Back to Journals » Cancer Management and Research » Volume 11
Comparison of transcriptional profiles in human lymphocyte cells irradiated with 12C ion beams at 0–2.0 Gy
Authors Zhang RF, Dang XH, Zhang ZX, Yuan YY, Ren Y, Duan ZK , Zuo YH
Received 27 September 2018
Accepted for publication 16 January 2019
Published 22 March 2019 Volume 2019:11 Pages 2363—2369
DOI https://doi.org/10.2147/CMAR.S188959
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Video abstract presented by Zhang et al.
Views: 186
Ruifeng Zhang, Xuhong Dang, Zhongxin Zhang, Yayi Yuan, Yue Ren, Zhikai Duan, Yahui Zuo
China Institute for Radiation Protection, Taiyuan, Shanxi 030006, China
Objective: Heavy ions have contributed to tumor site-specific radiotherapy and are a major health risk for astronauts. The purpose of this study was to investigate the changes in gene expression in peripheral lymphocytes of cancer patients and astronauts exposed to 12C ions, and identify suitable molecular biomarkers for health monitoring. We also aimed to observe the effects of treatment and the level of damage, by comparing the transcriptional profiles of human lymphocyte cell lines exposed to 12C ion beams at doses of 0–2.0 Gy.
Materials and methods: A human lymphocyte cell line was irradiated with 12C ion beams at 0, 0.1, 0.5, and 2.0 Gy and transcriptional profiles were evaluated using the Agilent human gene expression microarray at 24 hours after irradiation. Differentially expressed genes were identified using a fold change of ≥2.0. Representative genes were further validated by RT-PCR. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed to determine the roles of differentially expressed mRNAs.
Results: Based on the microarray assays, 1,113 genes were upregulated and 853 genes were downregulated in human lymphocyte cells irradiated with 0.1 Gy 12C ion beams compared with the control group, 1,095 genes were upregulated and 1,220 genes were downregulated in cells irradiated with 0.5 Gy 12C ion beams, and 1,055 genes were upregulated and 1,356 genes were downregulated in cells irradiated with 2.0 Gy. A total of 504 genes were differentially expressed in all irradiated groups, of which 88 genes were upregulated and 416 genes downregulated. Most of these altered genes were related to the cell cycle, apoptosis, signal transduction, DNA transcription, repair, and replication. The expression differences were further confirmed by RT-PCR for a subset of differentially expressed genes.
Conclusion: Differentially expressed genes between treatment and control groups at 24 hours post-irradiation increased as the radiation dose increased; upregulated genes gradually decreased and downregulated genes increased. Our data indicated that 12C ion beams could repress a number of genes in a dose-dependent manner, which might lead to the failure of multiple cellular biological functions.
Keywords: 12C ion beams, human lymphocyte cell line, microarray assay, gene expression
Introduction
Heavy ions, unlike photons, form a peak of energy deposition at the end of their path (known as a Bragg peak), with a steep decrease thereafter. Owing to the narrow width of the Bragg peak, spread-out Bragg peaks have been devised to obtain broad and uniform dose distributions at tumor sites, with little energy deposition in surrounding tissues.1,2 Accordingly, treatment with heavy ions may be particularly beneficial in malignancies that are difficult or hazardous to treat with surgery.3 Heavy ion radiotherapy for cancer contributes to the trend in tumor site-specific radiotherapy. In addition, space radiation is a major health risk for astronauts.4 To protect against irradiation during space travel, studies of the hazards of energetic heavy particles are needed.
Therefore, studies of the biological effects of heavy ions have broad practical applications. Radiation influences cell molecules (including water molecules and biomacromolecules, such as DNA, RNA, and proteins), and the effect of radiation on DNA is particularly important.5 Cellular DNA damage is the ultimate biological consequence of radiation, and leads to mutation, cell death, senescence, and carcinogenesis.6 Microarray technology is one approach to comprehensively characterize genome-wide expression differences and changes in transcriptional profiles induced by ionizing radiation. The objective of our study was to explore the molecular biological effect of heavy ions using microarray technology and to identify molecular biomarkers of health monitoring and to observe the effects of treatment and the level of damage.
Materials and methods
Cell culture
The human lymphocyte cell line Peng-EBV (provided by the Kunming Institute of Zoology, Chinese Academy of Science) was used. Human lymphocyte cells were grown in MEM supplemented with 20% FBS (complete medium, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Irradiation with carbon ion beams
The human lymphocyte cell line was exposed to 12C ion beams at doses of 0, 0.1, 0.5, and 2.0 Gy, with a dose rate of 0.3–0.5 Gy/min, 0 Gy was not irradiated. Irradiation with 12C ion beams was performed at the China Institute of Atomic Energy in Lanzhou. In this study, 165 MeV/u 12C ion beams were used. Immediately after irradiation, the medium was replaced with fresh complete medium and cells were incubated for 24 hours at 37°C in a humidified atmosphere containing 5% CO2.
RNA extraction, quantification, and quality control
Total RNA was extracted from all samples using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The yield of RNA was determined using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific), and the integrity of RNA was determined using an Agilent 2,100 Bioanalyzer and RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA).
lncRNA and mRNA microarray expression profiling
A whole genome expression analysis was performed by Shanghai OE Biotech Co., Ltd (Shanghai, China) according to the protocol for the one-color microarray-based gene expression analysis developed by Agilent Technologies. Sample labeling, microarray hybridization, and washing were performed according to the manufacturer’s standard protocols. Briefly, total RNAs were transcribed to double-stranded cDNA, synthesized into cRNA, and labeled with cyanine-3-CTP. The labeled cRNAs were hybridized onto the microarray. After washing, the arrays were scanned using the Agilent Scanner G2505C (Agilent Technologies).
Data analysis
Feature Extraction (version 10.7.1.1, Agilent Technologies) was used to analyze array images and obtain raw data. GeneSpring was employed for the basic data analysis. Raw data were normalized using the quantile algorithm. Differentially expressed genes were then identified based on fold change values, with a threshold fold change of ≥2.0. Afterward, a Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were used to determine the roles of these differentially expressed mRNAs. Finally, hierarchical clustering was performed to distinguish gene expression patterns among samples.
Reverse-transcriptase (RT) PCR
Differential gene expression between irradiation and control groups was verified for a subset of genes. Total RNA was extracted and reverse-transcribed to cDNA using reverse transcriptase (Takara, Kusatsu, Japan) and cDNA samples were subjected to real-time quantitative PCR using a Rotor-Gene 6000 PCR machine (Corbett Life Science, Mortlake, Australia). The relative expression values were normalized against the levels of internal control (β-actin). Primers are shown in Table 1.
  | Table 1 Primer sequences and amplification conditions for RT-PCR |
Statistical analysis
Data are expressed as mean and SDs. Independent sample t-tests were used to identify significant differences between control and treatment groups using SPSS 21.0. A P-value of <0.05 was considered statistically significant.
Results
Differential expression of mRNAs based on a microarray analysis
An optimized microarray platform was used to analyze and compare gene expression patterns between irradiated and control cells. In total, 504 mRNAs exhibited significantly different expression levels among the four groups (P≤0.05), including 88 upregulated and 416 downregulated genes (irradiated group vs control group). As shown in Table 2, the differentially expressed genes at 24 hours post-irradiation increased as the dose increased; upregulated genes gradually decreased, while downregulated genes increased. Further analysis of the differentially expressed genes indicated that the expression of some genes was dose-dependent, as shown in Tables 3 and 4.
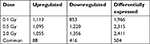  | Table 2 Differentially expressed genes in human lymphocyte cell line irradiated by 1 2C ions |
  | Table 3 Downregulated genes with dose-dependent expression |
  | Table 4 Upregulated genes with dose-dependent expression |
GO and KEGG pathway analysis
In the GO analysis, we found that in the 0.1 Gy dose group, the differentially expressed genes included a total of 580 altered genes that were involved in 185 biological process (BP) classifications, mainly including cell growth and development, cell differentiation, ion transport, cell adhesion, nervous system growth, and development. A total of 782 differentially expressed genes of the 0.5 Gy group were involved in 103 BP classifications, including transmembrane transport, ion transport, nervous system growth and development, positive correlation regulation of cell proliferation, prominent transmission. There were 802 differentially expressed genes in the 2.0 Gy group involved in 95 BP classifications, which were associated with regulation of DNA replication, signal transduction, protein phosphorylation, intracellular signaling, regulation of small GTPase signaling, the initiation of apoptosis, cell cycle arrest, and DNA damage-stimulated responses. Then we compared the GO results for differentially expressed genes with a dose-dependent relationship to 12C ion beam exposure. The results indicated that the major functions of upregulated genes with a dose-dependent relationship were transport, protein phosphorylation, and negative regulation of transcription chloride transport. The primary functions of downregulated genes were cell differentiation, ATP catabolic process, neuron–neuron synaptic transmission, and anterograde axon cargo transport, as shown in Figure 1.
The KEGG pathway analysis revealed that 83 differentially expressed genes exposed to 0.1 Gy 12C ion beams were associated with nine pathways, 99 differentially expressed genes at 0.5 Gy were associated with eight pathways, and 119 differentially expressed genes at 2.0 Gy were associated with ten pathways, as shown in Table 5. Table 6 shows five enrichment pathways for genes with dose-dependent relationships and the genes involved in the pathways.
  | Table 5 Pathways of differentially expressed genes after irradiation by 12C ions |
  | Table 6 Pathways of genes with dose-dependent relationships |
Validation by real-time quantitative PCR
To confirm the microarray results, qRT-PCR was performed for three randomly selected differentially expressed genes: ABCG1, APOL6, and LY6G5C. The qRT-PCR results were consistent with those of the microarray analysis; the three genes were differentially expressed with the same trends (upregulated or downregulated), as shown in Table 7.
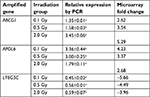  | Table 7 Validation of microarray results for three randomly chosen genes by qRT-PCR Note: aP<0.05, statistically significant. |
Discussion
Radiation damage is complex, involving interactions among multiple genes. Many regulatory genes are not independent factors, but are involved in networks of interacting genes. Microarray assay can be used to understand the role of radiation-induced genes in related functions, such as regulation of cell cycle, apoptosis and DNA repair. In our study, at a lower radiation dose (0.1 Gy), differential genes are mainly involved in cell growth and development, and cell differentiation. At 0.5 Gy, differential genes primarily include transmembrane transport, ion transport, nervous system growth and development, and positive correlation regulation of cell proliferation. There were additional differential genes associated with cell cycle and apoptosis in the 2.0 Gy dose group, which was not evident in the 0.1 Gy and 0.5 Gy dose groups. Moreover, the pathways associated with differentially expressed genes differed among dose groups. Pathways in the low-dose irradiation group were mainly inflammatory response-related regulatory and immunoregulation pathways, Notably, in the high-dose group we observed the enrichment of cancer regulation, lupus erythematosus, apoptosis, and p53 signaling pathways. These results provided convincing evidence that the degree of repair of heavy ion damage was dose-related.
In Long Xianhui’s study,7 human lymphocytoblast cells were irradiated with γ rays at 2 Gy and 10 Gy, transcriptional profiles were evaluated using gene expression microarray at 4 hours after irradiation. The differential genes associated with cell cycle, apoptosis, and DNA damage that were consistent with our results. Haruo Fujinag8 compared the biological characteristics of gene expression features provided by proton and X-ray irradiation on the PC cell line, MIAPaCa-2. The results showed there were similar cytocidal and tumoricidal effects, but the altered gene expression profile of irradiated MIAPaCa-2 cells was different.
In the current study, we identified various genes with dose-dependent expression that were closely associated with the occurrence, development, and treatment of cancer. The downregulated gene CSGalNAcT1 not only functions in cell recognition but is also involved in chondroitin sulfate proteoglycan, heparin, and proteoglycan biosynthetic processes. These materials can increase the biosynthesis of mRNA and DNA, promote metabolism, have anti-inflammatory activity, accelerate wound healing, and exhibit antineoplastic effects. CSGalNAcT1 is deleted on chromosome eight in oral squamous cell carcinoma.9 In addition, Hunter et al10 showed that CSGalNAcT1 is associated with tumor prognosis and malignant plasma cells. CSGalNAcT1 is involved in the regulation of metabolism with antineoplastic function. Our data showed that the downregulation of CSGalNAcT1 in cells irradiated by 12C ions increased as the dose increased. The role of CSGalNAcT1 downregulation in response to radiotherapy should be evaluated in future research.
We also identified two upregulated genes with dose-dependent expression that were related to cancer. ABCG1 is a member of the superfamily of ATP-binding cassette (ABC) transporters, which are putative drug transporters.11 Tumor cells acquire resistance to chemotherapeutic drugs by the expression of various multidrug resistance genes, and cancer stem cells can effectively increase drug resistance by the upregulation of drug efflux transporter genes.12–14 Patients with tumors with high expression of various ABC transporter pumps usually do not respond to chemotherapy because the ABC transporters located on the cytoplasmic side of resistant cells will efflux chemotherapeutic drugs out of tumor cells.15 Additionally, recent reports have established that the upregulation of key ABC transporter genes (ABCG1 and ABCB1) imparts resistance to 5-FU in U87-MCSF cells.16 Our data showed that the expression level of ABCG1 in cells irradiated by 12C ions increased as the dose increased. Since the increased expression of various ABC transporters may lead to chemo-resistance, the expression levels of ABC transporters should be detected before treatment with both chemotherapy and heavy ion radiotherapy.
Another upregulated gene, APOL6, belongs to a family of programmed cell death genes encoding proteins that can initiate host apoptosis or autophagic death. Each APOL gene putatively contains a BH3 protein domain; BH3-only proteins function as upstream activators of programmed cell death, responding to various stimuli, such as cell detachment, cytokine withdrawal, or DNA damage, before initiating cell death.17 Cancer cells frequently possess defects in the genetic and biochemical pathways of apoptosis. The full-length cDNA of ApoL6 has been identified, cloned, and functionally expressed in p53-null colorectal cancer cells (DLD-1). Furthermore, adenovirus harboring the full-length cDNA of ApoL6 induces marked apoptosis in a variety of cancer cell types.18 It is unclear whether the increased expression of ApoL6 is related to the effect of heavy ion radiotherapy. It is reasonable to speculate that the dysregulation of ApoL6 derails apoptosis and causes tumorigenesis.19
Ionizing radiation can induce nuclear DNA damage, which in turn results in changes in transcription, translation, and other processes, thereby affecting gene expression. Understanding the gene expression changes in response to heavy ion radiation has broad practical applications. More importantly, it can clarify the tumor responses to therapy at the molecular level, enabling the development of individualized treatment plans for patients.
Conclusion
The results of our study indicated that 12C ion beams could repress a number of genes in a dose-dependent manner, which might lead to the failure of multiple cellular biological functions. In the future, we aim to verify the functions of these genes for cancer patients, to observe the effect of treatment and the level of damage.
Disclosure
The authors report no conflicts of interest in this work.
References
Hamada N, Imaoka T, Masunaga S, et al. Recent advances in the biology of heavy-ion cancer therapy. J Radiat Res. 2010;51(4):365–383. | ||
Durante M, Orecchia R, Loeffler JS. Charged-particle therapy in cancer: clinical uses and future perspectives. Nat Rev Clin Oncol. 2017;14(8):483–495. | ||
Fokas E, Kraft G, An H. Engenhart-Cabillic R ion beam radiobiology and cancer: time to update ourselves. Biochim Biophys Acta. 2009;1796(2):216–229. | ||
Hellwig CE, Spitta LF, Koch K, et al. The role of the nuclear factor κB pathway in the cellular response to low and high linear energy transfer radiation. Int J Mol Sci. 2018;19(8):1–43. | ||
Shu-Zheng L. The effect of ionizing radiation on chromosomes. In: Medical Radiobiology. Beijing: Atomic Energy Press; 2006:98. | ||
Noda A. Radiation-induced unrepairable DSBs: their role in the late effects of radiation and possible applications to biodosimetry. J Radiat Res. 2018;59(suppl_2):ii114–ii120. | ||
Long XH, Xu QZ, He XP, et al. Comparison of transcription profiles between 2 Gy and 10 Gy irradiated human lymphoblastoid cells. China J Radiol Med Prot. 2006;26(2):110–113. | ||
Fujinaga H, Sakai Y, Yamashita T, et al. Biological characteristics of gene expression features in pancreatic cancer cells induced by proton and X-ray irradiation. Int J Radiat Biol. 2018:1–44. | ||
Yong ZW, Zaini ZM, Kallarakkal TG, et al. Genetic alterations of chromosome 8 genes in oral cancer. Sci Rep. 2014;4:1–9. | ||
Hunter AM, Leuchter AF, Power RA, et al. A genome-wide association study of a sustained pattern of antidepressant response. J Psychiatr Res. 2013;47(9):1157–1165. | ||
Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775(2):237–262. | ||
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. | ||
Hussein I, Waheed S, Ahmad KA, et al. Scutellaria baicalensis targets the hypoxia-inducible factor-1α and enhances cisplatin efficacy in ovarian cancer. J Cell Biochem. 2018;119(9):1–10. | ||
da Costa KM, Valente RC, Salustiano EJ, et al. Functional characterization of ABCC proteins from Trypanosoma cruzi and their involvement with thiol transport. Front Microbiol. 2018;9:1–19. | ||
Wu Q, Yang Z, Xia L, et al. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget. 2014;5(22):1–12. | ||
Chockalingam S, Ghosh SS. Amelioration of cancer stem cells in macrophage colony stimulating factor-expressing U87MG-human glioblastoma upon 5-fluorouracil therapy. PLoS One. 2013;8(12):e83877. | ||
Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–858. | ||
Liu Z, Lu H, Jiang Z, Pastuszyn A, Hu CA. Apolipoprotein L6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol Cancer Res. 2005;3:21–31. | ||
Aryee DN, Niedan S, Ban J, et al. Variability in functional p53 reactivation by PRIMA-1(Met)/APR-246 in Ewing sarcoma. Br J Cancer. 2013;109(10):2696–2704. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

