling. 2017; Available from: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
8. Swen J, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte— an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi:10.1038/clpt.2011.34
9. Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. 2017;92(11):1711–1722. doi:10.1016/j.mayocp.2017.09.001
10. Le K-Q, Prabhakar BS, Hong W-J, Li L-C. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol Sin. 2015;36(10):1212–1218. doi:10.1038/aps.2015.43
11. Wang D, Sadee W. CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet Genomics. 2016;26(1):40–43. doi:10.1097/FPC.0000000000000183
12. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi:10.1016/j.pharmthera.2012.12.007
13. Weinshilboum RM, Diane M, Otterness A, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39(1):19–52. doi:10.1146/annurev.pharmtox.39.1.19
14. Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc Natl Acad Sci U S A. 2005;102(26):9394. doi:10.1073/pnas.0502352102
15. DeLozier TC, Lee S-C, Coulter SJ, Goh BC, Goldstein JA. Functional characterization of novel allelic variants of CYP2C9 recently discovered in Southeast Asians. J Pharmacol Exp Ther. 2005;315(3):1085. doi:10.1124/jpet.105.091181
16. Wang ZH, Zhan YY, Li YX, et al. Effects of 24 CYP2D6 variants found in the chinese population on the metabolism of risperidone. Pharmacology. 2015;96(5–6):290–295. doi:10.1159/000441007
17. Flanagan SE, Patch A-M, Ellard S. Using SIFT and polyphen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14(4):533–537. doi:10.1089/gtmb.2010.0036
18. Devarajan S, Moon I, Ho M-F, et al. Pharmacogenomic next-generation DNA sequencing: lessons from the identification and functional characterization of variants of unknown significance in CYP2C9 and CYP2C19. Drug Metab Dispos. 2019;47(4):425.
19. Guo X, Chavez A, Tung A, et al. High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR-Cas9 in yeast. Nat Biotechnol. 2018;36(6):540–546. doi:10.1038/nbt.4147
20. Trubetskoy OV, Gibson JR, Marks BD. Highly miniaturized formats for in vitro drug metabolism assays using vivid® fluorescent substrates and recombinant human cytochrome P450 enzymes. J Biomol Screen. 2005;10(1):56–66. doi:10.1177/1087057104269731
21. Cheng Q, Sohl CD, Guengerich FP. High-throughput fluorescence assay of cytochrome P450 3A4. Nat Protoc. 2009;4(9):1258–1261. doi:10.1038/nprot.2009.123
22. Wadelius M. Warfarin pharmacogenetics: it matters if you’re black or white. Blood. 2014;124(14):2171. doi:10.1182/blood-2014-08-594119
23. Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2016;19:20. doi:10.1038/gim.2016.33
24. Karczewski KJ, Weisburd B, Thomas B, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45(D1):D840–D845. doi:10.1093/nar/gkw971
25. Lakiotaki K, Kanterakis A, Kartsaki E, Katsila T, Patrinos GP, Potamias G. Exploring public genomics data for population pharmacogenomics. PLoS One. 2017;12(8):e0182138. doi:10.1371/journal.pone.0182138
26. Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1):16–26. doi:10.1016/j.jaci.2013.10.040
27. Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30(1):163–165. doi:10.1093/nar/30.1.163
28. Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the association for molecular pathology. J Mol Diagn. 2018;20(3):269–276. doi:10.1016/j.jmoldx.2018.01.011
29. Eadon MT, Desta Z, Levy KD, et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin Pharmacol Ther. 2016;100(1):63–66. doi:10.1002/cpt.347
30. Athreya AP, Neavin D, Carrillo-Roa T, et al. Pharmacogenomics-driven prediction of antidepressant treatment outcomes: a machine learning approach with multi-trial replication. Clin Pharmacol Ther. 2019;106:855–865. doi:10.1002/cpt.v106.4
31. Athreya AP, Gaglio AJ, Cairns J, et al. Machine learning helps identify new drug mechanisms in triple-negative breast cancer. IEEE Trans Nanobioscience. 2018;17(3):251–259. doi:10.1109/TNB.2018.2851997
32. Moyer AM, Caraballo PJ. The challenges of implementing pharmacogenomic testing in the clinic. Expert Rev Pharmacoecon Outcomes Res. 2017;17(6):567–577. doi:10.1080/14737167.2017.1385395
33. Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin Pharmacol Ther. 2017;102(2):254–264. doi:10.1002/cpt.707
34. Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm. 2016;73(23):1967–1976. doi:10.2146/ajhp160030
35. Blagec K, Koopmann R, Crommentuijn – van Rhenen M, et al. Implementing pharmacogenomics decision support across seven European countries: the Ubiquitous Pharmacogenomics (U-PGx) project. J Am Med Inform Assoc. 2018;25(7):893–898. doi:10.1093/jamia/ocy005
36. van der Wouden CH, Cambon-Thomsen A, Cecchin E, et al. Implementing pharmacogenomics in Europe: design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin Pharmacol Ther. 2017;101(3):341–358. doi:10.1002/cpt.602
37. Wu RR, Myers RA, Hauser ER, et al. Impact of genetic testing and family health history based risk counseling on behavior change and cognitive precursors for type 2 diabetes. J Genet Couns. 2017;26(1):133–140. doi:10.1007/s10897-016-9988-z
38. Ritchie MD, Verma SS, Hall MA, et al. Electronic medical records and genomics (eMERGE) network exploration in cataract: several new potential susceptibility loci. Mol Vis. 2014;20:1281–1295.
39. Pulley JM, Denny JC, Peterson JF, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther. 2012;92(1):87–95. doi:10.1038/clpt.2011.371
40. Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi:10.1016/j.mayocp.2013.10.021
41. Bielinski SJ, St Sauver JL, Olson JE, et al. Cohort profile: the right drug, right dose, right time: using genomic data to individualize treatment protocol (RIGHT protocol). Int J Epidemiol. 2019. doi:10.1093/ije/dyz123
42. Manolio TA, Green ED. Leading the way to genomic medicine. Am J Med Genet C Semin Med Genet. 2014;166C(1):1–7. doi:10.1002/ajmg.c.31384
43. Moyer AM, Rohrer Vitek CR, Giri J, Caraballo PJ. Challenges in ordering and interpreting pharmacogenomic tests in clinical practice. Am J Med. 2017;130(12):1342–1344. doi:10.1016/j.amjmed.2017.07.012
44. Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet Genomics. 2017;27(11):387–393. doi:10.1097/FPC.0000000000000303
45. Sangkuhl K, Whirl-Carrillo M, Whaley RM, et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther. 2018;104(1):19–22.
46. Lee S-B, Wheeler MM, Patterson K, et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med. 2019;21(2):361–372. doi:10.1038/s41436-018-0054-0
47. Gandhi MJ, Ferriola D, Huang Y, Duke JL, Monos D. Targeted next-generation sequencing for human leukocyte antigen typing in a clinical laboratory: metrics of relevance and considerations for its successful implementation. Arch Pathol Lab Med. 2017;141(6):806–812. doi:10.5858/arpa.2016-0537-RA
48. Bousman CA, Dunlop BW. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharmacogenomics J. 2018;18(5):613–622. doi:10.1038/s41397-018-0027-3
49. Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for clinical CYP2C9 genotyping allele selection: a joint recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 2019. doi:10.1016/j.jmoldx.2019.04.003
50. Gaedigk A, Sangkuhl K, Whirl-Carrillo M, et al. The Evolution of PharmVar. Clin Pharmacol Ther. 2019;105(1):29–32. doi:10.1002/cpt.1275
51. Sim SC, Altman RB, Ingelman-Sundberg M. Databases in the area of pharmacogenetics. Hum Mutat. 2011;32(5):526–531. doi:10.1002/humu.21454
52. Pratt VM, Everts RE, Aggarwal P, et al. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT-RM collaborative project. J Mol Diagn. 2016;18(1):109–123. doi:10.1016/j.jmoldx.2015.08.005
53. Pratt VM, Zehnbauer B, Wilson JA, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and association for molecular pathology collaborative project. J Mol Diagn. 2010;12(6):835–846. doi:10.2353/jmoldx.2010.100090
54. Wu AHB. Genotype and phenotype concordance for pharmacogenetic tests through proficiency survey testing. Arch Pathol Lab Med. 2013;137(9):1232–1236. doi:10.5858/arpa.2012-0261-CP
55. Kelly MA, Caleshu C, Morales A, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by clingen’s inherited cardiomyopathy expert panel. Genet Med. 2018;20(3):351–359. doi:10.1038/gim.2017.218
56. Moon JY, Franchi F, Rollini F, et al. Role of genetic testing in patients undergoing percutaneous coronary intervention. Expert Rev Clin Pharmacol. 2018;11(2):151–164. doi:10.1080/17512433.2017.1353909
57. CPIC.org. 2019; Available from: https://cpicpgx.org/guidelines/.
58. Johansen Taber KA, Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers Med. 2014;7:145–162. doi:10.2147/PGPM
59. Maciel A, Cullors A, Lukowiak AA, Garces J. Estimating cost savings of pharmacogenetic testing for depression in real-world clinical settings. Neuropsychiatr Dis Treat. 2018;14:225–230. doi:10.2147/NDT.S145046
60. Myriad genetics genesight pgx test garners UnitedHealthcare coverage. Genomeweb 2019.
61. Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet Med. 2017. doi:10.1038/gim.2017.1181.
62. USFDA.org. FDA authorizes first direct-to-consumer test for detecting genetic variants that may be associated with medication metabolism. 2019; Available from: https://www.fda.gov/news-events/press-announcements/fda-authorizes-first-direct-consumer-test-detecting-genetic-variants-may-be-associated-medication.
63. Rosenblat JD, Lee Y, McIntyre RS. Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? A systematic review of clinical trials and cost-effectiveness studies. J Clin Psychiatry. 2017;78(6):720–729. doi:10.4088/JCP.15r10583
64. Goldberg JF. Do you order pharmacogenetic testing? Why? J Clin Psychiatry. 2017;78(8):1155–1156. doi:10.4088/JCP.17ac11813
65. Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82(4):388–394. doi:10.1111/j.1399-0004.2012.01908.x
66. Formea CM, Nicholson WT, McCullough KB, et al. Development and evaluation of a pharmacogenomics educational program for pharmacists. Am J Pharm Educ. 2013;77(1):10. doi:10.5688/ajpe778166
67. Haga SB, Mills R. Nurses’ communication of pharmacogenetic test results as part of discharge care. Pharmacogenomics. 2015;16(3):251–256. doi:10.2217/pgs.14.173
68. Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145(10):749–757. doi:10.7326/0003-4819-145-10-200611210-00007
69. Giri J, Curry TB, Formea CM, Nicholson WT, Rohrer Vitek CR. Education and knowledge in pharmacogenomics: still a challenge? Clin Pharmacol Ther. 2018;103(5):752–755. doi:10.1002/cpt.1019
70. Unertl KM, Jaffa H, Field JR, Price L, Peterson JF. Clinician perspectives on using pharmacogenomics in clinical practice. Per Med. 2015;12(4):339–347. doi:10.2217/pme.15.10
71. Roederer MW, Kuo GM, Kisor DF, et al. Pharmacogenomics competencies in pharmacy practice: a blueprint for change. J Am Pharm Assoc (2003). 2017;57(1):120–125. doi:10.1016/j.japh.2016.08.014
72. Higgs JE, Andrews J, Gurwitz D, Payne K, Newman W. Pharmacogenetics education in British medical schools. Genomic Med. 2008;2(3–4):101–105. doi:10.1007/s11568-009-9032-6
73. Swen JJ, Huizinga TW, Gelderblom H, et al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 2007;4(8):e209. doi:10.1371/journal.pmed.0040209
74. Ross CJ, Visscher H, Sistonen J, et al. The Canadian pharmacogenomics network for drug safety: a model for safety pharmacology. Thyroid. 2010;20(7):681–687. doi:10.1089/thy.2010.1642
75. Klein TE, Ritchie MD. PharmCAT: a pharmacogenomics clinical annotation tool. Clin Pharmacol Ther. 2018;104(1):19–22. doi:10.1002/cpt.928
76. Bank PCD, Swen JJ, Schaap RD, Klootwijk DB, Baak – Pablo R, Guchelaar H-J. A pilot study of the implementation of pharmacogenomic pharmacist initiated pre-emptive testing in primary care. Eur J Hum Genet. 2019;27(10):1532–1541. doi:10.1038/s41431-019-0454-x
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.