Back to Journals » Journal of Pain Research » Volume 15
Complement Receptor 3 Pathway and NMDA Receptor 2B Subunit Involve Neuropathic Pain Associated with Spinal Cord Injury
Authors Li Y, Fang SC, Zhou L, Mo XM, Guo HD, Deng YB, Yu HH, Gong WY
Received 29 March 2022
Accepted for publication 8 June 2022
Published 25 June 2022 Volume 2022:15 Pages 1813—1823
DOI https://doi.org/10.2147/JPR.S366782
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Qi Fang
Yong Li,1,* Sheng-Chun Fang,2,* Lan Zhou,1 Xue-Mei Mo,3 Hao-Dong Guo,3 Yan-Bo Deng,3 Hong-Hao Yu,1 Wei-Yi Gong3
1College of Biotechnology, Guilin Medical University, Guilin, Guangxi, 541100, People’s Republic of China; 2Department of Anesthesiology, Wuhan Children’s Hospital (Wuhan Maternal and Child Healthcare Hospital), Tongji Medical College of Huazhong University of Science and Technology, Wuhan, Hubei, 430015, People’s Republic of China; 3Department of Pain Management, Affiliated Hospital of Guilin Medical University, Guilin, Guangxi, 541000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong-Hao Yu, College of Biotechnology, Guilin Medical University, Guilin, Guangxi, 541100, People’s Republic of China, Email [email protected] Wei-Yi Gong, Department of Pain Management, Affiliated Hospital of Guilin Medical University, Guilin, 541000, Guangxi, People’s Republic of China, Email [email protected]
Background: Neuropathic pain (NP) after spinal cord injury (SCI-evoked NP) is clinically challenging; the underlying mechanisms are not fully understood, leading to a lack of promising treatment options. NP occurs in only a subset of patients with SCI. The injured spinal cord exhibits a series of histopathological changes, and the complement system has been shown to play an important role in these processes. In addition, NMDA receptor subunit 2B (NR2B) is involved in the development and maintenance of NP. This preliminary study was performed to investigate the correlations of the complement receptor 3/complement component 3 (CR3/C3) pathway and NR2B with SCI-evoked NP.
Methods: A trauma-induced SCI animal model was established and SCI-evoked NP was evaluated by behavioural analysis. Transcriptome analysis was performed to identify genes in the CR3/C3 pathway related to synaptic modification, while the expression and distribution of NR2B in the injured spinal cord, and the relation to NP, were examined by immunohistochemical analysis.
Results: Nine of seventeen SCI rats (52.9%) developed NP. C3 mRNA expression was significantly decreased in SCI-evoked NP rats and significantly increased in the non-NP SCI rats. C1q mRNA and CR3 mRNA expression were significantly increased in all SCI rats, but higher levels of expression were observed in the non-NP SCI rats. NR2B mRNA expression was significantly increased in the SCI-evoked NP rats and significantly decreased in the non-NP SCI rats. In addition, significantly elevated expression of NR2B-positive cells was seen in lamina II of the superficial dorsal horn in SCI-evoked NP rats in comparison with non-NP SCI rats.
Conclusion: NP occurred in only a subset of SCI rats, and the CR3/C3 pathway and NR2B were involved in SCI-evoked NP. Further studies are required to determine the mechanisms underlying the SCI-evoked NP associated with the CR3/C3 pathway and NR2B.
Keywords: spinal cord injury, neuropathic pain, complement receptor 3, complement component 3, NMDA receptor 2B subunit
Introduction
Spinal cord injury (SCI) results in a serious catastrophic events that leave patients with lifelong disabilities, including neuropathic pain (NP). An estimated 53% of SCI patients will develop NP,1,2 which adversely affects quality of life. Studies regarding the efficacy of current treatments, including medications, brain stimulation and biofeedback techniques, have yielded inconclusive results.3–7 Therefore, management of SCI-evoked NP is still a major clinical challenge.
A number of possible mechanisms of NP have been proposed, including peripheral, spinal and supraspinal mechanisms,8 but the mechanisms underlying SCI-evoked NP remain unclear. In addition, it is unclear why SCI-evoked NP occurs in only a subset of patients. Pathological changes, including abnormal synapses and synaptic dysfunction, are likely responsible for SCI-evoked NP. The injured spinal cord exhibits a series of histopathological changes, including acute injury, secondary injury and a glial scar surrounding and sequestering the damaged tissue. In the later phase of SCI, proliferating astrocytes, fibronectin and laminin deposition form the glial scar necessary for neural regeneration,9,10 which plays a key role in recovery of function following SCI. However, abnormal neural regeneration results in aberrant neural connectivity, which contributes to dysfunctions such as NP. In the healthy central nervous system (CNS), microglia-dependent synaptic pruning facilitates the construction of a normal neural network.11 Microglia engulf presynaptic inputs during peak retinogeniculate pruning, and engulfment is dependent upon neural activity and the microglia-specific phagocytic signalling pathway involving complement receptor 3/complement component 3 (CR3/C3).12,13 Furthermore, disrupting microglia-specific CR3/C3 signalling results in sustained deficits in synaptic connectivity. The N-methyl-
In this preliminary study, we established an animal model of trauma-induced SCI, evaluated NP after SCI by behavioural analysis, explored the expression of the key genes in the complement pathway related to synaptic modification and the NR2B gene related to NP by transcriptome analysis, and investigated the expression and distribution of the NR2B subunit in the spinal cord using immunohistochemical techniques. The results provide a basis for further research and will facilitate the development of therapeutic strategies for SCI-evoked NP.
Materials and Methods
Animals and Protocols
Sprague-Dawley rats weighing 250–350 g were housed under a 12-h light/dark cycle with controlled temperature and humidity (18°C–22°C, 50–60%), and were allowed free access to food and water. The experiments were conducted in accordance with institutional guidelines regarding the use of animals in research, and the protocol was approved by the Animal Care and Use Committee of Guilin Medical University (Approval No. 2019–0019).
All rats received health checks and were allowed to acclimatise to the laboratory conditions for 1 week before the experiments. Then, SCI models were established according to standard procedures.15 The Basso-Beattie-Bresnahan (BBB) Locomotor Rating Scale was used to confirm successful establishment of the SCI model,16 and the paw withdrawal threshold (PWT) was examined to evaluate SCI-evoked NP. Global transcriptome analysis was conducted to explore the target genes of the complement pathway and NR2B in association with SCI-evoked NP. In addition, immunofluorescence analysis was performed to determine the expression and distribution of NR2B in the superficial dorsal horn of the spinal cord of SCI-evoked NP and non-NP SCI rats. All behavioural measurements and morphological evaluations were conducted by experimenters blinded to the experimental protocol. The protocol is outlined in Figure 1.
Establishment and Assessment of SCI Model
Twenty rats were anaesthetised using 2–3% isoflurane mixed with air at a flow rate of 2 L/min. Laminectomy at T10 was performed and the spinal cord at T9 was exposed. A metal rod, 3 mm in diameter and weighing 10 g, was dropped from a height of 25 mm, and directly impacted the exposed spinal cord to create a contusive lesion. After contusion, the skin was closed with 4/0 sutures and the rat was placed in a heated cage to maintain the body temperature during recovery from anaesthesia. Manual abdominal massage and extrusion were performed twice daily until bladder reflex was established. The same surgical procedure was performed, but without injury, in three sham-operated rats. All rats received penicillin (30,000 U/kg, i.p. injection daily for 7 days) to prevent postoperative infection.
SCI rats were assessed using the BBB Locomotor Rating Scale, on which scores range from 0 (no observable locomotor movements) to 21 points (normal locomotor movements), to quantify multiple aspects including joint movement, toe clearance, paw placement, forelimb-hindlimb coordination, stepping, tail position and trunk stability.16 Before the tests, the rats were placed in a transparent plexiglass box (100 cm × 100 cm × 30 cm) with a pasteboard-covered anti-skid floor. The rats were checked individually for a total of 4 min by two observers before surgery. A BBB score of 0–2 for at least 3 days following lesion creation was considered to indicate successful SCI.
Pain Behaviour Test
The rats with successful SCI were evaluated repeatedly by the BBB Locomotor Rating Scale, at 7 days after SCI, and the PWT test17 was performed in rats with a BBB score > 8 at 7, 14, 21 and 28 days after SCI. At 28 days, rats with hyperalgesia were classified into the SCI-evoked NP group, while the others were classified into the non-NP SCI group.
PWT can be measured by the up-down method, using a calibrated von Frey hair to determine the threshold for sensing mechanical touch/pressure (range: 0.02–300 g).18,19 During testing, rats were restrained in a transparent plexiglass box (20 cm × 10 cm × 15 cm) with a wire mesh bottom, which allowed full access to the hind paw. The von Frey hair was pushed onto the skin of both hind paws until the filament bent. Each von Frey hair was applied to the mid-plantar hind paw for 2–3 s, with a 60-s interval between stimuli. The middle weight von Frey hair 2.0 (4.31) was first applied to the hind paw. A positive response was denoted by sharp withdrawal and/or licking of the test area immediately upon removal of the hair. If an ambiguous response occurred, the stimulus was repeated. When a positive response to a stimulus occurred, the next-smallest von Frey hair was applied. If an ambiguous response repeatedly occurred, the next-largest von Frey hair was applied. Testing was continued for five stimuli after the first response; the up-down method requires six responses in the immediate vicinity of the 50% von Frey threshold. A decrease of two steps was considered to indicate cutaneous hyperalgesia in this study.20
RNA-Seq Analysis
To elucidate the gene expression patterns, RNA-Seq analysis was performed in nine rats (SCI-evoked NP rats, non-NP SCI rats and sham rats, n = 3/group). Rats were anaesthetised (sodium pentobarbital, i.p., 50 mg/kg), and their spinal cords (T9) were excised and snap frozen in liquid nitrogen, and then subjected to RNA-Seq assay for transcriptome analysis. Briefly, 3 μg of total RNA was isolated and quantified, and reverse-transcribed into cDNA; adapters were ligated to each end of the cDNA. Sequencing was performed on the Illumina HiSeq™ platform (Illumina, San Diego, CA, USA), and then aligned to a reference genome database or assembled to obtain a genome-wide expression profile for further analysis. cDNA library construction and Illumina sequencing of these samples were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The relative NR2B, CR3, C3 and C1q mRNA expression levels in the spinal cord of rats were obtained via RNA-Seq analysis.
Immunohistochemistry
Rats were anaesthetised (sodium pentobarbital, i.p., 50 mg/kg) and perfused intracardially with 0.1 M phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde. Specimens were obtained 0.5 cm rostral to the injured spinal cord, at the epicentre of the spinal cord lesion, and 0.5 cm caudal to the injured spinal cord; they were then embedded in paraffin, cut into serial sections 3 μm thick and collected on polylysine-coated slides. The sections were then deparaffinised, rehydrated, subjected to antigen retrieval and finally subjected to immunohistochemical assay. Sections were incubated with primary rabbit monoclonal anti-NR2B antibody (ab65783; Abcam, Cambridge, UK) at 1:1000 dilution, and then with secondary antibody (Alexa Fluor 488-labeled goat anti-rabbit IgG).
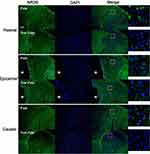
All sections were imaged at a magnification of 10×, with blue DAPI staining indicating cell nuclei and green Alexa Fluor 488 indicating NR2B. The photomicrographs were saved as TIF files and analysed quantitatively using ImageJ software (NIH, Bethesda, MD, USA). The NR2B-positive cells and total cells in laminin I–IV were measured in a blinded manner.
Data Presentation and Statistical Analysis
Statistical analysis was performed using SPSS software (version 22.0; SPSS Inc., Chicago, IL, USA). All data were tested for normality using the Kolmogorov–Smirnov test with Lilliefors correction, and for homogeneity of variance by Levene’s test. Differences between two groups of variables were analysed by Student’s t-test. Data are presented as the mean ± standard error of the mean (SEM). In all analyses, p < 0.05 was taken to indicate statistical significance.
Results
NP Only Occurred in a Subset of SCI Rats
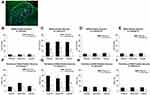
Based on the standard BBB score < 3 at 1–3 days and > 8 at 7 days after SCI, two SCI rats were excluded because the BBB score was > 3 at 1–3 days; another rat died after surgery. Finally, 17 SCI rats were included in the present study, 9 (52.9%) of which exhibited hyperalgesia associated with SCI. The body weight, BBB scores and PWT test results are shown in Figure 2.
Distinct Expression of C1q, C3, CR3 and NR2B Between SCI-Evoked NP and Non-NP SCI Rats
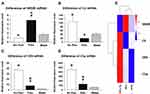
The RNA-Seq assay was performed to elucidate gene expression patterns. The fragments per kilobase of exon per million mapped fragments (FPKM) distribution and cluster analysis of differently expressed genes showed marked differences in gene expression levels among SCI-evoked NP rats, non-NP SCI rats and sham rats (Figure 3A and B). Further analysis identified a total of 4117 differentially expressed genes (DEGs), including 1729 that were upregulated and 2388 that were downregulated in SCI-evoked NP rats (Figure 3C). Venn diagram analysis identified 2633 DEGs between SCI-evoked NP and non-NP SCI rats; 1,304,41 and 1814 DEGs were specifically detected in SCI-evoked NP rats, non-NP SCI rats and sham rats, respectively, indicating different transcriptional changes among the three groups (Figure 3D). Gene Ontology (GO) analysis of DEGs between SCI-evoked NP and non-NP SCI rats was performed; the top 30 are listed in Figure 3E, including 11 associated with biological processes and 19 associated with cellular components. We also examined C1q, C3, CR3 and NR2B mRNA expression levels in spinal cord tissue. The data showed significant differences in the levels of NR2B, CR3, C3 and C1q transcripts among SCI-evoked NP rats, non-NP SCI rats and sham rats (Figures 4A–E).
Significant Increase in NR2B-Positive Cells in Lamina II of the Spinal Cord from SCI-Evoked NP Rats
Immunochemical analysis was performed to examine the expression and distribution of NR2B in the spinal cord of SCI rats. Representative images of NR2B expression in the spinal cord are shown in Figure 5. NR2B-positive cells were mainly present in lamina II of the superficial dorsal horn from SCI-evoked NP rats, with a few NR2B-positive cells scattered in the other laminae of the superficial dorsal horn from SCI-evoked NP rats and the whole superficial dorsal horn from non-NP SCI rats. The number and ratio of NR2B-positive cells were significantly higher in lamina II of the superficial dorsal horn from SCI-evoked NP than non-NP SCI rats, but there was no significant difference in the number or ratio of NR2B-positive cells in laminae I, III or IV between the two groups (Figure 6A–I).
Discussion
The occurrence of SCI-evoked NP in a subset of cases of SCI was investigated in an animal model mimicking clinical NP after SCI. Nine of the total of seventeen SCI rats (52.9%) showed NP; these animals also showed differential expression of C3, C1q, CR3 and NR2B mRNAs in the injured spinal cord, which may be involved in SCI-evoked NP. In addition, the number of NR2B-positive cells was significantly increased in lamina II of the superficial dorsal horn from the spinal cord of SCI-evoked NP rats in comparison to non-NP SCI rats.
Different C3, C1q and CR3 mRNA Expression Levels Have Important Implications for SCI-Evoked NP
Our data indicated differential expression of C3 mRNA between SCI-evoked NP rats and non-NP SCI rats, with significant upregulation of C3 mRNA seen in non-NP SCI rats. C3 is an immune protein cleaved by C3 convertase, and its breakdown products participate in the classical, alternative and lectin pathways of complement activation. Liu et al investigated gene expression in the injured spinal cord at 1 week after SCI, and demonstrated 36% upregulation of C3 mRNA expression.21 C3 exhibits neurotoxicity in various neurological diseases. In mouse models of amyloidosis and tauopathy, deletion of C3 rescues synapse loss, ameliorates neuron loss and brain atrophy, and improves neurophysiological and behavioural parameters.22 Aged mice exhibit region-specific and age-dependent synapse loss, which was not observed in C3 knockout mice.23 Mice deficient in C3 (C3−/−) showed reduced loss of synaptic terminals from injured motoneurons at 1 week after spinal nerve transection.24 Peterson et al investigated the potential role of C3 in the modulation of axon regeneration and neuronal survival after SCI. A twofold increase in sensory axon regeneration was observed in the spinal cord of C3−/− mice in comparison with wild-type controls.25 In vitro, the addition of C3 tripled both myelin-mediated neurite outgrowth inhibition and neuron loss versus myelin alone.25 Therefore, abnormalities of C3 quality and function have detrimental effects on synapse regeneration.
C1q plays an important role in synaptic pruning during CNS development, and is a causative factor in NP.26,27 Previous experimental studies demonstrated failure of synaptic pruning in mice deficient in C1q, and also that tagging of synapses by C3 is dependent on the upstream C1q. In C1q knockout mice proper synaptic connectivity cannot be established, and spontaneous epileptiform activity occurs.28 The absence of synaptic pruning can also result in lengthened dendrites, increased branching, and more dense dendritic spines.29 In the present study, upregulation of C1q mRNA was observed in all SCI animals, but the increase was greater in non-NP SCI than SCI-evoked NP rats. C1q may be involved in the synaptic pruning disorder of microglia associated with SCI-evoked NP, but further studies are required to clarify the underlying mechanisms.
The present study also demonstrated upregulation of CR3 mRNA in all SCI animals, although the level of expression was higher in non-NP SCI than SCI-evoked NP rats. CR3 is a heterodimer of α (CD11b) and β (CD18) transmembrane glycoproteins, which are mainly expressed in macrophages, monocytes, granulocytes and natural killer (NK) cells. CR3 plays prominent roles in the removal of invading pathogens and cell debris, and the induction of immune tolerance and synaptic pruning, and is also involved in the pathogenesis of numerous autoimmune and chronic inflammatory diseases. Widespread C3aR1 staining was observed at and around the site of SCI.30 At 35 days after SCI, when the lesion site is dominated by macrophages and a glial scar,31 C3aR1 expression remained high in GFAP+ astrocytes and Iba1+ microglia/macrophages.30
The differential levels of C3, C1q and CR3 mRNA expression in SCI-evoked NP and non-NP SCI rats in the present study suggest that the deficiency of C3 may be directly involved in the reduced synaptic pruning of microglia that contributes to the abnormal neural regeneration and neural connectivity associated with SCI-evoked NP. Synaptic pruning is an important function of microglia in CR3/C3-dependent signalling.32 CR3 is a phagocytic receptor expressed on the surface of microglia, and C3 is its ligand (localised to synaptically enriched regions). Microglia migrate to C3-enriched regions via CR3/C3-dependent signalling, and then engulf or prune immature synapses undergoing synaptic pruning. In contrast, deficiency of C3 and/or abnormal synaptic pruning of microglia result in an abundance of unmodified synapses that are enriched in the injured spinal cord.
Increased NR2B in Lamina II of Injured Spinal Cord is Implicated in SCI-Evoked NP
The results of the present study also demonstrated significantly increased NR2B mRNA expression in SCI-evoked NP rats, while non-NP SCI rats showed significantly reduced NR2B mRNA expression. In addition, NR2B-positive cells were significantly increased in lamina II of the superficial dorsal horn from the spinal cord of SCI-evoked NP rats. The NMDAR subunits are differentially distributed in the CNS, which may have significant functional implications. In the dorsal horn of the spinal cord, only NR2B was localised in laminae II–III, and NR2A and NR2C were not detected.33 Lamina II, which is dominated by nociceptive input from C fibre nociceptors and Aδ nociceptors, consists of excitatory glutamatergic interneurons. In addition, NR2B receptors are located primarily extrasynaptically.34,35 Activation of NR2B receptors by extrasynaptic glutamate would have a major influence on pain transmission in chronic pain states.36,37
Accumulating evidence suggests that NR2B, but not NR2A, is particularly important for the development and perception of persistent pain.38 Coincident with the allodynic phase after SCI, there was a significant increase in the NR2B mRNA level at 28 days, but not at 1 or 14 days.39 In the hemisection model, the nociceptive threshold and NR2B expression in the spinal cord were significantly increased; these effects were reversed by NR2B antagonists, including ifenprodil and Ro25-6981.40 Conantokin-G, an inhibitor of NR2B, was injected intrathecally into rats with spinal cord compression injury and dose-dependently attenuated nociceptive responses.41 Astaxanthin, a ketocarotenoid with anti-inflammatory effects, also reduced the expression of NR2B and improved NP following compression SCI.42 The present study also investigated the opposing changes in expression of NR2B mRNA in the injured spinal cord of SCI-evoked NP and non-NP SCI rats, and provided further evidence that the increased NR2B seen in lamina II of the injured spinal cord is involved in SCI-evoked NP.
Potential Mechanism of SCI-Evoked NP Involving the CR3/C3 Pathway and NR2B
The increased NR2B expression observed in lamina II of the injured spinal cord is a potential mechanism for the production of SCI-evoked NP, but is not the only such mechanism. Synapse regeneration, aberrant axonal connections and remodelling of neural circuits may be involved in SCI related-NP. In the late phase of SCI, nerve regeneration and remodelling of neural circuits are the foundation of neurofunctional recovery of the spinal cord. Normally, the balance between synaptic regeneration and synaptic pruning is crucial for axonal connections and neural circuits. However, impaired or supernumerary synapse regeneration within and outside of persistent scars may contribute to aberrant axonal connections and remodelling of neural circuits. Insufficient synaptic pruning of microglia because of a deficit of C3 may lead to an overabundance of synaptic connections. In contrast, excessive synaptic pruning of microglia induced by superfluous C3 and CR3 would result in synapse deficiency. The overabundance of synaptic connections caused by increased NR2B expression contributes to NP in a subset of SCI rats.
Conclusion
The present study suggested that NP only occurred in a subset of SCI rats. The genes involved in the CR3 pathway and encoding NR2B were differently expressed in the spinal cord of SCI-evoked NP and non-NP SCI rats, and a significant increase in the number of NR2B-positive cells was observed in lamina II of the spinal cord from SCI-evoked NP rats. We speculate that the CR3/C3 pathway and NR2B both contribute to SCI-evoked NP. Further studies are required to verify this hypothesis regarding the mechanism underlying SCI-evoked NP.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82060221), Guangxi Science and Technology Base and Talent Project (2019AC20329), Guangxi One Thousand Young and Middle-Aged College and University Backbone Teachers Cultivation Program.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Burke D, Fullen BM, Stokes D, Lennon O. Neuropathic pain prevalence following spinal cord injury: a systematic review and meta-analysis. Eur J Pain. 2017;21(1):29–44. doi:10.1002/ejp.905
2. Warner FM, Cragg JJ, Jutzeler CR, et al. Progression of neuropathic pain after acute spinal cord injury: a meta-analysis and framework for clinical trials. J Neurotrauma. 2019;36(9):1461–1468. doi:10.1089/neu.2018.5960
3. Song KS, Cho JH, Hong JY, et al. Neuropathic pain related with spinal disorders: a systematic review. Asian Spine J. 2017;11(4):661–674. doi:10.4184/asj.2017.11.4.661
4. Kumru H, Benito-Penalva J, Kofler M, Vidal J. Analgesic effect of intrathecal baclofen bolus on neuropathic pain in spinal cord injury patients. Brain Res Bull. 2018;140:205–211. doi:10.1016/j.brainresbull.2018.05.013
5. Brinzeu A, Berthiller J, Caillet JB, Staquet H, Mertens P. Ziconotide for spinal cord injury-related pain. Eur J Pain. 2019;23(9):1688–1700. doi:10.1002/ejp.1445
6. Karri J, Li S, Zhang L, Chen YT, Stampas A, Li S. Neuropathic pain modulation after spinal cord injury by breathing-controlled electrical stimulation (BreEStim) is associated with restoration of autonomic dysfunction. J Pain Res. 2018;11:2331–2341. doi:10.2147/JPR.S174475
7. Sato G, Osumi M, Morioka S. Effects of wheelchair propulsion on neuropathic pain and resting electroencephalography after spinal cord injury. J Rehabil Med. 2017;49(2):136–143. doi:10.2340/16501977-2185
8. Alles SRA, Smith PA, Isom LL. Etiology and pharmacology of neuropathic pain. Pharmacol Rev. 2018;70(2):315–347. doi:10.1124/pr.117.014399
9. Stroman PW, Khan HS, Bosma RL, et al. Changes in pain processing in the spinal cord and brainstem after spinal cord injury characterized by functional magnetic resonance imaging. J Neurotrauma. 2016;33(15):1450–1460. doi:10.1089/neu.2015.4257
10. Wang HF, Liu XK, Li R, et al. Effect of glial cells on remyelination after spinal cord injury. Neural Regen Res. 2017;12(10):1724–1732. doi:10.4103/1673-5374.217354
11. Bavencoffe A, Li Y, Wu Z, et al. Persistent electrical activity in primary nociceptors after spinal cord injury is maintained by scaffolded adenylyl cyclase and protein Kinase A and is associated with altered adenylyl cyclase regulation. J Neurosci. 2016;36(5):1660–1668. doi:10.1523/JNEUROSCI.0895-15.2016
12. Crown ED, Gwak YS, Ye Z, Johnson KM, Hulsebosch CE. Activation of p38 MAP kinase is involved in central neuropathic pain following spinal cord injury. Exp Neurol. 2008;213(2):257–267. doi:10.1016/j.expneurol.2008.05.025
13. Crown ED, Ye Z, Johnson KM, Xu GY, McAdoo DJ, Hulsebosch CE. Increases in the activated forms of ERK 1/2, p38 MAPK, and CREB are correlated with the expression of at-level mechanical allodynia following spinal cord injury. Exp Neurol. 2006;199(2):397–407. doi:10.1016/j.expneurol.2006.01.003
14. Xu F, Zhao X, Liu L, et al. Perturbing NR2B-PSD-95 interaction relieves neuropathic pain by inactivating CaMKII-CREB signaling. Neuroreport. 2017;28(13):856–863. doi:10.1097/WNR.0000000000000849
15. Vijayaprakash KM, Sridharan N. An experimental spinal cord injury rat model using customized impact device: a cost-effective approach. J Pharmacol Pharmacother. 2013;4(3):211–213. doi:10.4103/0976-500X.114607
16. Basso DM, Beattie MS, Bresnahan JC. A sensitive and reliable locomotor rating scale for open field testing in rats. J Neurotrauma. 1995;12(1):1–21. doi:10.1089/neu.1995.12.1
17. Nakae A, Nakai K, Yano K, Hosokawa K, Shibata M, Mashimo T. The animal model of spinal cord injury as an experimental pain model. J Biomed Biotechnol. 2011;2011:939023. doi:10.1155/2011/939023
18. Dixon WJ. Efficient analysis of experimental observations. Annu Rev Pharmacol Toxicol. 1980;20:441–462. doi:10.1146/annurev.pa.20.040180.002301
19. Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi:10.1016/0165-0270(94)90144-9
20. Leem JW, Willis WD, Weller SC, Chung JM. Differential activation and classification of cutaneous afferents in the rat. J Neurophysiol. 1993;70(6):2411–2424. doi:10.1152/jn.1993.70.6.2411
21. Liu S, Kang Y, Zhang C, et al. Isobaric Tagging for Relative and Absolute Protein Quantification (iTRAQ)-based quantitative proteomics analysis of differentially expressed proteins 1 week after spinal cord injury in a rat model. Med Sci Monit. 2020;26:e924266. doi:10.12659/MSM.924266
22. Wu T, Dejanovic B, Gandham VD, et al. Complement C3 is activated in human AD brain and is required for neurodegeneration in mouse models of amyloidosis and tauopathy. Cell Rep. 2019;28(8):2111–2123 e2116. doi:10.1016/j.celrep.2019.07.060
23. Shi Q, Colodner KJ, Matousek SB, et al. Complement C3-deficient mice fail to display age-related hippocampal decline. J Neurosci. 2015;35(38):13029–13042. doi:10.1523/JNEUROSCI.1698-15.2015
24. Berg A, Zelano J, Stephan A, et al. Reduced removal of synaptic terminals from axotomized spinal motoneurons in the absence of complement C3. Exp Neurol. 2012;237(1):8–17. doi:10.1016/j.expneurol.2012.06.008
25. Peterson SL, Nguyen HX, Mendez OA, Anderson AJ. Complement protein C3 suppresses axon growth and promotes neuron loss. Sci Rep. 2017;7(1):12904. doi:10.1038/s41598-017-11410-x
26. Cho K. Emerging roles of complement protein C1q in neurodegeneration. Aging Dis. 2019;10(3):652–663. doi:10.14336/AD.2019.0118
27. Asano S, Hayashi Y, Iwata K, et al. Microglia-astrocyte communication via C1q contributes to orofacial neuropathic pain associated with infraorbital nerve injury. Int J Mol Sci. 2020;21(18):6834. doi:10.3390/ijms21186834
28. Chu Y, Jin X, Parada I, et al. Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc Natl Acad Sci U S A. 2010;107(17):7975–7980. doi:10.1073/pnas.0913449107
29. Ma Y, Ramachandran A, Ford N, Parada I, Prince DA. Remodeling of dendrites and spines in the C1q knockout model of genetic epilepsy. Epilepsia. 2013;54(7):1232–1239. doi:10.1111/epi.12195
30. Brennan FH, Jogia T, Gillespie ER, et al. Complement receptor C3aR1 controls neutrophil mobilization following spinal cord injury through physiological antagonism of CXCR2. JCI Insight. 2019;4(9). doi:10.1172/jci.insight.98254
31. Brennan FH, Cowin GJ, Kurniawan ND, Ruitenberg MJ. Longitudinal assessment of white matter pathology in the injured mouse spinal cord through ultra-high field (16.4 T) in vivo diffusion tensor imaging. Neuroimage. 2013;82:574–585. doi:10.1016/j.neuroimage.2013.06.019
32. Schafer DP, Lehrman EK, Kautzman AG, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74(4):691–705. doi:10.1016/j.neuron.2012.03.026
33. Yung KK. Localization of glutamate receptors in dorsal horn of rat spinal cord. Neuroreport. 1998;9(7):1639–1644. doi:10.1097/00001756-199805110-00069
34. Zhou X, Ding Q, Chen Z, Yun H, Wang H. Involvement of the GluN2A and GluN2B subunits in synaptic and extrasynaptic N-methyl-D-aspartate receptor function and neuronal excitotoxicity. J Biol Chem. 2013;288(33):24151–24159. doi:10.1074/jbc.M113.482000
35. Pal B. Involvement of extrasynaptic glutamate in physiological and pathophysiological changes of neuronal excitability. CMLS. 2018;75(16):2917–2949. doi:10.1007/s00018-018-2837-5
36. Petrenko AB, Yamakura T, Baba H, Sakimura K. Unaltered pain-related behavior in mice lacking NMDA receptor GluRepsilon 1 subunit. Neurosci Res. 2003;46(2):199–204. doi:10.1016/S0168-0102(03)00061-0
37. Vizi ES, Kisfali M, Lorincz T. Role of nonsynaptic GluN2B-containing NMDA receptors in excitotoxicity: evidence that fluoxetine selectively inhibits these receptors and may have neuroprotective effects. Brain Res Bull. 2013;93:32–38. doi:10.1016/j.brainresbull.2012.10.005
38. Guo W, Zou S, Guan Y, et al. Tyrosine phosphorylation of the NR2B subunit of the NMDA receptor in the spinal cord during the development and maintenance of inflammatory hyperalgesia. J Neurosci. 2002;22(14):6208–6217. doi:10.1523/JNEUROSCI.22-14-06208.2002
39. Labombarda F, Coronel MF, Villar MJ, Nicola AF, Gonzalez SL. Neuropathic pain and temporal expression of preprodynorphin, protein kinase C and N-methyl-D-aspartate receptor subunits after spinal cord injury. Neurosci Lett. 2008;447(2–3):115–119. doi:10.1016/j.neulet.2008.09.062
40. Kim Y, Cho HY, Ahn YJ, Kim J, Yoon YW. Effect of NMDA NR2B antagonist on neuropathic pain in two spinal cord injury models. Pain. 2012;153(5):1022–1029. doi:10.1016/j.pain.2012.02.003
41. Hama A, Sagen J. Antinociceptive effects of the marine snail peptides conantokin-G and conotoxin MVIIA alone and in combination in rat models of pain. Neuropharmacology. 2009;56(2):556–563. doi:10.1016/j.neuropharm.2008.10.008
42. Fakhri S, Dargahi L, Abbaszadeh F, Jorjani M. Astaxanthin attenuates neuroinflammation contributed to the neuropathic pain and motor dysfunction following compression spinal cord injury. Brain Res Bull. 2018;143:217–224. doi:10.1016/j.brainresbull.2018.09.011
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.