Back to Archived Journals » Hypoxia » Volume 2
Complex role of HIF in cancer: the known, the unknown, and the unexpected
Authors Tiburcio P, Choi H, HuangLE
Received 22 March 2014
Accepted for publication 22 April 2014
Published 18 June 2014 Volume 2014:2 Pages 59—70
DOI https://doi.org/10.2147/HP.S50651
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Patricia Denise Tiburcio,1,2 Hyunsung Choi,1 L Eric Huang1,2
1Department of Neurosurgery, Clinical Neurosciences Center, University of Utah, Salt Lake City, UT, USA; 2Department of Oncological Sciences, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA
Abstract: Tumor hypoxia has long been recognized as a driving force of malignant progression and therapeutic resistance. The discovery of hypoxia-inducible transcription factors (HIFs) has greatly advanced our understanding of how cancer cells cope with hypoxic stress by maintaining bioenergetics through the stimulation of glycolysis. Until recently, however, it remained perplexing why proliferative cancer cells opt for aerobic glycolysis, an energy-inefficient process of glucose metabolism. Furthermore, the role of HIF in cancer has also become complex. In this review, we highlight recent groundbreaking findings in cancer metabolism, put forward plausible explanations to the complex role of HIF, and underscore remaining issues in cancer biology.
Keywords: cancer biology, hypoxia, metabolism, oncogenic signaling
Introduction
Cellular adaptation to O2 deprivation – hypoxia – is a physiological response during the course of normal development and aging in metazoans, and is critical for continued growth and survival of the organism. Central to this adaptive response is hypoxia-inducible factor 1 (HIF-1), a transcription factor that orchestrates global responses to hypoxia in gene expression.1 HIF-1 is a heterodimer composed of HIF-1α (gene symbol HIF1A) and aryl hydrocarbon receptor nuclear translocator (gene symbol ARNT, and also known as HIF-1β).2 Both subunits possess a basic helix-loop-helix domain capable of dimerization and binding to the promoters of hundreds of target genes through the recognition of hypoxia-responsive elements that contain the core sequence RCGTG.3–7
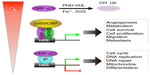
As the regulatory subunit of HIF-1, the O2-labile HIF-1α stabilizes in response to hypoxia, leading to heterodimerization, DNA binding, and recruitment of the transcription co-activators p300 and CBP.8–10 HIF-1α uses a hydrophobic interface including Cys800 to interact with p300 and CBP.11–14 Interestingly, in normoxia, HIF-1 target genes are transcriptionally active but paused for RNA polymerase II elongation; hypoxia-activated HIF-1α employs CDK8-Mediator and the super elongation complex to alleviate the pausing for elongation.15 Numerous HIF-1 target genes are activated by this canonical mechanism of transcription, such as those involved in angiogenesis, metabolic reprogramming, cell survival and proliferation, and migration and metastasis (Figure 1).16–18 Also contributing to these cancer biological processes is HIF-2α (gene symbol EPAS1),19,20 a paralog of HIF-1α, which shares similar biochemical properties, regulatory mechanisms, and target genes, even though distinct, and even opposing, functions of HIF-2α have begun to be appreciated.21
HIF-1α is sensitive to O2, owing to the presence of an O2-dependent degradation domain that mediates ubiquitin-proteasomal proteolysis.22 The von Hippel-Lindau (VHL) protein is a part of the E3 ubiquitin ligase,23,24 which recognizes two highly conserved proline residues (Pro402 and Pro564 in HIF-1α, Pro405 and Pro531 in HIF-2α) for polyubiquitination; however, hydroxyl modification of these proline residues is a prerequisite for VHL recognition.25–27 Prolyl hydroxylation is catalyzed by three prolyl hydroxylase domain-containing (PHD) proteins of the Fe(II)- and 2-oxoglutarate (2OG)-dependent dioxygenase family: PHD1, PHD2, and PHD3 (gene symbols EGLN2, EGLN1, and EGLN3, respectively), which use molecular O2 for sensing and signaling.28–34
Likewise, the factor inhibiting HIF-1 (FIH1, gene symbol HIF1AN)35 is another type of hydroxylase that modifies Asn803 in the HIF-1α transactivation domain to block p300 interaction, thereby providing an additional mechanism for modulating HIF-1α activity.36,37 Although FIH1 is a dioxygenase that uses molecular O2 for substrate modification, its in vivo role for HIF-1α regulation requires further investigation because mice with a Hif1an-null mutation demonstrated no alterations of Hif-1 function but exhibited a hypermetabolic state with hyperventilation and decreased body mass, which was regulated systemically by the nervous system.38
Besides hydroxylation, numerous modulatory mechanisms have been identified, adding the complexity of HIF regulation. These include oncogenes and tumor-suppressor genes, acetylation and SUMOylation, reactive oxygen species and nitric oxide, and microRNAs. These regulatory mechanisms have been extensively reviewed.39
In addition, both HIF-1α and HIF-2α employ crosstalk mechanisms to regulate cell proliferation, DNA repair, mitochondrial biology, and cell stemness.40,41 These noncanonical mechanisms of HIF-1α and HIF-2α actions seem independent of ARNT and direct DNA binding; rather, they require protein–protein interactions (Figure 1). HIF-1α and HIF-2α regulate cell-cycle genes in an opposing fashion; HIF-1α inhibits cell-cycle progression by suppressing c-Myc activity, whereas HIF-2α does the reverse.42–44 By similar mechanisms, HIF-1α represses, whereas HIF-2α stimulates, DNA repair genes.45–47 HIF-1α engages in the Notch signaling pathway to block cell differentiation, and the β-catenin signaling pathway to regulate embryonic and adult cell proliferation, albeit in an opposite way.48–51 Finally, HIF-1α also inhibits DNA replication through direct interaction with the adenosine triphosphatase Cdc6.52 Further studies are required to elucidate detailed mechanisms of noncanonical actions of HIF-1α and HIF-2α.
HIF-1α, HIF-2α, and cancer
Solid cancers frequently harbor hypoxic regions where HIF-1α and HIF-2α become overexpressed and activated.53 Furthermore, oncogenic signaling and growth factor stimulation lead to the activation of the phosphotidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) and mitogen-activated protein kinase (MAPK) pathways, thereby stimulating HIF-1α and HIF-2α synthesis.16,17,54–56 As a result, increased expression of both HIF-1α and HIF-2α has been observed in a wide variety of human cancers and, in general, is associated with poor prognosis.17,57 Likewise, numerous studies support the notion that HIF-1α and HIF-2α promote tumor angiogenesis and growth.17,18,57 Furthermore, rare somatic, gain-of-function mutations in EPAS1 have been identified to be associated with paraganglioma with polycythemia58 and pheochromocytoma.59
It has been recognized, however, that HIF-1α expression is also associated with favorable prognosis, eg, in patients with neuroblastoma and renal cell carcinomas.21,60 Forced expression of HIF-1α in renal cell carcinoma cell lines retarded tumor growth in xenografts,61,62 suggesting a tumor-suppressing role for HIF-1α. Conversely, it is suggested that Hif-1α is not required for renal cyst development in conditional Vhl-null mice.63 Furthermore, rare somatic, loss-of-function HIF1A mutations have been identified in the specimens of clear-cell renal carcinoma,64,65 and common focal, homozygous deletions have been detected in cell lines of VHL-deficient renal cell carcinomas.66 Despite this cogent evidence, the mechanism by which HIF-1α acts as a tumor suppressor remains unclear, and how cancer cells might escape from HIF-1α suppression needs to be addressed, especially in majority of renal cell carcinomas where HIF-1α is commonly overexpressed. What may explain retarded tumor growth in the xenograft studies, however, is that HIF-1α inhibits cell proliferation.40 Consistently, renal cell carcinomas with HIF-1α expression are statistically much smaller than those without, and intriguingly seem more metastatic.47 Therefore, the role of HIF-1α in malignant progression and metastasis cannot be ruled out in VHL-deficient renal cell carcinomas.
HIF-2α, on the other hand, is believed to be a major contributor to VHL-deficient renal carcinogenesis.33 However, in other experimental settings, HIF-2α seems to be a tumor suppressor; HIF-2α increases apoptosis in glioma67 and inhibits oncogenic signaling and activates a tumor-suppressor gene in non-small-cell lung cancer.68 Clearly, the role of HIF-1α and HIF-2α in cancer biology is complex, and their biological functions are likely context dependent.
Critical role of HIF-1α in metabolic switch from oxidative phosphorylation to anaerobic glycolysis
Among the first recognized biological functions of HIF-1α was metabolic adaptation to decreased O2 availability, resulting from transcriptional upregulation of multiple genes involved in glucose transport and glycolysis, such as those encoding aldolase A (ALDOA), phosphoglycerate kinase 1 (PGK1), lactate dehydrogenase A (LDHA), and glucose transporters (SLC2A1 and SLC2A3).1 Genetic deletion of Hif1a gene in mouse embryonic cells confirmed the critical role for HIF-1α in glycolysis and lactate production.69,70
Under normal O2 tensions, glycolysis catabolizes glucose to pyruvate, which is converted to acetyl-CoA by pyruvate dehydrogenase for oxidative phosphorylation in the tricarboxylic acid (TCA) or Krebs cycle. Under hypoxia, cells decrease oxidative phosphorylation in the mitochondria and adopt anaerobic glycolysis and conversion of pyruvate to lactate. As a key regulator of this process, HIF-1α actively suppresses mitochondrial O2 consumption by transcriptionally upregulating both pyruvate dehydrogenase kinases 1 and 3 (PDK1 and PDK3, respectively),71–73 which subsequently inactivate pyruvate dehydrogenase to block pyruvate conversion and flux of acetyl-CoA into the TCA cycle. Moreover, HIF-1 has been shown to inhibit oxidative phosphorylation by inhibiting mitochondrial biogenesis.74
In addition to the critical role in the metabolic switch from oxidative phosphorylation to anaerobic fermentation – the Pasteur effect, HIF also stimulates energy storage for hypoxic cell survival through the induction of glycogen and lipid synthesis.75 Primarily, HIF-1α promotes glycogen accumulation through transcriptional activation of several genes involved in glycogen biosynthesis, such as GYS1, PPP1R3C, and PGM1.76–78 HIF-1α also upregulates expression of the peroxisome proliferator-activated receptor γ (gene symbol PPARG), thereby increasing fatty acid uptake and triglyceride biosynthesis in cardiac hypertrophy.79 Additional HIF-1α target genes responsible for lipid accumulation include HILPDA and LPIN1.80,81 Interestingly, conditional deletion of murine Vhl in hepatocytes indicates a role for Hif-2α, rather than Hif-1α, in lipid synthesis, oxidation, and storage,82 another example of context dependence for HIF function.
HIF-1α diverts glycolytic metabolites into biosynthesis by blunting pyruvate production
In comparison to oxidative phosphorylation, glycolysis is inefficient in energy generation. Consistent with the role of HIF-1α in energy maintenance and conservation,83 it stands to reason that cancer cells adopt glycolysis under low O2 tensions for survival. However, cancer cells generally manifest characteristics of increased proliferation associated with high glucose uptake and lactate production even in the presence of O2 – aerobic glycolysis or the Warburg effect.84–86 Until recently, the significance of aerobic glycolysis remained debatable because it was unclear why cancer cells prefer energy-inefficient glycolysis to support proliferation, notwithstanding the recognition of the role of HIF-1α in aerobic glycolysis.87
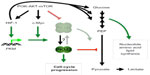
A reinterpretation of the Warburg effect is that aerobic glycolysis is not merely employed for bioenergetics but, more importantly, for biosynthesis of macromolecules (nucleotides, amino acids, and lipids) necessary for cell proliferation.85,86,88 This view has integrated oncogenic signaling (PI3K-AKT-mTOR and c-Myc) with regulation of metabolic pathways, biosynthesis, and cell proliferation. Recent studies have pointed out the M2 splice isoform of pyruvate kinase (PKM2, a splice variant of the PKM gene) as a metabolic switch for aerobic glycolysis and tumorigenesis.89,90 In normal adult tissues, the splice variant PKM1 catalyzes the final step of glycolysis by transferring the phosphate from phosphoenolpyruvate to adenosine diphosphate, thereby yielding pyruvate and adenosine triphosphate, whereas PKM2 is expressed primarily in lung tissues and proliferating cells including stem cells and cancer cells.91 The transcription factor c-Myc promotes PKM2 splicing by inducing the expression of three heterogeneous nuclear ribonucleoproteins (hnRNPs).92 It has been shown that oncogenic signaling converges on the activation of mTOR to increase HIF-1α levels for PKM transcription and concomitantly c-Myc levels for PKM2 splicing (Figure 2).90
In addition to being a glycolytic enzyme, PKM2 can function as a protein kinase for gene transcription in the nucleus.93–95 Activation of epithelial growth factor receptor induces extracellular signal-regulated kinase 2 (gene symbol MAPK1)-mediated phosphorylation and translocation of PKM2 into the nucleus. Nuclear PKM2 interacts with β-catenin to activate target genes such as CCND1 and MYC through phosphorylation of histone H3, which leads to the dissociation of HDAC3 and acetylation of histone H3. Accordingly, nuclear PKM2 promotes cell-cycle progression and cell proliferation by stimulating cyclin D1 and c-Myc expression. c-Myc further enhances PKM2 expression in a positive-feedback loop and also induces glycolytic gene expression.93–95 PKM2 can also phosphorylate signal transducer and activator of transcription 3 (gene symbol STAT3) to drive gene expression.96 In addition, PKM2 is a transcriptional coactivator of HIF-1α transcriptional activity.97 PKM2 can be hydroxylated by PHD3, and hydroxylated PKM2 enhances HIF-1α activity through direct interaction. Taken together, PKM2 stimulation of c-Myc and HIF-1α further promotes glycolysis.
So, is PKM2 essential to tumor growth and lactate production? On the contrary, mice with a conditional allele that abolishes Pkm2 but not Pkm1 expression show accelerated mammary tumor formation and heterogeneous Pkm1 expression.98 Pkm1 was detected in non-proliferating cells but not in those in proliferation, which suggests that active pyruvate kinase is required for non-proliferating, but not necessarily for proliferating, tumor cells. Interestingly, PKM2 found in tumor cells is usually dimeric with low enzymatic activity for glycolysis, whereas the tetrameric form in normal cells has a high activity.91 Furthermore, oncogenic signaling inhibits the tetrameric form of PKM2 through direct and selective binding to tyrosine-phosphorylated peptides, thereby enabling anabolism from glucose metabolites for tumor growth.99
Thus, HIF-1α-mediated PKM2 expression provides a regulated, bifunctional protein that can act on the one hand as a tetrameric glycolytic enzyme for bioenergetics and lactate production but on the other as a dimeric transcriptional coactivator/protein kinase for cell proliferation. Most importantly, loss of glycolytic activity resulting from either a dimer or a tyrosine-phosphorylated peptides-bound tetramer diverts glycolytic metabolites into the anabolic pathways (Figure 2).
Hereditary mutations in fumarate hydratase and succinate dehydrogenase
Neither HIF-1α nor HIF-2α is known to transcriptionally regulate genes in the TCA cycle; rather, it is the intermediate metabolites including fumarate and succinate that have been shown to increase the activities of these transcription factors.100 Whereas germline mutation in the FH gene (encoding fumarate hydratase) predisposes individuals to hereditary leiomyomatosis and renal cell carcinoma, hereditary mutations in the genes encoding four subunits of succinate dehydrogenase (SDHA, SDHB, SDHC, and SDHD) and one cofactor (SDHAF2) are linked to familial pheochromocytomas and paragangliomas.100–102 These tumors are characterized by the induction of pseudo-hypoxia, ie, increased HIF activities and target gene expression in normoxia. At the molecular level, inactivation of fumarate hydratase and succinate dehydrogenase leads to the accumulation of fumarate and succinate, respectively,103 resulting in allosteric inhibition of prolyl hydroxylase PHDs and induction of HIF-1α and HIF-2α.104–106 Targeted inactivation of mouse Fh1 confirmed the resultant activation of Hif-1α and Hif-2α and development of proliferative renal cysts.107 FH-deficient renal cancer cells also exhibit aerobic glycolysis and increased expression of HIF-1α.108 Additionally, accumulated fumarate produces succinated glutathione, resulting in increased mitochondrial reactive oxygen species and HIF-1α activation.109 In fact, it has been shown that fumarate can modify numerous proteins including mitochondrial aconitase 2 for the inhibition of aconitase activity in Fh1-null mouse embryonic fibroblasts,110 even though reduced cytosolic, but not mitochondrial, aconitase activity is observed in FH-deficient renal cancer cells associated with iron deficiency.108
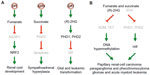
Despite these findings, whether activation of HIF pathway has a causal effect on renal cyst formation was questioned.111 Indeed, in mouse genetic studies, Fh1−/−-associated renal cyst formation was independent of Hif-1α and Hif-2α, and was further exacerbated by inactivation of Hif-1α but not Hif-2α.112 In fact, FH-deficient cysts and tumors were associated with upregulation of the KEAP1-NRF2 antioxidant pathway (Figure 3A), resulting from derepression of nuclear factor erythroid 2-like 2 (NRF2, gene symbol NFE2L2) upon fumarate inhibition of the negative regulator kelch-like ECH-associated protein 1 (gene symbol KEAP1) by succination. Consistently, upregulation of antioxidant response genes through the KEAP1-NRF2 axis is a distinct feature of type 2 papillary renal cell carcinoma caused by FH mutation, in contrast to those of clear cell carcinomas arising from SDH or VHL mutation.113 Furthermore, NRF2 apparently decreases HIF-1α levels by reducing reactive oxygen species in FH-deficient cells.109 The NRF2-mediated antioxidant and detoxification program is involved in oncogene-induced tumorigenesis.114 Intriguingly, NRF2 has also been shown to promote biosynthesis by redirecting glucose and glutamine into the anabolic pathways in the presence of activated PI3K-AKT signaling.115 Despite the lack of involvement of Hif-1α in Fh1−/−-associated renal cyst development, upregulation of HIF-1α remains essential to glycolytic metabolism and oncogenic growth of FH-deficient renal cancer,108 consistent with the aforementioned role of HIF-1α in glycolysis and anabolism.
Although mutations in SDHD is linked to hereditary paraganglioma,116 heterozygous deletion of Sdhd in mice develops no tumor but subtle glomus cell hypertrophy and hyperplasia.117 Succinate accumulation has been shown to specifically inhibit PHD3 apoptotic activity, thereby blocking the apoptosis of sympathetic neuronal precursor cells during development and contributing to the pathogenesis of familial pheochromocytoma (Figure 3A).118 Interestingly, mice with germline deletion of Egln3 also exhibited reduced apoptosis and increased cell numbers in the superior cervical ganglia, adrenal medulla, and carotid body.119 Furthermore, a combined heterozygous deletion of Epas1 rendered sympathetic neurons more sensitive to induced apoptosis, indicating an anti-apoptotic role of HIF-2α in this setting. Conversely, as mentioned above, rare somatic, gain-of-function mutations in EPAS1,58,59 as well as germline mutations in FH120 have been associated with malignant paragangliomas and pheochromocytomas. Furthermore, hypermethylation has also been identified in tumors with SDH and FH mutations. Methylome analysis of a large cohort of paraganglioma patients has revealed a hypermethylator phenotype similar to that in gliomas.121 Sdhb deletion in mouse chromaffin cells led to DNA hypermethylation, resulting from succinate inhibition of the JmjC domain-containing histone demethylases and DNA hydroxylases (Figure 3B). Finally, DNA hypermethylation has also been identified in tumors with FH mutations.109,121
Somatic mutations in isocitrate dehydrogenase
There has been rapid expansion of knowledge in recent years of single somatic mutations in cytosolic isocitrate dehydrogenase 1 (gene symbol IDH1), and these mutations were identified initially in >70% WHO grade II–III gliomas and secondary glioblastomas.122 Tumors lacking mutations in IDH1 often had single mutations in IDH2, a mitochondrial gene in the TCA cycle for interconversion of isocitrate and 2OG. Similar mutations have also been found in <23% acute myeloid leukemia, albeit mainly in IDH2.123–125 Interestingly, all the mutations are heterozygous and affect the same active site of the enzyme at a single arginine residue (Arg132 of IDH1 and Arg172 of IDH2), thereby reducing the intracellular concentration of 2OG,122 a cosubstrate of the HIF prolyl hydroxylases. Accordingly, it was found that mutant IDH1 forms a catalytically inactive heterodimer with the wild type, resulting in a decrease of PHD activity and an increase of HIF-1α levels in human gliomas harboring an IDH1 mutation.126
This loss-of-function theory was only part of the story, however, because a wild type allele of IDH1 or IDH2 always remains in these tumors, raising the possibility of gain of function as result of mutations. Indeed, both mutant IDH1 and mutant IDH2 acquire the ability to catalyze the nicotinamide adenine dinucleotide phosphate-dependent reduction of 2OG to (R)-enantiomer of 2-hydroxyglutarate ((R)-2HG).124,127,128 Accordingly, elevated levels of (R)-2HG are detectable in IDH mutant gliomas and acute myeloid leukemias.
So, is (R)-2HG an oncometabolite stimulating hypoxic signaling for tumorigenesis? On the contrary, it has been suggested that (R)-2HG downregulates HIF-1α and HIF-2α levels by increasing PHD1 and PHD2 activity to promote transformation of immortalized human astrocytes (Figure 3A).129 This finding was supported by the evidence that (R)-2HG can substitute 2OG for PHD enzymatic activity in a cell-free reaction, and this finding is correlated with diminished expression of HIF-1α and HIF-2α in IDH1 mutant cells and reduced HIF target gene expression in IDH mutant gliomas. Furthermore, decreased HIF-1α expression is conducive to transformation by mutant IDH1. Moreover, (R)-2HG is sufficient to promote leukemogenesis by inducing cytokine independence and blocking hematopoietic differentiation, whereas the (S)-enantiomer of 2-hydroxyglutarate, despite being a more potent inhibitor of 2OG-dependent dioxygenases,129–131 fails to do so owing to its inhibition of PHD activity to increase, rather than decrease, HIF-1α levels.132
The notion that (R)-2HG inhibits HIF signaling seems at odds with the genetic evidence of mouse studies; increased Hif-1α protein levels and target gene transcription were observed in the embryo of brain Idh1 (R132H) knock-in mice where high levels of (R)-2HG were produced,133 and no alteration of Hif-1α signaling was found in the hematopoietic stem cells and progenitor cells from the knock-in mice of the myeloid lineage.134 Although the mechanisms by which HIF-1α prevents astrocyte transformation and leukemogenesis remain unclear, a lower level of HIF-1α expression in IDH1 mutant cells might be conducive to tumor initiation because HIF-1α also upregulates histone demethylases,135,136 which have been shown to be inhibited by (R)-2HG for transformation.
In addition to PHDs, (R)-2HG is a potent inhibitor of other 2OG-dependent dioxygenases,137 including the TET family of 5-methylcytosine hydroxylases, which convert 5-methyl-cytosine to 5-hydroxymethylcytosine for DNA demethylation.138–140 In fact, IDH1 somatic mutations in glioblastomas and lower-grade gliomas have a CpG island methylator phenotype, displaying DNA hypermethylation at a large number of loci.141,142 Similarly, acute myeloid leukemias with IDH1 or IDH2 mutations also exhibit genome-wide DNA hypermethylation with a specific signature shared with those harboring mutations in TET2,125 a member of the TET protein family. Consistently, expression of IDH mutants was found to impair TET2 catalytic function, and mutations in IDH1 and IDH2 were mutually exclusive with those in TET2 in a large cohort of acute myeloid leukemias.125
Although both forms of 2-HG have been shown to be competitive inhibitors of multiple 2OG-dependent dioxygenases, including the JmjC domain-containing histone demethylases and the TET family of 5-methylcytosine hydroxylases as well as PHDs,130 the half-maximal inhibitory concentration of (R)-2HG for histone demethylases is 200-fold less than for PHD2, indicating more important effects of IDH mutations on chromatin remodeling.131 In keeping with this, IDH mutation has been shown to inhibit histone demethylation and induce DNA hypermethylation in cell culture and animal model, thereby blocking cell differentiation.134,143,144 Furthermore, targeted inhibition of mutant IDH2 with the small molecule AGI-6780 induces differentiation of established erythroleukemia cells and primary human acute myeloid leukemia cells in vitro.145 The small-molecule inhibitor of mutant IDH1 (AGI-5198) also promotes differentiation of glioma cells harboring IDH1 mutation and inhibits the growth of tumor xenografts.146 Interestingly, AGI-5198 induces histone demethylation and expression of genes associated with glial differentiation without appreciable changes in genome-wide DNA methylation, suggesting additional mechanisms of IDH1 mutation for glioma growth.
Conclusion
It took nearly two decades to appreciate that HIF-1α stimulation of glycolysis is not merely to maintain bioenergetics for cell survival, but equally importantly is to promote biosynthesis of macromolecules for cell proliferation. HIF-1α does the latter by blunting the glycolytic pathway through the induction of PKM2. PKM2 engages in various mechanisms as a dimer to drive cell proliferation147 and as a glycolytic enzyme in tetramer is further suppressed by oncogenic signaling. Although how PKM2 oscillates between dimer and tetramer is not well understood, receptor tyrosine kinase-mediated activation of the PI3K-AKT-mTOR pathway seems essential in orchestrating cellular biosynthesis of nucleotides, amino acids, and lipids that involves multiple pathways including HIF-1α and c-Myc signaling, and glutamine-dependent anaplerosis for cell proliferation.148
Yet, rapid cell proliferation induces hypoxia, which is known to suppress mTORC1 through multiple mechanisms.21 In fact, HIF-1α is known to inhibit mTORC1 activity through transcriptional upregulation of DDIT4 (encoding REDD1), a negative regulator of mTORC1 activity,149,150 whereas HIF-2α, unrelated to aerobic glycolysis, stimulates mTORC1 activity by transcriptionally upregulating SLC7A5 (encoding an amino acid transporter).151 So, how do cancer cells escape from these mechanisms for proliferation? One possible answer to this conundrum is to inactivate HIF-1α, as mentioned above, albeit rarely, which implies the benefit of keeping HIF-1α expression in cancer cells. Alternatively, cancer cells may decrease expression of the stress-sensor protein ATM (ataxia telangiectasia mutated, gene symbol ATM), which is required for transcriptional activation of REDD1 through HIF-1α phosphorylation,152 or they may attempt to maintain mTORC1 activity by relying on exogenous desaturated lipids from serum for survival.153 Lastly, they may adopt a “stop-and-go” mechanism by entering cell-cycle arrest while promoting an angiogenic response to alleviate hypoxic stress for continued proliferation.154
By altering the intracellular or intercellular context, cancer cells can adopt aberrant ways of maintaining survival and proliferation. Therefore, the selection of NRF2 signaling for detoxification and anabolism, rather than anti-proliferative HIF-1α signaling, might be beneficial to renal cyst development at the early stage in FH-mutant cells. Similarly, an initial decrease in HIF-1α levels may facilitate DNA hypermethylation in cells harboring FH, SDH, or IDH mutations, because HIF-1α does the opposite by inducing histone demethylases and DNA demethylation, as also observed in metastasis of VHL-deficient renal cancer.155 It is also noteworthy that none of the FH, SDH, and IDH mouse models has recapitulated a corresponding phenotype of the human diseases, suggesting the requirement of additional genetic/epigenetic alterations for tumorigenesis. Further studies are warranted to elucidate the mechanisms underlying these changes and the logistics of utilizing various signaling pathways to cancer’s best advantage. Understanding of these dynamic processes is expected to provide more unexpected answers to the mystery of cancer.
Acknowledgments
This work was supported in part by an NIH Grant CA084563 from the National Cancer Institute. We thank K Kraus for editorial assistance.
Disclosure
The authors have no conflicts of interest in this work.
References
Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. | |
Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. | |
Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. | |
Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37(14):4587–4602. | |
Mole DR, Blancher C, Copley RR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284(25):16767–16775. | |
Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38(7):2332–2345. | |
Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–17. | |
Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia- inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271(50):32253–32259. | |
Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93(23):12969–12973. | |
Kallio PJ, Okamoto K, O’Brien S, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17(22):6573–6586. | |
Ema M, Hirota K, Mimura J, et al. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18(7):1905–1914. | |
Gu J, Milligan J, Huang LE. Molecular mechanism of hypoxia- inducible factor 1alpha -p300 interaction. A leucine-rich interface regulated by a single cysteine. J Biol Chem. 2001;276(5):3550–3554. | |
Freedman SJ, Sun Z-YJ, Poy F, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99(8):5367–5372. | |
Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002;99(8):5271–5276. | |
Galbraith MD, Allen MA, Bensard CL, et al. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell. 2013;153(6):1327–1339. | |
Harris AL. Hypoxia – a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. | |
Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. | |
Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975. | |
Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. | |
Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA. 1997;94(9):4273–4278. | |
Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. | |
Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998; 95(14):7987–7992. | |
Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. | |
Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19(16):4298–4309. | |
Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. | |
Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. | |
Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A. 2001;98(17):9630–9635. | |
Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. | |
Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. | |
Huang LE, Bunn HF. Hypoxia-inducible Factor and Its Biomedical Relevance. Journal of Biological Chemistry. 2003;278(22):19575–19578. | |
Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. | |
Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. | |
Kaelin WG. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8(11):865–873. | |
Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol (Lond). 2013;591(Pt 8):2027–2042. | |
Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15(20):2675–2686. | |
Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295(5556):858–861. | |
Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16(12):1466–1471. | |
Zhang N, Fu Z, Linke S, et al. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11(5):364–378. | |
Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. | |
Huang LE. Carrot and stick: HIF-alpha engages c-Myc in hypoxic adaptation. Cell Death Differ. 2008;15(4):672–677. | |
Lendahl U, Lee KL, Yang H, Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009;10(12):821–832. | |
Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23(9):1949–1956. | |
Gordan JD, Bertout JA, Hu C-J, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–347. | |
Hammer S, To KK-W, Yoo Y-G, Koshiji M, Huang LE. Hypoxic suppression of the cell cycle gene CDC25A in tumor cells. Cell Cycle. 2007;6(15):1919–1926. | |
Koshiji M, To KK-W, Hammer S, et al. HIF-1α Induces Genetic Instability by Transcriptionally Downregulating MutSα Expression. Mol Cell. 2005;17(6):793–803. | |
To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. EMBO J. 2006;25(20):4784–4794. | |
Gordan JD, Lal P, Dondeti VR, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14(6):435–446. | |
Gustafsson MV, Zheng X, Pereira T, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005; 9(5):617–628. | |
Mazumdar J, O’Brien WT, Johnson RS, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12(10):1007–1013. | |
Lim J-H, Chun Y-S, Park J-W. Hypoxia-inducible factor-1alpha obstructs a Wnt signaling pathway by inhibiting the hARD1-mediated activation of beta-catenin. Cancer Res. 2008;68(13):5177–5184. | |
Kaidi A, Williams AC, Paraskeva C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9(2):210–217. | |
Hubbi ME, Kshitiz, Gilkes DM, et al. A Nontranscriptional Role for HIF-1 as a Direct Inhibitor of DNA Replication. Sci Signal. 2013;6(262):ra10–ra10. | |
Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11(3):293–299. | |
Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2(10):803–811. | |
Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441(7092):437–443. | |
Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. | |
Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15(4):678–685. | |
Zhuang Z, Yang C, Lorenzo F, et al. Somatic HIF2AGain-of-Function Mutations in Paraganglioma with Polycythemia. N Engl J Med. 2012;367(10):922–930. | |
Favier J, Buffet A, Gimenez-Roqueplo A-P. HIF2A mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367(22):2161–author reply 2161–2162. | |
Qing G, Simon MC. Hypoxia inducible factor-2alpha: a critical mediator of aggressive tumor phenotypes. Curr Opin Genet Dev. 2009;19(1):60–66. | |
Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1(3):237–246. | |
Raval RR, Lau KW, Tran MGB, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25(13):5675–5686. | |
Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66(5):2576–2583. | |
Morris MR, Hughes DJ, Tian Y-M, et al. Mutation analysis of hypoxia-inducible factors HIF1A and HIF2A in renal cell carcinoma. Anticancer Res. 2009;29(11):4337–4343. | |
Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. | |
Shen C, Beroukhim R, Schumacher SE, et al. Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1(3):222–235. | |
Acker T, Diez-Juan A, Aragonés J, et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8(2):131–141. | |
Mazumdar J, Hickey MM, Pant DK, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A. 2010;107(32):14182–14187. | |
Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–162. | |
Seagroves TN, Ryan HE, Lu H, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21(10):3436–3444. | |
Kim J-W, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. | |
Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. | |
Lu C-W, Lin S-C, Chen K-F, Lai Y-Y, Tsai S-J. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem. 2008;283(42):28106–28114. | |
Zhang H, Gao P, Fukuda R, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. | |
Brahimi-Horn MC, Bellot G, Pouysségur J. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev. 2011;21(1):67–72. | |
Pescador N, Villar D, Cifuentes D, et al. Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS ONE. 2010;5(3):e9644. | |
Shen GM, Zhang FL, Liu XL, Zhang JW. Hypoxia-inducible factor 1- mediated regulation of PPP1R3C promotes glycogen accumulation in human MCF-7 cells under hypoxia. FEBS Lett. 2010;584(20):4366–4372. | |
Pelletier J, Bellot G, Gounon P, Lacas-Gervais S, Pouysségur J, Mazure NM. Glycogen Synthesis is Induced in Hypoxia by the Hypoxia-Inducible Factor and Promotes Cancer Cell Survival. Front Oncol. 2012;2:18. | |
Krishnan J, Suter M, Windak R, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9(6):512–524. | |
Gimm T, Wiese M, Teschemacher B, et al. Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J. 2010;24(11):4443–4458. | |
Mylonis I, Sembongi H, Befani C, Liakos P, Siniossoglou S, Simos G. Hypoxia causes triglyceride accumulation by HIF-1-mediated stimulation of lipin 1 expression. J Cell Sci. 2012;125(Pt 14):3485–3493. | |
Rankin EB, Rha J, Selak MA, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29(16):4527–4538. | |
Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability – a calculated mechanism underlying tumor progression. J Mol Med. 2007;85(2):139–148. | |
Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. | |
DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. | |
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. | |
Lum JJ, Bui T, Gruber M, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21(9):1037–1049. | |
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. | |
Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. | |
Sun Q, Chen X, Ma J, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proceedings of the National Academy of Sciences. 2011;108(10):4129–4134. | |
Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15(4):300–308. | |
David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–368. | |
Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. | |
Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. | |
Yang W, Xia Y, Hawke D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. | |
Gao X, Wang H, Yang JJ, Liu X, Liu Z-R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. | |
Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. | |
Israelsen WJ, Dayton TL, Davidson SM, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155(2):397–409. | |
Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. | |
Gottlieb E, Tomlinson IPM. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–866. | |
Hao H-X, Khalimonchuk O, Schraders M, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325(5944):1139–1142. | |
Dahia PLM. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer. 2014;14(2):108–119. | |
Pollard PJ. Accumulation of Krebs cycle intermediates and over-expression of HIF1 in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14(15):2231–2239. | |
Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7(1):77–85. | |
Isaacs JS, Jung YJ, Mole DR, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8(2):143–153. | |
Brière J-J, Favier J, Benit P, et al. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum Mol Genet. 2005;14(21):3263–3269. | |
Pollard PJ, Spencer-Dene B, Shukla D, et al. Targeted inactivation of fh1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell. 2007;11(4):311–319. | |
Tong W-H, Sourbier C, Kovtunovych G, et al. The Glycolytic Shift in Fumarate-Hydratase-Deficient Kidney Cancer Lowers AMPK Levels, Increases Anabolic Propensities and Lowers Cellular Iron Levels. Cancer Cell. 2011;20(3):315–327. | |
Sullivan LB, Martinez-Garcia E, Nguyen H, et al. The Proto-oncometabolite Fumarate Binds Glutathione to Amplify ROS-Dependent Signaling. Mol Cell. 2013;51(2):236–248. | |
Ternette N, Yang M, Laroyia M, et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Reports. 2013;3(3):689–700. | |
Ratcliffe PJ. Fumarate hydratase deficiency and cancer: activation of hypoxia signaling? Cancer Cell. 2007;11(4):303–305. | |
Adam J, Hatipoglu E, O’Flaherty L, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20(4):524–537. | |
Ooi A, Wong J-C, Petillo D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20(4):511–523. | |
DeNicola GM, Karreth FA, Humpton TJ, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. | |
Mitsuishi Y, Taguchi K, Kawatani Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. | |
Baysal BE, Ferrell RE, Willett-Brozick JE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–851. | |
Piruat JI, Pintado CO, Ortega-Saenz P, Roche M, Lopez-Barneo J. The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol Cell Biol. 2004;24(24):10933–10940. | |
Lee S, Nakamura E, Yang H, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8(2):155–167. | |
Bishop T, Gallagher D, Pascual A, et al. Abnormal sympathoadrenal development and systemic hypotension in PHD3−/− mice. Mol Cell Biol. 2008;28(10):3386–3400. | |
Castro-Vega LJ, Buffet A, de Cubas AA, et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet. 2014;23(9):2440–2446. | |
Letouzé E, Martinelli C, Loriot C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23(6):739–752. | |
Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. | |
Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. | |
Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. | |
Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. | |
Zhao S, Lin Y, Xu W, et al. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1α. Science. 2009;324(5924):261–265. | |
Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. | |
Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. | |
Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. | |
Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. | |
Chowdhury R, Yeoh KK, Tian Y-M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. | |
Losman J-A, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625. | |
Sasaki M, Knobbe CB, Itsumi M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–2049. | |
Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. | |
Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proceedings of the National Academy of Sciences. 2009;106(11):4260–4265. | |
Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30(1):344–353. | |
Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36(1):7–18. | |
Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. | |
Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. | |
Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13(1):28–35. | |
Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. | |
Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. | |
Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. | |
Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. | |
Wang F, Travins J, Delabarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. | |
Rohle D, Popovici-Muller J, Palaskas N, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013;340(6132):626–630. | |
McKnight SL. Please Keep Me 2uned to PKM2. Mol Cell. 2014;53(5):683–684. | |
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. | |
Shoshani T, Faerman A, Mett I, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22(7):2283–2293. | |
Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. | |
Elorza A, Soro-Arnáiz I, Meléndez-Rodríguez F, et al. HIF2α acts as an mTORC1 activator through the amino acid carrier SLC7A5. Mol Cell. 2012;48(5):681–691. | |
Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell. 2010;40(4):509–520. | |
Young RM, Ackerman D, Quinn ZL, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27(10):1115–1131. | |
Koshiji M, Huang LE. Dynamic balancing of the dual nature of HIF-1alpha for cell survival. Cell Cycle. 2004;3(7):853–854. | |
Vanharanta S, Shu W, Brenet F, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19(1):50–56. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.