Back to Journals » International Journal of General Medicine » Volume 16
Comprehensive Analysis of the Expression, Prognosis, and Biological Significance of PLOD Family in Bladder Cancer
Authors Chen R, Jiang M, Hu B, Fu B, Sun T
Received 14 December 2022
Accepted for publication 20 February 2023
Published 24 February 2023 Volume 2023:16 Pages 707—722
DOI https://doi.org/10.2147/IJGM.S399875
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Ru Chen,1,2,* Ming Jiang,1,* Bing Hu,1 Bin Fu,1 Ting Sun1
1Department of Urology, the First Affiliated Hospital of Nanchang University, Nanchang City, People’s Republic of China; 2Department of Urology, the First Hospital of Putian City, Putian City, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bin Fu; Ting Sun, Email [email protected]; [email protected]
Background: Large numbers of studies have identified that procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD) family members play important roles in tumorigenesis and tumor progression in various cancers. However, the expression pattern, clinical value and function of PLOD family have yet to be analyzed systematically and comprehensively in bladder urothelial carcinoma (BLCA).
Methods: We investigated the transcriptional levels, genetic alteration, biological function, immune cell infiltration, data on survival of PLODs in patients with BLCA based on UALCAN, the Cancer Genome Atlas (TCGA) database, Gene Expression Profiling Interactive Analysis (GEPIA), TIMER, STRING, cBioPortal and GSCALite databases. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed in R software using the Cluster Profiler Bioconductor package. Protein–protein interaction (PPI) network was established by STRING and visualized by using R version (3.6.3) software. Survival analysis was performed using the packages “survminer”.
Results: The mRNA and protein expression patterns of PLOD family members were noticeably increased in BLC compared with normal tissue. The mRNA expression levels of PLOD1-2 genes were significantly correlated with histological subtypes and PLOD1 was significantly correlated with pathological stage. Furthermore, the high expression levels of PLOD1-2 were remarkably associated with poor overall survival (OS) in BLCA patients, meanwhile high expression levels of PLOD1 and PLOD3 were markedly associated with poor progression-free interval (PFI). In co-expression gene analysis, 50 genes were primarily associated with the differentially expressed PLODs in BLCA. Functional enrichment analysis revealed that protein hydroxylation, collagen fibril organization, and lysine degradation were key biological functions of PLODs in BLCA. Moreover, PLOD family genes were identified as being associated with the activities of tumor-infiltrating immune cells and closely associated with immune responses in BLCA.
Conclusion: PLOD family members might serve as potential therapeutic targets and prognostic markers for BLCA patients’ survival.
Keywords: bladder cancer, PLODs, immune infiltration, prognosis, biomarker
Introduction
Globally, BLCA is the leading cause of cancer death and the common malignant tumor of the urinary system.1 In the United States, there are expected to be approximately 82,290 new cases of bladder cancer and 16,710 new deaths in 2022, according to research.2 Generally, bladder cancer is classified as NMIBC or MIBC.3 There is a high risk of distant metastases and death in 15% to 25% of the cases of NMIBC which progress to MIBC.4 A third of newly diagnosed patients with urothelial carcinomas of the bladder have MIBC, which can be localized or metastatic.5 Recently, the overall survival rate of patients bearing bladder cancer has improved due to early diagnosis, robotic surgery, and immunotherapy.6 However, the prognosis of patients with bladder cancer remains unsatisfactory due to the low use of neoadjuvant (NAC) chemotherapy.7 Nevertheless, conventional NAC adoption continues to give unpredicted over staging at final pathological report after radical cystectomy with curative intent. This is due to still limited ability of accurate and tailored management. Thus, presence of pN-positive disease and positive surgical margins (and their location) significantly affected survival rates.8–10 Despite radical surgery and radiotherapy, one-quarter of muscle-invasive bladder cancer patients still have a poor prognosis.11 Furthermore, tumor microenvironment which consists of cancer cells, immune cells, and extracellular matrix in BLCA is highly heterogeneous,12,13 which may result in different clinical responses to the same treatments.14,15 The recurrence rate in NMIBC ranges from 50% to 70%, whereas the occurrence of distant metastases in MIBC ranges from 50% to 80%.16 Because of the high incidence and recurrence rate of bladder cancer, it is urgent to explore the molecular mechanism of the occurrence and development of bladder cancer. Over conventional pathological markers, immunohistochemical components have been advocated and explored together with clinical ready-to-use biomarkers.17–20 In spite of this, the lack of high-quality data and prospective validation still hampers the adoption of these tools. Molecular biomarkers represent an intriguing weapon in this scenario both considering diagnostic, neoadjuvant and adjuvant setting.21–24 Therefore, identification of reliable markers for improving the prognosis of urothelial carcinomas of the bladder are urgently required.
PLOD proteins are the lysyl hydroxylase involved in the lysyl hydroxylation of collagen.25 In collagen cross-linking and deposition, lysyl hydroxylation is catalyzed by PLOD, which is composed of three members including PLOD1, PLOD2, and PLOD3.26 Aberrant expressions of PLOD1/2/3 which are regulated by multiple cytokines, transcription factors, as well as miRNA species might participate in tumorigenesis and tumor development,25,27 indicating that targeting PLOD family members might act as a potential strategy for cancer therapy. There is growing evidence that collagen deposition and cross-linking promote the progression of cancer by enhancing cancer cell proliferation, invasion, and migration.28 Further understanding of how PLOD is regulated during cancer development may identify signaling pathways that can be targeted to inhibit its activity that are involved in cell growth and survival. Recent studies have shown that members of the PLOD family demonstrated a critical role in the development and progression of cancer including pancreatic, lung, and gastric cancer.29–31 To data, there is no research about the effect of PLOD family in BLCA.
With the use of online databases, platforms, and a variety of data sets, we investigated the expression levels and prognosis of PLODs in BLCA in the current study. It is hoped that this study will give insight into the molecular mechanism of BLCA and uncover potential new therapeutic targets and prognostic biomarkers.
Materials and Methods
Ethics Statement
According to the First Affiliated Hospital of Nanchang University Ethics Committee, this study was approved (approval number 2020157). The datasets were obtained from public databases, and all data were collected with written consent.
Patients and Sample Collection
The comparison of BLCA tissues with corresponding non-cancer tissues was carried out on 20 pairs of radical cystectomy patients in the general surgery department of the first Affiliated Hospital of Nanchang University (Nanchang, China) from September 2021 to September 2022.
Cell Lines and Cell Culture
Both cell lines (T24 and BIU), which were obtained from the ATCC, were cultured in DMEM and MEM media, respectively. The remaining one cell line SV-HUC-1 was maintained in F-12K medium. In addition to FBS (5%), penicillin (100 IU/mL) and streptomycin (100 mg/mL), the media were supplemented with these substances. In an incubator, cell lines were maintained at 37°C with 5% CO2 in a humidified atmosphere.
Quantitative Reverse Transcription PCR (qRT-PCR)
The expression level of PLODs’ mRNA was quantified using qRT-PCR. The primers were listed as follows: Human PLOD1_F:GCCGTTTGTGTCCCTGTTCTTC; Human PLOD1_R:ATGCTGTGCCAGGAACTCTTCC; Human PLOD2_F:GACAGCGTTCTCTTCGTCCTCA; Human PLOD2_R:CTCCAGCCTTTTCGTGGTGACT; Human PLOD3_F:CGAGTGTGAGTTCTACTTCAGCC; Human PLOD3_R:CCAGAAGTTGGACCACAGCTTG; GAPDH forward GTCTCCTCTGACTTCAACAGCG, GAPDH reverse: ACCACCCTGTTGCTGTAGCCAA. The first step was to generate the cDNA by extracting RNA from 20 pairs of bladder cancer and adjacent normal tissues and bladder cancer cell lines (T24 and BIU) by Trizol Reagent (Cowin Biotech Co., Ltd., China) according to the manufacturer’s protocol. All qRT-PCRs were conducted under the constant settings (95 °C for 10 min, 40 cycles of 95°C for 15s and 60 °C for 1 min) using the PerfectStart Green qPCR SuperMix kit (TransGen Biotech Co, Ltd, China).
Data Acquisition and Processing
In order to investigate the distribution of the PLOD genes in normal bladder tissues, we used the Genotype-Tissue Expression (GTEx) database, which can be found at https://www.gtexportal.org/.A database of 33 types of cancer was downloaded from The Cancer Genome Atlas (TCGA), which includes gene expression data (HTSeq-FPKM), clinicopathological data, immune subtypes, survival data, and stemness scores (RNA-based). In order to perform the difference analysis, we used the Limma package from Bioconductor. A gene with an average count value greater than 1 was excluded. p<0.05 was considered, as well as |log2 (FC)|>1.0.
UALCAN
The database (http://ualcan.path.uab.edu) collected RNA-seq and clinical data of 31 cancer types from TCGA,32 which provided a useful platform for analyzing gene expression in tumor and normal tissue. We used this database to examine the relationship between gene expression levels and clinicopathological characteristics in bladder cancer. p-value <0.05 (*), p-value <0.01 (**) and p-value <0.001 (***).
GEPIA Dataset
A new interactive web server has been developed for the analysis of RNA sequencing data derived from 9736 tumors and 8587 healthy samples from the TCGA and GTEx datasets.33 In our study, we obtained data about PLOD1/2/3 mRNA expressions and clinical prognosis from the GEPIA database.
Human Protein Atlas
Immunohistochemistry-based expression data are collected in Human Protein Atlas (https://www.proteinatlas.org), which confirm whether the expression at the mRNA and protein levels matched.34 HPA was used to collect representative IHC images of PLOD family in patients with BLCA and normal tissues.
Timer
TIMER (cistrome.shinyapps.io/timer) is an effective tool to show a comprehensive understanding of tumor-immune interactions.35 In BLCA samples, we investigated the associations among tumor purity, CD4+ T, CD8+ T immune cell infiltration, and mRNA expression levels of real hub genes based on the TIMER database.
String
With STRING, it is possible to search online for known protein interactions. We used STRING to create a network of mutations for PLODs and their neighboring genes that are frequently altered.
GSCALite
GSCALite (bioinfo.life.hust.edu.cn/web/GSCALite/) provides a platform for analyzing genomic cancer data.36 Based on an online database, we examined the interaction between miRNA and PLODs. Then we analyzed the relationship between miRNAs and clinicopathological characteristics. Kaplan–Meier Plotter was utilized to illustrate the correlation between the hub miRNAs expression between high- and low-risk groups and disease progression of BLCA via R software from the R package survminer.
Results
Aberrant Expression of PLOD Family Genes in Patients Bearing BLCA
To begin, the mRNA expression levels of the three PLOD family genes were analyzed in the BLCA samples based on TCGA dataset (https://portal.gdc.cancer.gov/). As showcased in Figure 1A-C, the transcription levels of PLOD1/2/3 were significantly higher in patients bearing BLCA compared with normal samples. Our next step was to analyze the mRNA transcription levels of PLOD family members in BLCA patients based on the UALCAN database. As showcased in Figure 1D-F, comparing BLCA tissues with normal tissues, PLOD1 and PLOD3 mRNA levels were significantly elevated, while there was no significant difference in the expression of PLOD2. In order to further validate this conclusion, we further carried out a quantitative real-time PCR (RT-qPCR) analysis of 20 pairs of cancerous and adjacent normal tissues. As showcased in Figure 1G-I, according to the results, BLCA tissue displayed significantly increased transcriptional levels of PLOD1/2/3 (p < 0.05) compared to normal tissue. As shown in Figure S1, the transcriptional levels of PLOD1/2/3 (p < 0.05) were also significantly elevated in T24 and BIU bladder cancer cell lines compared with that in SV-HUC-1 cell line. Based on the HPA database, the protein levels of PLOD1/2/3 members were analyzed in BLCA patients following transcriptional analysis. As shown in Figure 2A-F, the expression levels of PLOD1–3 proteins were dramatically higher in BLCA tissues than in adjacent normal bladder tissues. Based on the above findings, we concluded that the expression of PLOD1/2/3 was significantly elevated in BLCA tissue compared with normal tissue at both mRNA and protein levels.
The Association Between PLOD Family Genes and the Clinicopathological Parameters in Patients with BLCA
We investigated the correlation between the expression of PLODs’ mRNA and clinicopathological parameters in this study, including histological subtypes and pathological stage using the UALCAN data and GEPIA database. As showcased in Figure 3A-C, we further confirm the correlation between three PLOD family members gene expressions and histological subtypes using the UALCAN web tool. The expression of PLOD1-2 expressed higher in non-papillary tumor than that in papillary tumor, while there was no difference for PLOD3 between non-papillary tumor and papillary tumor. As showcased in Figure 3D-F, the higher the clinical stage, the more the PLOD1 gene expression. The mRNA expressions of PLOD2 and PLOD3 were not associated with patients’ pathological stages. Taken together, results showed that mRNA expression levels of PLOD1-2 were significantly correlated with histological subtypes and PLOD1 was significantly correlated with pathological stage.
The Prognostic Values of mRNA Expression of PLOD in Patients Bearing BLCA
With the help of Kaplan–Meier survival curves, we evaluated the association between differentially expressed PLOD members and BLCA prognosis according to the TCGA BLCA data. The OS curves of the PLOD1/2/3 are demonstrated in Figure 4A, C and E, respectively. Our results showed that higher mRNA expression PLOD1 (HR = 1.75, 95% CI: 1.31–2.36, and p < 0.001) and PLOD2 (HR = 1.52, 95% CI: 1.14–2.04, and p = 0.005) were markedly associated with poorer OS in BLCA patients, while the mRNA expression in PLOD3 was not associated with OS of BLCA patients. A study was also conducted to investigate the prognostic roles of differentially expressed PLOD members in the PFI of patients with BLCA. The results showed that higher mRNA expression of PLOD1 (HR = 1.62, 95% CI: 1.21–2.18, and p = 0.001) and PLOD3 (HR = 1.52, 95% CI: 1.12–2.06, and p = 0.007) were associated with shorter PFI, while the mRNA expression of PLOD2 had no effect on the PFI of ccRCC patients (Figure 4B, D and F). Taken together, they might be used as indicators for predicting clinical outcome in BLCA.
Independent Prognostic Value of mRNA Expression Levels of PLOD Family Members in Terms of OS in Patients Bearing BLCA
The independent prognostic value of these genes was assessed via the TCGA database based on Cox proportional hazards model.37 Using univariate Cox regression analysis, we found that higher mRNA expression of PLOD1 (HR = 1.380, 95% CI: 1.152–1.654, and p < 0.001) was significantly associated with shorter OS. Multivariate analysis for OS revealed that higher mRNA expressions of PLOD1 (HR = 1.328, 95% CI: 1.099–1.605, and p = 0.003) showed an independent association with shorter OS of patients with BLCA. In total, the results indicated that PLOD1 might act as an independent prognostic factor for cancer survival and suggested that the increased expression of PLOD1 might play a significant role in the progression of BLCA (Supplementary Table 1).
Alteration in the Frequency of PLOD Family Genes in BLCA Patients
Data from the cBioPortal database were used to identify the frequency of genetic changes in each of the three PLOD genes among BLCA patients. Genetic alterations of PLODs are shown in Figure 5A, and the frequency of genetic alterations according to the cBioPortal database is shown in Figure 5B. The BLCA dataset indicated that the percentage changes of DNA alterations of PLODs were 0.6% (PLOD1), 3% (PLOD2), and 2.9% (PLOD3), respectively. Our next step was to examine the relationship between changes in PLOD gene expressions and prognosis of BLCA based on the cBioPortal database. In this study, Kaplan–Meier curves were used to determine the overall survival as well as the progression-free survival (PFS) of BLCA patients with altered or unaltered mRNA expression levels of PLOD1/2/3. As shown in Figure 5C, the alteration of PLOD genes in BLCA patients were closely related to OS (p = 0.0395), and patients bearing BLCA with altered PLOD gene expression showed a markedly shorter PFS (p = 0.007293) compared to those with unaltered PLOD gene expression (Figure 5D).
Predicted Functions and Pathways of the Changes in PLODs and the 50 Most Frequently Altered Neighbor Genes in Patients with BLCA
In order to better understand the underlying mechanisms of PLOD family members in BLCA, we built a network of PLOD family members and their functionally related genes. Based on the cBioPortal database, we identified the top 50 genes associated with PLOD family members and PPI network was built using STRING database (Figure 6A). The results showed that the protein hydroxylation-related genes including PLOD1/PLOD2/P4HA2/PLOD3/P3H4/P3H1 were significantly related to PLOD mutations in BLCA patients. With the use of R packages ggplot2 v3.3.2, a gene ontology enrichment analysis was conducted on three aspects: biological process, cell composition, and molecular function.38 Biological processes (BP) such as GO:0018126 (protein hydroxylation), GO:0030199 (collagen fibril organization), GO:0032963 (collagen metabolic process), GO:0030198 (extracellular matrix organization) and GO:0043062 (extracellular structure organization) were significantly regulated by PLOD alterations in BLCA. Cellular components (CC), including GO:0005788 (endoplasmic reticulum lumen), GO:0062023 (collagen-containing extracellular matrix), GO:0005791 (rough endoplasmic reticulum), GO:0030867 (rough endoplasmic reticulum membrane) and GO:0030131 (clathrin adaptor complex) were significantly associated with the PLOD alterations. In addition, PLOD mutations also affected the molecular functions (MF), such as GO:0031418 (L-ascorbic acid binding), GO:0016706 (oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors), GO:0048029 (monosaccharide binding), GO:0051213 (dioxygenase activity) and GO:0019842 (vitamin binding). An analysis of KEGG pathways revealed that PLOD mutations in BLCA were associated with an enrichment in lysine degradation (Figure 6B and Supplementary Table 2).
The Expression of PLOD Family Members is Correlated with Immune Infiltration Levels in BLCA
With the help of TIMER database, we examined the relationship between immune infiltration and PLOD expression in BLCA to explore the immune microenvironment. As shown in Figure 7A-C, all members of the PLOD family were associated with negative tumor purity, according to the results. The abundance of CD8 + T cell, macrophages, neutrophil cell and dendritic cells showed positive association with the expressions of PLOD family members. A negative correlation was observed between PLOD1 expression and B cell infiltration, whereas there was no correlation between PLOD2 and PLOD3 expression and B cell infiltration. Significantly positive correlation was found between PLOD2/3 expression and CD4+T cell infiltration, while PLOD1 expression showed no correlation with the CD4 + T cells. With the TIMER, we analyzed the relationship between different somatic copy number alterations and immune cell infiltration in BLCA samples. As showcased in Figure 7D-F, results revealed the SCNA of PLOD1/3 was significantly correlated to the infiltrating levels of CD8+ T cells, CD4+ T cells, neutrophils, and dendritic cells, while that of PLOD2 was in significant connections with the infiltrating levels of CD8+ T and CD4+ T cells.
Possible Regulatory Mechanisms of PLODs’ mRNA Expression in BLCA Patients
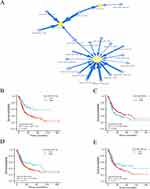
Individual miRNAs can regulate a wide range of genes, and an individual target gene can be co-regulated by several miRNAs. miRNAs are also known to regulate gene expression. According to the GSCALite database, we showed the related miRNAs lists that were related to PLODs. As shown in Figure 8A, there were four miRNAs (miR-145-5p, miR-124-3p, miR-548t-5p and miR-7-5p) regulating POLD1, 15 miRNAs (miR-145-5p, miR-26a-5p, miR-3157-5p, miR-520h, miR-514a-3p, miR-1197, miR-1267, miR-514b-3p, miR-3166, miR-96-5p, miR-330-3p, miR-382-5p, miR-513a-3p, miR-548d-3p and miR-369-3p) regulating PLOD2, and 2 (miR-491-5p and miR-369-3p) regulating PLOD3.Then, we continue to investigate the relationship between these miRNAs and the prognosis of bladder cancer. Based on the data, four (miR-145-5p, miR-96-5p, miR-382-5p, miR-369-3p) of them have a significant correlation with overall survival in patients with BLCA (Figure 8B-E). According to the results above, miRNA-PLODs regulatory networks may be important regulators of PLODs expression and may also be valuable prognostic markers and therapeutic targets for patients with BLCA.
Discussion
Due to the critical role in collagen synthesist, PLOD family genes have gained increased attention. There is increasing evidence that PLODs play a key role in tumorigenesis and the prognosis of cancer, yet functions of PLODs in BLCA remain still unclear.39–41 We analyzed the expression level, genetic alteration, potential function, immunity-related evaluations and prognostic value of PLOD family members in BLCA, in order to propose potential therapeutic and prognostic strategies for BLCA patients. In this work, our results showed that PLOD1–3 in BLCA showed a high level of expression.
PLOD1 belongs to the PLOD family of proteins that is responsible for lysyl hydroxylation which acts specifically as a telopeptide lysyl hydroxylase. PLOD1 has been studied extensively in cancer studies over the past few years. A previous study found that the expression of PLOD1 was increased in gastric cancer tissues and higher expression of PLOD1 was significantly associated with both shorter PFS and OS.42 Yuan et al found that high expression of PLOD1 could increase the proliferation and colony formation of U87 cells by activating the HSF1 signaling pathway, suggesting PLOD1/HSF1 as an effective therapeutic target for gliomas.43 Studies have reported that PLOD1 promoted lung cancer through E2F1 activation and proposed a rationale for targeting the PLOD1/E2F1 axis to treat lung cancer.44 The previous study revealed that PLOD1 promoted tumorigenesis and metastasis in osteosarcomas, providing a potential therapeutic strategy for treatment.45 A recent study found that PLOD1 expression was directly regulated by miR-140-5p in bladder cancer cells, and aberrant expression of PLOD1 significantly contributed to bladder cancer development.46 In our study, we further proved that the high expression level of PLOD1 was significantly correlated with higher histological subtypes and pathological stage. In addition, PLOD1 over-expression was associated with shorter OS and PFS, suggesting PLOD1 might act as a tumor-promoting oncogene in BLCA patients. Multivariate Cox regression analyses showed that high PLOD1 expression independently predicted a poor prognosis in BLCA patients. According to our results, PLOD1–3 was closely related to CD8+ T cells, neutrophils, macrophages, and dendritic cells. Researchers previously found that neutrophil adhesion to fibronectin could result in hydroxylysine which was a product of PLOD being released into the extracellular space further to affect the morphology and cytoskeleton of attached cells.47,48 Therefore, we hypothesized that PLOD1 expression was related to immune infiltration in BLCA and could function as a potential marker of the tumor immune microenvironment. Taken together, the present study showed that the PLOD1 might be a promising prognostic and therapeutic target for patients bearing BLCA.
PLOD2, which is a gene that encodes an enzyme responsible for catalyzing the post-translational modification of collagens, is the main enzyme that mediates stabilized collagen crosslinks.49 PLOD2 overexpression has been linked to poor outcomes in various cancers in several studies. Some scholars had found that PLOD2 expression was upregulated and positively related to the metastasis of breast cancer, thus providing a potential therapeutic target of breast cancer.50 Song et al found an association between high PLOD2 expression and poor outcomes in glioma patients.51 Li et al found that PLOD2 was significantly upregulated in cervical cancer (CESC) and positively associated with poor prognosis, indicating that PLOD2 might act as a novel diagnostic and prognostic marker for CESC patients.52 In addition, many articles have reported that PLOD2 is regulated by many other factors. For example, PLOD2 was regulated by hypoxia-inducible factor-1α (HIF-1α) to promote sarcoma metastasis and by miRNA-26a-5p and miR-26b-5p to inhibit bladder cancer cell aggressiveness.53,54 In our study, compared to normal tissues, BLCA tissue expressed higher levels of PLOD2. Furthermore, a high PLOD2 expression was significantly correlated with poor OS and markedly correlated with higher histological subtypes, suggesting an oncogenic role of PLOD2. Recent studies found a significant correlation between PLOD2 and infiltrating immune cells including neutrophils in several cancers,30,52,55 suggesting that PLOD2 acts as an important modulator of the tumor immune microenvironment. In osteosarcoma, PLOD2 was highly expressed, which was related to poor prognosis and associated with immune cell infiltration.56 These observations were in accordance with our findings that PLOD2 was closely related to the infiltration of immune cells (CD8+ T cells, neutrophils, macrophages, and dendritic cells), suggesting that PLOD2 might be potentially utilized as a new target for immunotherapy of BLCA.
PLOD3 is a membrane-bound homodimeric enzyme that hydroxylates lysyl residues in collagen-like peptides, which involved in the biosynthesis of several collagens and glycosylation activity.57 Emerging evidence suggests that PLOD3 is involved in tumorigenesis in various cancer types. PLOD3 knockdown suppressed renal cell carcinoma malignance via inhibiting TWIST1-mediated activation of β-catenin and AKT signaling and also inhibited tumor growth in lung cancer through regulating the PKC-delta signaling pathway.57,58 The PLOD3 gene is upregulated in gastric cancer, which promotes its progression.59 Through p53-independent regulation of the p21 pathway in gliomas, PLOD3 silencing suppressed cell proliferation and induced G1 phase arrest.60 In our study, PLOD3 is overexpressed in BLCA. The expression of PLOD3 was significantly related to shorter PFI and markedly correlated with higher histological subtypes. There was a significant correlation between PLOD3 mRNA levels and immune cell infiltrations including CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell, suggesting that PLOD3 might regulate the tumor microenvironment.
Interestingly, we found a positive correlation between PLOD gene expression levels and immune cell infiltration and tumor-immune interactions, indicating the immune infiltration possibly slowed down the growth of tumors and metastases through its specific mechanisms, which might provide insight into biomarkers for immune therapy in BLCA. In the current research, there is a close relationship between PLODs’ alteration and the prognosis of patients bearing BLCA, but future experimental researches are needed to confirm the mechanisms of PLODs in cancer development. We also observed the genetic alterations of PLODs had significant relevance to poor OS and PFS in these patients. Based on the GO and KEGG pathway enrichment analysis, the biological functions of PLODs were mainly involved in protein hydroxylation, dioxygenase activity and lysine degradation, providing insights into a potential underlying mechanism of carcinogenesis.
Conclusions
The study explored the mRNA and protein expression and prognostic value of PLODs in BLCA. Our results indicated that the mRNA expressions of PLOD1–3 were upregulated and found to be significantly correlated with poor prognosis in patients bearing BLCA. PLOD1–3 mRNA expressions were found to be significantly related to clinical cancer stages and histological subtypes in BLCA. PLOD1 expression might serve as an independent prognostic factor for BLCA based on univariate and multivariate Cox regression analyses. Combined with immune-cell infiltration analysis, we found that many immune cells in BLCA were associated with PLOD1–3 expression. These results suggested that PLOD family members might be able to serve as potential therapeutic targets and prognostic markers for patients with BLCA.
Data Sharing Statement
All relevant data are within the manuscript.
Ethics Approval and Consent to Participate
According to the Declaration of Helsinki’s ethical principles, the study was conducted. The fresh bladder cancer tissues and adjacent normal tissues were collected from the First Affiliated Hospital of Nanchang University, which was approved by the Human Research Ethics Committee of this hospital. The written informed consent was collected from each patient.
Acknowledgments
The authors would like to thank the patients who participated in this study, and two independent senior pathologists for technical support. Ru Chen and Ming Jiang are co-first authors for this study.
Funding
This work was supported by Grant 2021QNA070 for Youth Research Fund from Fujian Provincial Health Commission, China.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Anonymous, Erratum: global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2020;70(4):313. doi:10.3322/caac.21609
2. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi:10.3322/caac.21763
3. Breyer J, Wirtz RM, Otto W, et al. In stage pT1 non-muscle-invasive bladder cancer (NMIBC), high KRT20 and low KRT5 mRNA expression identify the luminal subtype and predict recurrence and survival. Virchows Arch. 2017;470(3):267–274. doi:10.1007/s00428-017-2064-8
4. Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: prostate and Bladder Tumours. Eur Urol. 2016;70:106–119. doi:10.1016/j.eururo.2016.02.028
5. Prasad SM, Decastro GJ, Steinberg GD. Medscape, Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat Rev Urol. 2011;8(11):631–642. doi:10.1038/nrurol.2011.144
6. Cumberbatch MGK, Noon AP. Epidemiology, aetiology and screening of bladder cancer. Transl Androl Urol. 2019;8:5–11. doi:10.21037/tau.2018.09.11
7. Reardon ZD, Patel SG, Zaid HB, et al. Trends in the use of perioperative chemotherapy for localized and locally advanced muscle-invasive bladder cancer: a sign of changing tides. Eur Urol. 2015;67(1):165–170. doi:10.1016/j.eururo.2014.01.009
8. Afferi L, Lonati C, Montorsi F, et al. Selecting the Best Candidates for Cisplatin-based Adjuvant Chemotherapy After Radical Cystectomy Among Patients with pN+ Bladder Cancer. Eur Urol Oncol. 2022;5(6):722–725. doi:10.1016/j.euo.2022.04.001
9. Claps F, van de Kamp MW, Mayr R, et al. Risk factors associated with positive surgical margins’ location at radical cystectomy and their impact on bladder cancer survival. World J Urol. 2021;39(12):4363–4371. doi:10.1007/s00345-021-03776-5
10. Mathieu R, Lucca I, Roupret M, Briganti A, Shariat SF. The prognostic role of lymphovascular invasion in urothelial carcinoma of the bladder. Nat Rev Urol. 2016;13(8):471–479. doi:10.1038/nrurol.2016.126
11. Sternberg CN, Skoneczna I, Kerst JM, et al. Immediate versus deferred chemotherapy after radical cystectomy in patients with pT3-pT4 or N+ M0 urothelial carcinoma of the bladder (EORTC 30994): an intergroup, open-label, randomised Phase 3 trial. Lancet Oncol. 2015;16(1):76–86. doi:10.1016/S1470-2045(14)71160-X
12. da Costa JB, Gibb EA, Nykopp TK, et al. Molecular tumor heterogeneity in muscle invasive bladder cancer: biomarkers, subtypes, and implications for therapy. Urol Oncol. 2022;40(7):287–294. doi:10.1016/j.urolonc.2018.11.015
13. Warrick JI, Sjödahl G, Kaag M, et al. Intratumoral Heterogeneity of Bladder Cancer by Molecular Subtypes and Histologic Variants. Eur Urol. 2019;75(1):18–22. doi:10.1016/j.eururo.2018.09.003
14. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi:10.1038/nature12625
15. Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16(1):41. doi:10.1186/s12943-017-0600-4
16. Meng Q, Lei T, Zhang M, et al. Identification of proteins differentially expressed in Adriamycin-resistant (pumc-91/ADM) and parental (pumc-91) human bladder cancer cell lines by proteome analysis. J Cancer Res Clin Oncol. 2013;139(3):509–519. doi:10.1007/s00432-012-1350-8
17. Mertens LS, Claps F, Mayr R, et al. Prognostic markers in invasive bladder cancer: FGFR3 mutation status versus P53 and KI-67 expression: a multi-center, multi-laboratory analysis in 1058 radical cystectomy patients. Urol Oncol. 2022;40(3):110 e111–110 e119. doi:10.1016/j.urolonc.2021.10.010
18. Mori K, Janisch F, Mostafaei H, et al. Prognostic value of preoperative blood-based biomarkers in upper tract urothelial carcinoma treated with nephroureterectomy: a systematic review and meta-analysis. Urol Oncol. 2020;38(5):315–333. doi:10.1016/j.urolonc.2020.01.015
19. Claps F, Rai S, Mir MC, et al. Prognostic value of preoperative albumin-to-fibrinogen ratio (AFR) in patients with bladder cancer treated with radical cystectomy. Urol Oncol. 2021;39(12):835 e839–835 e817. doi:10.1016/j.urolonc.2021.04.026
20. Schuettfort VM, D`Andrea D, Quhal F, et al. Impact of preoperative serum albumin-globulin ratio on disease outcome after radical cystectomy for urothelial carcinoma of the bladder. Urol Oncol. 2021;39(4):235 e235–235 e214. doi:10.1016/j.urolonc.2020.11.005
21. Laukhtina E, Pradere B, Lemberger U, et al. Molecular biomarkers to help select neoadjuvant systemic therapy for urothelial carcinoma of the bladder. Curr Opin Urol. 2022;32(5):561–566. doi:10.1097/MOU.0000000000001013
22. Claps F, Mir MC, Zargar H. Molecular markers of systemic therapy response in urothelial carcinoma. Asian J Urol. 2021;8(4):376–390. doi:10.1016/j.ajur.2021.05.001
23. Mir MC, Campi R, Loriot Y, et al. Adjuvant systemic therapy for high-risk muscle-invasive bladder cancer after radical cystectomy: current options and future opportunities. Eur Urol Oncol. 2022;5(6):726–731. doi:10.1016/j.euo.2021.04.004
24. de Kruijff IE, Beije N, Martens JWM, et al. Liquid biopsies to select patients for perioperative chemotherapy in muscle-invasive bladder cancer: a systematic review. Eur Urol Oncol. 2021;4(2):204–214. doi:10.1016/j.euo.2020.01.003
25. Qi Y, Xu R. Roles of PLODs in collagen synthesis and cancer progression. Front Cell Dev Biol. 2018;6:66. doi:10.3389/fcell.2018.00066
26. Valtavaara M, Szpirer C, Szpirer J, Myllyla R. Primary structure, tissue distribution, and chromosomal localization of a novel isoform of lysyl hydroxylase (lysyl hydroxylase 3). J Biol Chem. 1998;273(21):12881–12886. doi:10.1074/jbc.273.21.12881
27. Kurozumi A, Kato M, Goto Y, et al. Regulation of the collagen cross-linking enzymes LOXL2 and PLOD2 by tumor-suppressive microRNA-26a/b in renal cell carcinoma. Int J Oncol. 2016;48(5):1837–1846. doi:10.3892/ijo.2016.3440
28. Provenzano PP, Inman DR, Eliceiri KW, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6(1):11. doi:10.1186/1741-7015-6-11
29. Zhang J, Tian Y, Mo S, Fu X. Overexpressing PLOD family genes predict poor prognosis in pancreatic cancer. Int J Gen Med. 2022;15:3077–3096. doi:10.2147/IJGM.S341332
30. Qi Q, Huang W, Zhang H, et al. Bioinformatic analysis of PLOD family member expression and prognostic value in non-small cell lung cancer. Transl Cancer Res. 2021;10(6):2707–2724. doi:10.21037/tcr-21-73
31. Li SS, Lian YF, Huang YL, Huang YH, Xiao J. Overexpressing PLOD family genes predict poor prognosis in gastric cancer. J Cancer. 2020;11:121–131. doi:10.7150/jca.35763
32. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
33. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
34. Uhlen M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
35. Li B, Severson E, Pignon J-C, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
36. Liu CJ, Hu -F-F, Xia M-X, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772. doi:10.1093/bioinformatics/bty411
37. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416 e411. doi:10.1016/j.cell.2018.02.052
38. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi:10.1089/omi.2011.0118
39. Li L, Wang W, Li X, Gao T. Association of ECRG4 with PLK1, CDK4, PLOD1 and PLOD2 in esophageal squamous cell carcinoma. Am J Transl Res. 2017;9:3741–3748.
40. Gjaltema RAF, de Rond S, Rots MG, Bank RA. Procollagen lysyl hydroxylase 2 expression is regulated by an alternative downstream transforming growth factor beta-1 activation mechanism. J Biol Chem. 2015;290:28465–28476. doi:10.1074/jbc.M114.634311
41. Jover E, Silvente A, Marin F, et al. Inhibition of enzymes involved in collagen cross-linking reduces vascular smooth muscle cell calcification. FASEB J. 2018;32(8):4459–4469. doi:10.1096/fj.201700653R
42. Wang D, Zhang S, Chen F. High expression of PLOD1 drives tumorigenesis and affects clinical outcome in gastrointestinal carcinoma. Genet Test Mol Biomarkers. 2018;22:366–373. doi:10.1089/gtmb.2018.0009
43. Yuan B, Xu Y, Zheng S. PLOD1 acts as a tumor promoter in glioma via activation of the HSF1 signaling pathway. Mol Cell Biochem. 2022;477:549–557. doi:10.1007/s11010-021-04289-w
44. Li B, Yang H, Shen B, Huang J, Qin Z. Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 increases cellular proliferation and colony formation capacity in lung cancer via activation of E2F transcription factor 1. Oncol Lett. 2021;22:851. doi:10.3892/ol.2021.13112
45. Jiang H, Guo W, Yuan S, Song L. PLOD1 is a prognostic biomarker and mediator of proliferation and invasion in osteosarcoma. Biomed Res Int. 2020;2020:3418398. doi:10.1155/2020/3418398
46. Yamada Y, Kato M, Arai T, et al. Aberrantly expressed PLOD 1 promotes cancer aggressiveness in bladder cancer: a potential prognostic marker and therapeutic target. Mol Oncol. 2019;13(9):1898–1912. doi:10.1002/1878-0261.12532
47. Galkina SI, Fedorova NV, Ksenofontov AL, et al. Neutrophils as a source of branched-chain, aromatic and positively charged free amino acids. Cell Adh Migr. 2019;13(1):98–105. doi:10.1080/19336918.2018.1540903
48. Risteli M, Ruotsalainen H, Salo AM, et al. Reduction of lysyl hydroxylase 3 causes deleterious changes in the deposition and organization of extracellular matrix. J Biol Chem. 2009;284(41):28204–28211. doi:10.1074/jbc.M109.038190
49. van der Slot AJ, Zuurmond A-M, Bardoel AFJ, et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278(42):40967–40972. doi:10.1074/jbc.M307380200
50. Hu HL, Wang C-F, Wei X-H, et al. Correlation between procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 and breast cancer. Int J Clin Exp Pathol. 2019;12(3):1015–1021.
51. Song Y, Zheng S, Wang J, et al. Hypoxia-induced PLOD2 promotes proliferation, migration and invasion via PI3K/Akt signaling in glioma. Oncotarget. 2017;8(26):41947–41962. doi:10.18632/oncotarget.16710
52. Li G, Wang X, Liu G. PLOD2 Is a Potent Prognostic Marker and Associates with Immune Infiltration in Cervical Cancer. Biomed Res Int. 2021;2021:5512340. doi:10.1155/2021/5512340
53. Eisinger-Mathason TS, Zhang M, Qiu Q, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3(10):1190–1205. doi:10.1158/2159-8290.CD-13-0118
54. Miyamoto K, Seki N, Matsushita R, et al. Tumour-suppressive miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by regulating PLOD2 in bladder cancer. Br J Cancer. 2016;115(3):354–363. doi:10.1038/bjc.2016.179
55. Yang B, Zhao Y, Wang L, et al. Identification of PLOD Family Genes as Novel Prognostic Biomarkers for Hepatocellular Carcinoma. Front Oncol. 2020;10:1695. doi:10.3389/fonc.2020.01695
56. Wang Z, Fan G, Zhu H, et al. PLOD2 high expression associates with immune infiltration and facilitates cancer progression in osteosarcoma. Front Oncol. 2022;12:980390. doi:10.3389/fonc.2022.980390
57. Xie D, Li J, Wei S, et al. Knockdown of PLOD3 suppresses the malignant progression of renal cell carcinoma via reducing TWIST1 expression. Mol Cell Probes. 2020;53:101608. doi:10.1016/j.mcp.2020.101608
58. Baek JH, Yun HS, Kwon GT, et al. PLOD3 suppression exerts an anti-tumor effect on human lung cancer cells by modulating the PKC-delta signaling pathway. Cell Death Dis. 2019;10(3):156. doi:10.1038/s41419-019-1405-8
59. Wang B. PLOD3 is upregulated in gastric cancer and correlated with clinicopathologic characteristics. Clin Lab. 2019;65. doi:10.7754/Clin.Lab.2018.180541
60. Tsai CK, Huang L-C, Tsai W-C, et al. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget. 2018;9:15705–15720. doi:10.18632/oncotarget.24594
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.