Back to Journals » International Journal of General Medicine » Volume 15
Comprehensive Analysis of Prognostic Value and Immune Infiltration of NLRC4 and CASP1 in Colorectal Cancer
Authors Peng L, Zhu N, Wang D, Zhou Y, Liu Y
Received 25 December 2021
Accepted for publication 16 May 2022
Published 3 June 2022 Volume 2022:15 Pages 5425—5440
DOI https://doi.org/10.2147/IJGM.S353380
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Li Peng,* Ni Zhu,* Dan Wang,* Yanhong Zhou, Yifei Liu
School of Stomatology and Ophthalmology, Xianning Medical College, Hubei University of Science and Technology, Xianning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yanhong Zhou; Yifei Liu, Email [email protected]; [email protected]
Introduction: Nod-like receptor C4 (NLRC4) is a member of the Nod-like receptor (NLR) family, and its expression mediates the activation of caspase-1 (CASP1). Abnormal expression of NLRC4 and CASP1 is associated with multiple tumors. However, the expression differences, prognostic value and immune correlation of NLRC4 and CASP1 in colorectal cancer (CRC) remain to be determined.
Methods: In this study, TCGA, CCLE, HPA, PrognoScan, STRING and GeneMANIA databases were used to analyze differences in expression, prognostic value, genetic alterations and immune cell infiltration of NLRC4 and CASP1 in CRC patients. Then, we further validated the expression of NLRC4 and CASP1 in CRC using immunohistochemistry (IHC).
Results: NLRC4 and CASP1 were expressed low in CRC tissues and CRC cell lines. The expression of NLRC4 was significantly related to the patient’s gender and lymph node metastasis. NLRC4 and CASP1 down-regulated expression was observably correlated with poor survival and diverse immune cells infiltration in CRC patients. NLRC4 and CASP1 have a gene mutation alteration. NLRC4 and CASP1 had a significant positive correlation in CRC.
Conclusion: This study will provide new ideas for the prognosis and treatment in CRC. NLRC4 and CASP1 are expected to be novel biomarkers and potential immunotherapy targets in CRC patients.
Keywords: Nod-like receptor C4, caspase-1, colorectal cancer, bioinformatics, immunohistochemistry
Introduction
Colorectal cancer (CRC) is a highly aggressive tumor of the digestive system that is difficult to diagnose and treat and is found worldwide, its morbidity and mortality have gradually increased in recent years.1,2 Due to the difficulty of early diagnosis and poor treatment effect, the overall 5-year survival rate of CRC is still very low. Although CRC treatment has improved, most patients are diagnosed with CRC in the middle and late stages or have distant metastases because the early clinical symptoms of CRC are not obvious, and there are no sensitive biomarkers for early diagnosis.3
As we all know, long-term chronic inflammation is associated with the development of cancer.4 Inflammation are thought to affects multiple stage of tumorigenesis, it mediates host resistance to pathogenic microorganisms and defense tissue homeostasis, their dysregulation may lead to the occurrence of inflammatory diseases and cancer.5 NLRC4 is an important component of inflammasomes, the imbalance of NLRC4 is closely related to the occurrence of colitis-associated CRC.6 And a key pathway of inflammation is the activation of caspase-1 (CASP1).7 As a downstream signaling pathway molecule of NLRC4, CASP1 also plays an essential role in the onset and development of tumors. It is of great significance to study the clinical significance of NLRC4 and CASP1 in CRC – NLRC4 and CASP1 are expected to be effective biomarkers for the early diagnosis and prognosis of CRC – and develop new strategies for targeted therapies for CRC.
Nod-like receptor C4 (NLRC4) is a member of the Nod-like receptor (NLR) family that assembles into the NLRC4 inflammasome when stimulated by pathogen-related molecular patterns (PAMPs) and damage-related molecular patterns (DAMPs). NLRC4 mediates the maturation and release of CASP1, and further promotes the release of various inflammatory factors, including interleukin 1 beta (IL-1β) and interleukin-18 (IL-18), and plays a vital role in a variety of tumors.8 Depending on the type of cancer, NLRC4 may function as a tumor promoter or suppressor. For example, NLRC4 promotes tumor progression in breast cancer,9 glioma cancer10 and liver cancer11 but has a tumor suppressor role in melanoma12 and CRC.13 According to research reports, CASP1 is highly expressed in normal human tissues but downregulated in some malignant tumor tissues, such as prostate cancer,14 breast cancer,15 non-small cell lung cancer,16 and ovarian cancer.17 NLRC4 and CASP1 may play a potential role in the molecular mechanism of tumorigenesis and development, but their correlation and clinical significance in CRC still need further discussion.
The establishment and rapid development of various databases further promoted extensive bioinformatics applications and provided a reliable platform for current tumor research. Therefore, in our study, NLRC4 and CASP1 in CRC were analyzed comprehensively in terms of differential expression, clinical relevance, prognostic value, gene mutation and immune cell infiltration through a combined method of bioinformatics and IHC that could provide a theoretical basis for early diagnosis, prognostic evaluation and immune targeted therapy of CRC.
Materials and Methods
Gene Expression Analysis
Multiple bioinformatics databases were visited to study the expression levels of NLRC4 and CASP1 in CRC tissues and CRC cell lines.
The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) is a public website that contains sequencing and clinical data for over 30 human cancers. It aims to help humans have a better understanding of cancer and improve the ability of cancer prevention, diagnosis and treatment.18 The clinical information of 644 tissue samples from CRC patients was downloaded from the TCGA database. Using this database, we analyzed the differential mRNA expression of NLRC4 and CASP1 in CRC tissues and normal tissues, and further explored the association between NLRC4 and CASP1 and revealed their diagnostic value in CRC.
Cancer Cell Line Encyclopedia (CCLE, www.broadinstitute.org/ccle) collected the public information on gene expression, chromosome copies, and massively parallel sequencing data of more than 947 tumor cell lines.19 In this study, we downloaded different tumor cell lines from the CCLE database to analyze the mRNA expression levels of NLRC4 and CASP1.20,21
The Human Protein Atlas (HPA, www.proteinatlas.org) is a great large-scale protein research project, which aims to perform IHC of normal and tumor tissues by using a large number of antibodies, and records protein staining intensity and cell location in the tissues, obtained related images.22 In our study, we online analyzed the protein expression of NLRC4 and CASP1 in normal and CRC tissues by IHC.
Immune Infiltrating Analysis
In this study, we downloaded 698 patient CRC samples from the TCGA database and analyzed the correlation among NLRC4 and CASP1 mRNA expression and immune cell infiltration like macrophages, neutrophils, Th1 cells, DC, iDC, aDC, T helper cells, T cells, cytotoxic cells and Th2 cells in CRC. p<0.05 was indicated to be statistically significant.
Gene Mutation Analysis
In our research, 620 CRC patients with gene mutation data and transcriptome data were downloaded from the TCGA database, and somatic mutations in CRC patients were analyzed by maftools package in R software.
Survival Analysis
PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) aims to compare the relationship between gene expression and patient prognosis in widely published cancer microarray datasets and provide systematic analysis for gene prognostic value, including OS, DSS and DFS.23,24 This study used the PrognoScan database to investigate the correlation between the expression of NLRC4 and CASP1 and survival time of CRC patients to evaluate the prognostic significance of the two genes, and statistical significance if p<0.05.
Co-Expression Network Analysis
GeneMANIA (http://www.genemania.org) is a helpful and flexible forecasting website that can identify genes with similar functions based on gene-related effects and map genetic interaction networks.25 In our research, GeneMANIA was used to study the association of NLRC4 in terms of coexpression, colocalization and pathways. STRING (https://string-db.org/) is an powerful online tool for studying protein interaction relationships, the website possess a big collection of consolidated protein–protein interaction data.26 In this study, the STRING database was used to assess correlations involving NLRC4.
Immunohistochemistry
The colorectal cancer tissue chips (including 35 cases of cancer tissue and 35 cases of adjacent colorectal cancer tissues) were purchased from Wuhan Aiweier Biotechnology Corporation, China (CRC-1402) (Table 1).
 |
Table 1 Clinical Information of 35 Patients with Colorectal Cancer |
Tissue chips were immunostained with antibodies against NLRC4 (1:300, ABclonal, China) and CASP1 (1:800, Proteintech, USA) following the standard immunohistochemical kit of the Dako REAL EnVision Detection System (Dako, USA) procedure. The staining score mainly includes two aspects: the staining intensity and the proportion of positive cells. The cells not stained or very lightly stained were scored as 0, the cells stained light yellow were scored as 1, the cells stained brownish-yellow were scored as 2, and the cells stained dark brown were scored as 3. The number of positive cells <5% was scored as 0, the number of positive cells from 6% to 25% were scored as 1, the number of positive cells from 26% to 50% were scored as 2, the number of positive cells from 51% to 75% were scored as 3, and the number of positive cells ≥76% were scored as 4. The two scores were added together. A total score ≤3 was considered negative, and a score >3 was considered positive.
Statistical Analysis
The statistical analyses and graphs created in the TCGA were performed by R software (version: 3.6.3). We used the ggplot2 software package to analyze the mRNA differential expression of NLRC4 and CASP1 in different tumor cell lines and CRC tissues and normal tissues. And the association between NLRC4 and CASP1 also used by ggplot2 software package. The maftools software package was used to analyze somatic mutations in CRC patients. And the association between NLRC4 and CASP1 mRNA expression and immune cell infiltration in CRC was assessed by GSVA package, the immune infiltration algorithm was ssGSEA, correlation analysis was spearmen. The pROC package was used to evaluate the diagnostic value of NLRC4 and CASP1 in CRC. Gene expression of unpaired samples from TCGA database used the Wilcoxon rank sum test and paired samples used the Wilcoxon signed-rank test.
SPSS 26.0 software was used for IHC data analysis. The χ2 test was used to analyze the expression differences and relationship with the clinical characteristics of CRC of NLRC4 and CASP1 in tumor tissues and adjacent tissues. The correlation between NLRC4 and CASP1 was assessed with Spearman correlation measures (p<0.05 was considered statistically significant).
Results
The mRNA Expression Levels of NLRC4 and CASP1 in CRC Tissues and Cell Lines
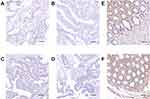
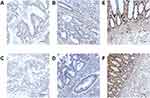
To explore the potential role of NLRC4 and CASP1 in CRC, we compared the expression level of NLRC4 and CASP1 between CRC tissues and normal tissues by using the TCGA database. The results showed that the mRNA expression levels of NLRC4 and CASP1 were lower in cancer tissues than in normal tissues (Figure 1). To validate these findings, we explored the mRNA expression of NLRC4 and CASP1 in CRC cell lines from the CCLE data set. As expected, the results indicated that NLRC4 and CASP1 were expressed low in CRC cell lines (Figure 2). To verify these results, we further explored the IHC staining results of NLRC4 and CASP1 from the HPA database. The protein expression of NLRC4 in CRC tissues and normal colon tissues was compared by antibody HPA006592. The expression of NLRC4 protein is down-regulated in CRC samples compared with normal tissues (Figure 3A). Correspondingly, the protein expression of CASP1 in CRC tissues and normal rectum tissues was compared by antibody HPA003056. The CASP1 protein was expressed lower in CRC tissues than in normal tissues (Figure 3B).
Prognostic Value of NLRC4 and CASP1 in Patients with CRC
To evaluate the prognostic value of NLRC4 and CASP1 at diverse transcription levels in CRC, we explored the relationship between NLRC4 and CASP1 at different expression levels and clinical outcomes using the PrognoScan website. We discovered that lower NLRC4 expression was significantly correlated with poorer OS in patients with CRC (HR=0.24, Cox p=0.037). In addition, CRC patients with low mRNA levels of NLRC4 had shorter DFS and DSS, but the difference was not statistically significant (p>0.05) (Figure 4A–C). Moreover, CRC patients with lower CASP1 expression experienced significantly shorter OS (HR=0.75, COX p=0.043), DFS (HR=0.60, COX p=0.015) and DSS (HR=0.71, COX p=0.015) than high CASP1 expression patients (Figure 4D–F). The findings indicate that low NLRC4 and CASP1 expression levels were significantly associated with poor prognosis in CRC patients.
Gene Mutation Alteration of NLRC4 and CASP1 in Patients with CRC
In this study, 536 CRC patients containing the whole exome sequencing data in the TCGA database were included for subsequent analysis, the results suggest that 4% of CRC patients have NLRC4 somatic mutations (Figure 5A), and 1% of CRC patients have CASP1 somatic mutations as well (Figure 5B), the most important gene mutations are known as missense mutation, it indicated that missense mutation may be an important pathological factor in CRC.
 |
Figure 5 NLRC4 and CASP1 gene alterations in CRC in the TCGA database. (A) Genetic alteration frequencies of NLRC4 in CRC. (B) Genetic alteration frequencies of CASP1 in CRC. |
Correlation Between NLRC4 and CASP1 Expression and Immune Infiltration in CRC
Immune infiltration in the tumor microenvironment plays a crucial role in tumor progression, immunotherapy effectiveness, and disease prognosis. Therefore, we analyzed the correlation between NLRC4 and CASP1 and immune infiltration in CRC using the TCGA databases. We found that both NLRC4 and CASP1 are significantly positively associated with a variety of immune cell infiltration, the most significant immune cells with NLRC4 including macrophages, neutrophils, Th1 cells, DC, iDC and T helper cells (Figure 6A). In addition, CASP1 expression was significantly positively correlated with infiltrating levels of T-cells, aDC, cytotoxic cells, Th2 cells, Th17 cells and Th1 cells (Figure 6B).
Interaction Network of NLRC4 and CASP1 at the Gene and Protein Levels
In addition, using STRING and GeneMANIA database, we constructed a protein–protein interaction and gene–gene interaction PPI network of NLRC4. Network construction identified 10 genes significantly coregulated with NLRC4, namely CASP1, NLRP3, NLRP6, NLRP1, AIM2, NAIP, PYCARD, GSDMD, BECN1, CASP8. The significant biological processes were pyroptosis, activation of cysteine-type endopeptidase activity involved in apoptotic process, defense response to bacterium, negative regulation of inflammatory response, activation of innate immune response, positive regulation of apoptotic signaling pathway (Figure 7A). Moreover, the gene–gene interaction network of NLRC4 was constructed through GeneMANIA, and the results showed that the central nodes of NLRC4 were surrounded by multiple genes that were closely correlated with NLRC4. The top five primary genes included CASP1, NLRP1, PYCARD, NLRP4 and NOD2. Functional analysis demonstrated that these genes are mainly associated with apoptosis, innate immune response, regulation of inflammatory response, regulation of IL-1β production, and positive regulation of defense response (Figure 7B). Due to the close relationship of NLRC4 and CASP1, we further used the Spearman method to analyze the correlation between NLRC4 and CASP1 in CRC and pROC package to evaluate their clinical diagnostic value from TCGA database. The results suggested that the two genes had a significant positive connection in CRC (Figure 8A). The ROC curve and area under the curve (AUC) were calculated to assess their clinical diagnostic value. The analysis showed that the AUCs were 0.701 and 0.589, respectively (Figure 8B).
 |
Figure 7 Gene and protein interaction network of NLRC4 in the STRING and GeneMANIA databases. (A) Protein–protein interaction network of NLRC4. (B) Gene–gene interaction network of NLRC4. |
 |
Figure 8 The correlation and diagnostic value of NLRC4 and CASP1 in CRC. (A) The correlation between NLRC4 and CASP1 in CRC. (B) The diagnostic value of NLRC4 and CASP1 in CRC. |
NLRC4 Protein Expression in CRC and Adjacent Normal Tissues Determined by IHC
To validate the expression of NLRC4 in CRC, we used IHC to detect the protein expression of NLRC4 in 35 CRC tissue samples and 35 adjacent normal tissue samples. The protein expression levels of NLRC4 in CRC were significantly lower than those in normal tissues adjacent to the cancer (Table 2 and Figure 9). These results were almost consistent with those obtained from the bioinformatics analysis.
 |
Table 2 The Protein Expression Levels of NLRC4 and CASP1 in CRC and Adjacent Tissues by IHC |
CASP1 Protein Expression in CRC and Adjacent Normal Tissues Determined by IHC
Furthermore, the IHC results also indicated the levels of CASP1 protein expression were highly downregulated in CRC tissues compared to normal tissues (Table 2 and Figure 10).
Association Between NLRC4 and CASP1 Protein Expression and Clinicopathological Characteristics of CRC Patients
We assessed the correlation between the protein expression difference of NLRC4 and CASP1 and the clinical features of CRC patients. The protein expression level of NLRC4 was significantly related to patient sex (χ2=6.386, p=0.012) and lymph node metastasis (χ2=6.733, p=0.035). Nevertheless, CASP1 had no apparent correlation with clinicopathological characteristics (p>0.05) (Table 3).
 |
Table 3 Association between NLRC4 and CASP1 and clinical Features in CRC Determined by IHC |
Correlation of NLRC4 and CASP1 Expression in CRC Tissues
Last, we performed a correlation analysis on the expression of NLRC4 and CASP1 in CRC tissues, the expression of NLRC4 was positively correlated with CASP1 in CRC patients (r=0.470, p=0.002) (Table 4). The correlation results of NLRC4 and CASP1 expression in CRC tissues are consistent with the results of bioinformatics analysis.
 |
Table 4 Correlation of NLRC4 and CASP1 in CRC |
Discussion
CRC is a malignant tumor in the digestive system and is a neoplasm cancer found worldwide that has severely threatened human health and life.27 Nevertheless, the potential biological mechanism of CRC remains elusive. Thus, it is vital to confirm molecular biomarkers for the early diagnosis, prognosis evaluation and tumor therapy of CRC patients. This study systematically analyzed the roles of NLRC4 and CASP1 in CRC to provide more data for further research on CRC.
As significant innate immune receptors, inflammasomes were proposed to be involved in either tumor progression or suppression.28 NLRC4 is considered to be a member of the NLR family.29 The role of NLRC4 in CRC has been reported in several studies. A previous study discovered that NLRC4 mRNA expression was significantly down-regulated in CRC tissues, and its low expression may be significantly related to the occurrence of CRC.30 Siegel et al reported that NLRC4 could eliminate damaged intestinal epithelial cells through P53-mediated apoptosis and restrict further abnormal proliferation and transformation into tumors.31 In our study, we found that NLRC4 is down-regulated in CRC, and lower expression was positive significantly related to patients’ lymph node metastasis, this result is consistent with the above research results. It proved that the presence of NLRC4 is a key pathway in inhibiting the progression of CRC. However, Allen et al reported no significant difference in disease progression of CRC in NLRC4-/- mice compared with similarly treated WT-/- mice.32 It may be that the differences in gut microbiota influence the outcome of NLRC4 deficiency.
In addition, to determine the feasibility of NLRC4 as a prognostic factor for CRC patients, we evaluated the prognostic value of abnormal NLRC4 expression in CRC. Our result revealed that lower NLRC4 transcription levels lead to worse OS in CRC patients, but DFS and DSS were not statistically significant. One possible explanation for these observations is the recording difficulty on DFS and DSS due to the bias in definition differences, interference from patient comorbidities, uncertainty in cause of death. Even so, our results have confirmed that NLRC4 expression is associated with OS in CRC, NLRC4 still has guiding value for the overall survival time of CRC patients, it might has potential as a prognostic marker to predict the survival rate of patients.
CASP1 plays a key role in pyroptosis, and its tumor suppression function in various cancers has been widely reported.33 The relationship between CASP1 and CRC has been proved by researchers. Domblides et al found a strong downregulation at the protein level of CASP1 in CRC epithelial cells compared with normal epithelial cells, and its lower expression level correlated with worse clinical outcomes for CRC patients.34 Consistent with previous research results, our research indicated that the expression of CASP1 in CRC was markedly lower than those in normal tissues. It proves that the decrease of CASP1 expression is closely related to the occurrence of CRC, CASP1 may have a protective effect in CRC. Next, we further explored the prognostic significance of abnormally expressed CASP1 in CRC patients, we found that CRC patients with higher mRNA levels of CASP1 had a better clinical prognosis. Thus, these data suggest that CASP1 expression level could be a new early diagnostic and predictive prognostic marker in CRC.
At present, the tumor microenvironment plays an increasingly important role in tumor cell survival or death.35,36 Immune cell infiltration in the tumor microenvironment significantly influences tumor growth and is thought to be a vital factor in both the clinical prognosis and immunotherapy effect of cancer patients.37 Up to now, some immunotherapy molecules, such as PD-1 and PD-L1 have been extensively studied,38–40 but the relationship between NLRC4, CASP1 and immunotherapy has rarely been reported. This study was the first to illustrate that the expression of NLRC4 and CASP1 was closely associated with the infiltration of various immune cells such as T-cells, DC, macrophages, neutrophils, and Th1 cells. Immune responses mediated by multiple immune cells are thought to be a critical component of cell-mediated antitumor effect.41 And some research has confirmed that NLRC4 and CASP1 have significant significance in the innate and adaptive immune response.42,43 So, we speculate that the antitumor effects of NLRC4 and CASP1 are attributable to changes in the immune microenvironment. Targeting inflammasome signaling represents a promising and novel therapeutic strategy against colitis and CRC.
In addition, to assess the possible functions of NLRC4 and CASP1 in CRC, we constructed a PPI network. The results indicated that NLRC4 and CASP1 are observably associated with multiple processes of cell apoptosis, pyroptosis, innate immune response, regulation of inflammatory response, and regulation of defense response. These results showed that NLRC4 and CASP1 may be involved in cell death, innate immune regulation, and regulating the release of inflammatory factors to control the progression of CRC. Gene mutation is still an important biological process in tumorigenesis, our results suggest that both NLRC4 and CASP1 have different genetic changes, so it could be thought that the mutation or aberrant deficiency of the two genes in CRC cells may be an important pathological factor in CRC.
Although studies have confirmed that activation of NLRC4 activates the expression of CASP1, but their exact relationship in CRC remains unclear. Our study indicated that NLRC4 and CASP1 have a significant positive correlation in CRC. The data confirmed that NLRC4 and CASP1 may interact to influence the progression of CRC, but the specific mechanism needs to be further clarified.
Nevertheless, there were several limitations to our research that need to be mentioned. First, the limited small sample size may cause some restriction to the research. Second, the underlying biological mechanism controlling its expression levels remains unclear. To solve this problem, larger sample cases and further in vivo and in vitro studies are necessary to improve the reliability of the findings.
Conclusion
The results strongly demonstrated that the expression of NLRC4 and CASP1 might play an essential role in tumor progression and immune responses in CRC, which may be beneficial for the development of diagnosis, prognosis and treatment for CRC patients to improve their outcomes.
Abbreviations
NLRC4, nod-like receptor C4; NLR, Nod-like receptor; CASP1, caspase-1; CRC, colorectal cancer; IHC, immunohistochemistry; PAMPs, pathogen-related molecular patterns; DAMPs, damage-related molecular patterns; IL-1β, interleukin 1 beta; IL-18, interleukin-18; TCGA, the cancer genome atlas; CCLE, Cancer Cell Line Encyclopedia; HPA, the Human Protein Atlas; AUC, area under the curve.
Ethics Approval and Consent to Participate
This study was conducted with approval from the Ethics Committee of Hubei University of Science and Technology. TCGA belongs to public databases. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data and therefore has no other conflicts of interest.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was funded with grants from the Medical School of Facial Features of Hubei University of Science and Technology (No. 2020XZ35), the High-level Talent Research Project of Xianning City (No. 202001), the Xianning Municipal Science and Technology Plan Project (No.2021ZRKX025), and the Hubei Provincial Colleges and Universities Provincial Teaching Research Project (No. 2020641).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pilleron S, Soto-Perez-de-Celis E, Vignat J, et al. Estimated global cancer incidence in the oldest adults in 2018 and projections to 2050. Int J Cancer. 2021;148(3):601–608. doi:10.1002/ijc.33232
2. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi:10.3322/caac.21601
3. Wu C, Li M, Meng H, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62(5):640–647. doi:10.1007/s11427-018-9461-5
4. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer related inflammation. Nature. 2008;454:436–444. doi:10.1038/nature07205
5. Chen GY, Núñez G. Inflammasomes in intestinal inflammation and cancer. Gastroenterology. 2011;141(6):1986–1999. doi:10.1053/j.gastro.2011.10.002
6. Carvalho FA, Nalbantoglu I, Aitken JD, et al. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol. 2012;5(3):288–98. doi:10.1038/mi.2012.8
7. Karki R, Man SM, Kanneganti TD. Inflammasomes and Cancer. Cancer Immunol Res. 2017;5(2):94. doi:10.1158/2326-6066.CIR-16-0269
8. Sundaram B, Kanneganti TD. Advances in understanding activation and function of the NLRC4 inflammasome. Int J Mol Sci. 2021;22(3):1048. doi:10.3390/ijms22031048
9. Jin H, Kim HJ. NLRC4, ASC and caspase-1 are inflammasome components that are mediated by P2Y2R activation in breast cancer cells. Int J Mol Sci. 2020;21(9):3337. Published 2020 May 8. doi:10.3390/ijms21093337
10. Lim J, Kim MJ, Park Y, et al. Upregulation of the NLRC4 inflammasome contributes to poor prognosis in glioma patients. Sci Rep. 2019;9(1):7895. doi:10.1038/s41598-019-44261-9
11. Sonohara F, Inokawa Y, Kanda M, et al. Association of inflammasome components in background liver with poor prognosis after curatively-resected hepatocellular carcinoma. Anticancer Res. 2017;37(1):293–300. doi:10.21873/anticanres.11320
12. Janowski AM, Colegio OR, Hornick EE, et al. NLRC4 suppresses melanoma tumor progression independently of inflammasome activation. J Clin Invest. 2016;126(10):3917–3928. doi:10.1172/JCI86953
13. Hu B, Elinav E, Huber S, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A. 2010;107(50):21635–21640. doi:10.1073/pnas.1016814108
14. Winter RN, Kramer A, Borkowski A, Kyprianou N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res. 2001;61(3):1227–1232.
15. Sun Y, Guo Y. Expression of caspase-1 in breast cancer tissues and its effects on cell proliferation, apoptosis and invasion. Oncol Lett. 2018;15(5):6431–6435. doi:10.3892/ol.2018.8176
16. Huang T, Zhang P, Li W, et al. G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 2017;8(4):e2726. doi:10.1038/cddis.2017.65
17. Feng Q, Li P, Salamanca C, et al. Caspase-1alpha is down-regulated in human ovarian cancer cells and the overexpression of caspase-1alpha induces apoptosis. Cancer Res. 2005;65(19):8591–8596. doi:10.1158/0008-5472.CAN-05-0239
18. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68–A77. doi:10.5114/wo.2014.47136
19. Caponigro G, Stransky N, et al. Addendum: the Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2019;565(7738):E5–E6. doi:10.1038/s41586-018-0722-x
20. Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–508. doi:10.1038/s41586-019-1186-3
21. Zhou T, Cai Z, Ma N, et al. A novel ten-gene signature predicting prognosis in hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:629. Published 2020 Jul 14. doi:10.3389/fcell.2020.00629
22. Navani S. Manual evaluation of tissue microarrays in a high-throughput research project: the contribution of Indian surgical pathology to the Human Protein Atlas (HPA) project. Proteomics. 2016;16(8):1266–1270. doi:10.1002/pmic.201500409
23. Saindane M, Rallabandi HR, Park KS, et al. Prognostic significance of prostaglandin-endoperoxide synthase-2 expressions in human breast carcinoma: a Multiomic Approach. Cancer Inform. 2020;19:1176935120969696. doi:10.1177/1176935120969696
24. Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi:10.1053/j.gastro.2009.11.005
25. Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–W64. doi:10.1093/nar/gky311
26. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
27. Sun J, Zhao H, Lin S, et al. Integrative analysis from multi-centre studies identifies a function-derived personalized multi-gene signature of outcome in colorectal cancer. J Cell Mol Med. 2019;23(8):5270–5281. doi:10.1111/jcmm.14403
28. Petrilli V. The multifaceted roles of inflammasome proteins in cancer. Curr Opin Oncol. 2017;29(1):35–40. doi:10.1097/CCO.0000000000000346
29. Lee C, Do HTT, Her J, Kim Y, Seo D, Rhee I. Inflammasome as a promising therapeutic target for cancer. Life Sci. 2019;231:116593. doi:10.1016/j.lfs.2019.116593
30. Liu R, Truax AD, Chen L, et al. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget. 2015;6(32):33456–33469. doi:10.18632/oncotarget.5587
31. Siegel SJ, Rakoff-Nahoum S. Innate immune pattern recognition and the development of intestinal cancer. Microbiome Cancer. 2019;299–316. doi:10.1007/978-3-030-04155-7_14
32. Allen IC, TeKippe EM, Woodford RM, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207(5):1045–1056. doi:10.1084/jem.20100050
33. Eren E, Ozoren N. Caspase-1: past and future of this major player in cell death and inflammation. Eur J Biol. 2019;79(1):51–61. doi:10.26650/EurJBiol.2020.0038
34. Domblides C, Soubeyran I, Lartigue L, et al. Prognostic role of inflammasome components in human colorectal cancer. Cancers (Basel). 2020;12(12):3500. doi:10.3390/cancers12123500
35. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
36. Matarrese P, Mattia G, Pagano MT, et al. The sex-related interplay between TME and cancer: on the critical role of estrogen, microRNAs and autophagy. Cancers (Basel). 2021;13(13):3287. doi:10.3390/cancers13133287
37. Sun J, Zhang Z, Bao S, et al. Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J Immunother Cancer. 2020;8(1):e000110. doi:10.1136/jitc-2019-000110
38. Spranger S, Spaapen RM, Zha Y, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. doi:10.1126/scitranslmed.3006504
39. Muenst S, Schaerli AR, Gao F, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Tr. 2014;146(1):15–24. doi:10.1007/s10549-014-2988-5
40. Wang Z, Chen L, Ma Y, et al. Peptide vaccine-conjugated mesoporous carriers synergize with immunogenic cell death and PD-L1 blockade for amplified immunotherapy of metastatic spinal. J Nanobiotechnology. 2021;19(1):243. doi:10.1186/s12951-021-00975-5
41. Gao X, Wang X, Yang Q, et al. Tumoral expression of IL-33 inhibits tumor growth and modifies the tumor microenvironment through CD8+ T and NK cells. J Immunol. 2015;194(1):438–445. doi:10.4049/jimmunol.1401344
42. Nordlander S, Pott J, Maloy KJ. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol. 2014;7(4):775–785. doi:10.1038/mi.2013.95
43. Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13(4):325–332. doi:10.1038/ni.2231
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.