Back to Journals » Infection and Drug Resistance » Volume 13
Comparison of in vitro Susceptibility of Mycobacteria Against PA-824 to Identify Key Residues of Ddn, the Deazoflavin-Dependent Nitroreductase from Mycobacterium tuberculosis
Authors Zhang F, Li S , Wen S, Zhang T, Shang Y , Huo F, Xue Y, Li L, Pang Y
Received 1 December 2019
Accepted for publication 23 January 2020
Published 11 March 2020 Volume 2020:13 Pages 815—822
DOI https://doi.org/10.2147/IDR.S240716
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Fuzhen Zhang, 1, 2,* Shanshan Li, 2,* Shuan Wen, 2 Tingting Zhang, 2 Yuanyuan Shang, 2 Fengmin Huo, 2 Yi Xue, 2 Ling Li, 1, 3 Yu Pang 2
1Biosafety Level 3 Laboratory, School of Public Health, Southern Medical University, Guangzhou, People’s Republic of China; 2National Clinical Laboratory on Tuberculosis, Beijing Key Laboratory on Drug-Resistant Tuberculosis Research, Beijing Chest Hospital, Capital Medical University, Beijing Tuberculosis and Thoracic Tumor Institute, Beijing, People’s Republic of China; 3Experimental Teaching Center, School of Basic Medical Science, Southern Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yu Pang
Beijing Chest Hospital, Capital Medical University, No. 97, Machang, Tongzhou District, Beijing 101149, People’s Republic of China
Tel/Fax +86 10 8950 9359
Email [email protected]
Ling Li
Southern Medical University, No. 1023-1063, Shatai South Road, Baiyun District, Guangzhou 510515, People’s Republic of China
Tel +86 18680280210
Email [email protected]
Objective: PA-824 (Pretomanid), a bicyclic nitroimidazole drug, exhibits significant bactericidal activity toward Mycobacterium tuberculosis (MTB) in vitro and in vivo, but not against Mycobacterium smegmatis. Through catalytic bioreduction, deazaflavin-dependent nitroreductase (Ddn) within MTB directly converts PA-824 to potent bactericidal products. This study aimed to identify key MTB Ddn residues involved in PA-824 conversion toward development of in vitro surrogate markers for detection of mycobacterial resistance to PA-824.
Methods: We evaluated in vitro activity of PA-824 toward MTB and nontuberculous mycobacterial species using antimicrobial susceptibility testing. Ddn amino acid sequence alignments and phylogenetic analysis revealed putative key enzyme active site residues. Candidate MTB Ddn residues required for PA-824 conversion activity were evaluated for loss-of-function using recombinantly cloned Ddn mutant proteins expressed in Mycobacterium smegmatis.
Results: PA-824 minimum inhibitory concentrations of 90% of bacterial growth (MIC 90s) against MTB and Mycobacterium kansasii were 0.12 mg/L and 8 mg/L, respectively, but > 32 mg/L for Mycobacterium spp. M. avium, M. intracellulare, M. abscessus and M. fortuitum. MTB Ddn and M. kansasii Ddn homologous sequences shared the greatest similarity (89.3% amino acid identity). M. smegmatis expressing Ddn proteins with Y65L, A76V or Y133F substitutions (but not V75L, Q125K or V148I) were resistant to PA-824.
Conclusion: Our data demonstrated that PA-824 exhibited excellent and moderate levels of in vitro activity against MTB and M. kansasii, respectively. Substitutions of Ddn residues Y65, A76 or Y133 conferred mycobacterial resistance to PA-824.
Keywords: mycobacteria, PA-824, susceptibility, Ddn
Introduction
Tuberculosis (TB) remains a major global health threat, especially to individuals infected with drug-resistant Mycobacterium tuberculosis (MTB). In response to changing environmental influences and widespread anti-tuberculosis drug use, MTB adaptation involves processes that generate mutations to safeguard genetic information,1 perpetuating the emergence of multidrug-resistant tuberculosis (MDR-TB) cases. According to the World Health Organization (WHO) Global Tuberculosis Report 2019, 0.484 million incident MDR-TB cases were detected in 2018, of which 6.2% were extensive drug resistance (XDR) infection cases.2 Therefore, research efforts to discover new anti-mycobacterial drugs have accelerated in recent years. PA-824 (Pretomanid), a promising bicyclic nitroimidazole drug, has shown a significant inhibitory effect toward MTB in vitro and in vivo,3,4 with activity against both replicating and nonreplicating MTB.3 Currently, PA-824, a member of a new class of anti-tuberculosis chemotherapy drugs (nitroimidazoles), has been approved by the US Food and Drug Administration for use in the treatment of non-responsive multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) cases within the general population.5
Two mechanisms involved in anti-MTB activity of PA-824 include the inhibition of cell wall formation (by blocking mycolic acid biosynthesis) and respiratory poisoning (as shown in a microarray study6), both of which abrogate MTB growth.3 Meanwhile, F420-dependent reduction of PA-824 has been shown to produce a mixture of three metabolites, including two reactive nitrogen species and one stable product. In addition, PA-824 itself is converted by a cofactor F420-dependent deazaflavin nitroreductase (Ddn) to toxic products, including nitrous oxide (NO) and others, that accumulate within bacteria and obstruct normal electron flow and homeostasis even during bacterial latency.7 F420 synthesis requires FbiA, FbiB and FbiC activities, while redox cycling of F420 requires functional F420-dependent glucose-6-phosphate dehydrogenase (Fgd1 or FGD).3,8,9 Loss of any of these five enzymes leads to high levels of resistance to PA-824. Ddn, F420 and Fgd1 are commonly produced by bacterial members of the Mycobacterium tuberculosis complex (MTBC), thus rendering members of this group susceptible to PA-824. Mycobacterium leprae, which lacks a Ddn-encoding gene, thus is naturally resistant to PA-824,10 as further evidence supporting an important role of Ddn in PA-824 activation. Meanwhile, multiple mutated sites within the gene encoding Ddn (ddn) have been identified in PA-824-resistant MTB isolates, suggesting that ddn mutations may be the major mycobacterial mechanism for the development of PA-824 resistance.11 Although some researchers have searched for Ddn active sites involved in catalysis of PA-824 via in vitro enzyme activity experiments,11,12 data generated by these efforts are still lacking because Ddn sequences are not highly conserved among mycobacteria.
Nontuberculous mycobacteria (NTM), a heterogeneous group of widely distributed environmental mycobacterial species, can cause various human illnesses in both immunocompetent and immunocompromised patients.13 NTM include M. avium, M. intracellulare, M. kansasii, M. abscessus and M. fortuitum as the most prevalent species. Although F420 is found in some mycobacteria,14,15 FGD is found only in species of Mycobacterium, Nocardia and in Gordonia amarae.16 Therefore, in this study, we evaluated in vitro activity and MIC values of PA-824 toward clinical strains of MTB and NTMs and conducted amino acid sequence alignments of MTB Ddn and its homologues of NTMs to reveal putative Ddn active sites. Expression of recombinant Ddn mutants derived from MTB Ddn in M. smegmatis, a mycobacterial species that naturally cannot convert PA-824 to antimycobacterial products, permitted identification of MTB Ddn active site residues required for enzymatic PA-824 conversion.
Materials and Methods
Mycobacterium Strains
A total of 227 mycobacterial isolates were randomly selected from the Tuberculosis BioBank maintained at the National Clinical Laboratory on Tuberculosis, Beijing Chest Hospital between 2010 and 2018. These isolates included 72 isolates of MTB, 22 isolates of M. avium, 34 isolates of M. intracellulare, 33 isolates of M. kansasii, 36 isolates of M. abscessus and 30 isolates of M. fortuitum. All mycobacterial strains were identified at the species level using multi-locus sequence analysis of 16S rRNA, hsp65, rpoB and 16S-23S rRNA internal transcribed spacer (ITS) gene sequences.13,17 M. smegmatis MC2 155 was obtained from our laboratory.
Antimicrobial Susceptibility Testing
The microplate alamarBlue assay (MABA), which is based on colorimetric conversion of alamarBlue reagent, was used to conduct MTB and NTM isolate growth measurements in the presence of PA-824.18 Pure PA-824 powder was synthesized and purified by HanXiang Biotech Co., Ltd. (Shanghai, China), with confirmation of the structure of the synthesized agent was confirmed using nuclear magnetic resonance-based analysis. Susceptibilities of MTB isolates to PA-824 were tested via the broth microdilution method using 7H9 broth with 10% Middlebrook oleic acid-albumin-dextrose-catalase complex (OADC), as previously described. Susceptibilities of NTM isolates to PA-824 were tested via a broth microdilution method using cation-adjusted Mueller-Hinton broth according to the guidelines from the Clinical and Laboratory Standards Institute (CLSI).19 Minimum inhibitory concentrations for inhibition of 90% of bacterial growth (MIC90 values) were calculated based on comparisons to bacterial growth in wells without PA-824. Bacterial cultures with or without PA-824 were incubated at 37°C, with cultures of rapidly growing mycobacteria (RGM) incubated for 3 days and cultures of MTB and slowly growing mycobacteria (SGM) incubated for 7 days. On the last day of culture, 70 μL of diluted alamarBlue solution was added to each well; then, plates were incubated for an additional 24 h at 37°C and color changes were read by visual inspection (as per manufacturer’s instructions provided with alamarBlue). Each MIC90 value was determined by identifying the well containing the lowest concentration of antimicrobial agent solution that did not change color from blue to pink. MIC90s were used for PA-824 drug sensitivity testing of M. smegmatis transformed with recombinant plasmids containing cloned inserts for achieving expression of MTB Ddn or mutant Ddn proteins in M. smegmatis. All experiments were performed in triplicate. For isolate tests producing discordant MIC results, experiments were repeated until three identical MIC values were obtained.
Multiple Sequence Alignments and Phylogenetic Analysis
Initially, we searched the protein database at the National Center for Biotechnology Information for MTB Ddn amino acid sequences. In addition, using a published MTB Ddn sequence (CCP46369.1) as query sequence, we performed searches of the RefSeq database using Blastp to obtain homologous amino acid sequences of Ddn from M. avium (WP_023864176.1), M. intracellulare (WP_064935907.1), M. kansasii (ORB84327.1), M. abscessus (EUA78268.1), M. fortuitum (WP_065019583.1) and M. smegmatis (WP_058127102.1). ClustalW software (http://www.clustal.org/) was used to align sequences and reconstruct a phylogenetic tree based on sequence similarities between MTB Ddn and its homologues of NTMs.
Cloning of MTB Ddn and Ddn Mutants
Ddn mutant sites were selected based on Ddn sequence alignment results. DNA samples of H37Rv ddn insert and no-load plasmid pMV261 were sent to TSINGKE Biotechnology Co., Ltd. (Beijing, China) for plasmid construction and attachment of 3×Flag-encoding sequences to MTB ddn sequences to add protein tags to C-termini of expressed Ddn proteins for use in protein immunoaffinity purification. In brief, ddn and mutated ddn DNA inserts were cloned into no-load plasmid pMV261 via homologous recombination then recombinant plasmids were transformed into E. coli DH5α competent cells that were subsequently cultured to generate suitable plasmid quantities needed for plasmid sequence confirmation.
Electroporation
Cultures of E. coli DH5α containing plasmid encoding Ddn or mutants were incubated for 12–16 hrs at 37°C with continuous shaking at 200 rpm. Recombinant plasmids were extracted using a plasmid extraction kit (Axygen, Union City, CA, USA). Electrocompetent M. smegmatis cells were prepared as previously described.20 Approximately 500 ng of plasmid was added to 200 µL of cell suspension and mixed by gentle pipetting then dispensed into a prechilled electroporation cuvette (with a 0.2 cm electrode gap) followed by incubation on ice. The Bio-Rad Gene Pulser used for electroporation was set at 2.5 kV, 1000 Ω and 25 μF. Cell suspensions were immediately diluted with 2 mL of 7H9 broth then transferred to 15 mL snap-cap plastic tubes and incubated at 37°C with shaking at 200 rpm for 2 h. Finally, cells were plated onto 7H10 agar containing 50 μg/mL kanamycin then incubated for 3–4 d at 37°C. Monoclonal M. smegmatis was selected for verification.
DNA Isolation, PCR and Sequencing
Bacterial suspensions of M. smegmatis containing MTB Ddn plasmid or mutant-Ddn plasmids were centrifuged at 12,000 rpm for 2 min then pellets were each resuspended in 200 μL of sterile water and heated in a 100°C water bath for 10 min. After centrifugation, supernatants were collected for use as templates for PCR amplification.
For PCR, DNA encoding the MTB Ddn open reading frame was amplified using the forward primer 5ʹ-CGGGATCCCGATGCCGAAATCACCGCCGCGGTTTC-3ʹ and reverse primer 5ʹ-CCCAAGCTTGGGTCAGGGTTCGCAAACCACGATCGGG-3ʹ. A 25 μL PCR reaction mixture was prepared containing 12.5 μL 2× Taq PCR Master Mix, 1 μL of DNA template, 0.5 μL of each primer and 10.5 μL of ddH2O. Amplification was conducted using an initial denaturation step at 95°C for 5 min followed by 35 amplification cycles of (denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec), with a final extension at 72°C for 10 min. PCR products were sequenced by Ruibio BioTech Company (Beijing, China).
Western Blot Analysis
Protein expression of Ddn and its mutants were verified by Western blotting using mouse anti-Flag antibody (#F3165, Sigma-Aldrich, 1:5000). M. smegmatis transformed with MTB Ddn plasmid or mutant plasmid cultures were centrifuged at 12,000 rpm for 2 min then pellets were each resuspended in 500 μL of 1× PBS buffer and 5 μL of MCE protease inhibitor “Cocktail”. After blending, ultrasonication was performed for 2 min using a low-frequency setting then cell lysates were prepared as previously described.21 Finally, an enhanced chemiluminescence detection system (iBright CL 1500 Imaging System; Thermo Fisher Scientific, Inc.) was used to detect protein levels using antibody specific for GroEL1 and rabbit anti-His (#12698s, Cell Signaling, 1:1000 for immunoblot analysis), with M. smegmatis GroEL1 used as an internal reference.22
Growth Curve Analysis
Wild-type M. smegmatis was cultured in liquid Middlebrook 7H9 with 0.2% glycerol and 0.05% Tween 80. For cultures of M. smegmatis transformed with Ddn plasmid or mutant plasmids, 50 μg/mL kanamycin was added. Optical density at 600 nm was measured every 12 h using an ultraviolet spectrophotometer until bacterial growth reached stationary stage. All experiments were performed in triplicate.
Results
MICs of PA-824 Against Mycobacterial Isolates
Comparisons of in vitro antibacterial activities of PA-824 against various mycobacterial isolates are summarized in Table 1. The MIC90 of PA-824 for MTB was 0.12 mg/L, with only two (2.78%) isolates possessing slightly higher MICs. For the major group of slowly growing mycobacteria, PA-824 showed poor inhibitory activity, with MIC90 values of >32 mg/L observed for M. avium and M. intracellulare isolates. By contrast, PA-824 showed moderately good inhibitory activity toward M. kansasii growth (MIC90 of 8 mg/L), with MICs for 42.42% of isolates (14 isolates) of ≤4 mg/L. Meanwhile, PA-824 MIC90 values against rapidly growing mycobacterial M. abscessus and M. fortuitum isolates were >32 mg/L, showing low growth inhibition. Notably, we observed that the MIC distribution of PA-824 inhibition of growth of M. fortuitum isolates differed from that of M. abscessus isolates, with MIC values ˂32 mg/L for 60% of M. fortuitum isolates (18 isolates) (Table 1).
 |
Table 1 MIC Distributions for Various Mycobacteria Species Against PA-824 |
Identification of MTB Ddn and Ddn Homologous Sequences of NTMs
As shown in Figure 1A, Ddn of M. tuberculosis shared 89.3% amino acid sequence identity with Ddn homologue of M. kansasii, 66.6% with that of M. avium and 63.8% with that of M. intracellulare, with low sequence similarity observed among Ddn homologous sequences of M. fortuitum, M. smegmatis and M. abscessus. As shown in Figure 1B, M. tuberculosis Ddn is more closely related to Ddn homologue of M. kansassii. However, M. avium and M. intracellulare Ddn homologues are closely related (with homology of 79.9%), M. fortuitum and M. smegmatis Ddn homologues are closely related (with homology of 81.5%), while the M. abscessus Ddn homologous sequence significantly differed from other mycobacterial Ddn homologous sequences. On the basis of sequence alignment results across mycobacterial species, we selected conserved sites between Ddn active site domains of M. tuberculosis and M. kansasii and excluded sequences from other mycobacterial species from this analysis. Ultimately, candidate Ddn residues Y65, V75, A76, Q125, Y133, V148 were analyzed in this study.
Growth of M. smegmatis Containing Plasmids Enabling Expression of MTB Ddn or Mutant Ddn Proteins
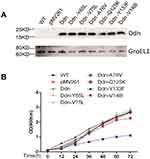
We introduced plasmids encoding MTB Ddn or mutant proteins into M. smegmatis, with recombinant plasmids constructed as indicated in Figure 2A. Next, we assessed whether the introduction of exogenous genes had any effect on the growth of M. smegmatis. As shown in Figure 2B, we found no change or only a slight decrease in growth rate in 7H9 broth for most recombinant strains as compared with their wild-type parents, except for M. smegmatis expressing Ddn-Y133F, which showed a significant decrease in growth rate compared with wild-type M. smegmatis.
MICs of M. smegmatis Expressing MTB Ddn or Ddn Mutant Proteins
Antibacterial activities of PA-824 against M. smegmatis recombinant strains expressing MTB Ddn mutant proteins are listed in Table 2. Bacterial sensitivity to PA-824 significantly increased after expression of MTB Ddn in M. smegmatis, with MIC decreasing from 384 mg/L to 24 mg/L (a 16-fold decrease). Notably, M. smegmatis expressing Ddn proteins with amino acid substitutions Y65L, A76V or Y133F exhibited high-level resistance to PA-824, with MIC90s of 384 mg/L, 192 mg/L and 192 mg/L observed, respectively. However, bacteria expressing Ddn mutant proteins with substitutions of residues V75L, Q125K or V148I did not exhibit any difference in PA-824 resistance than in bacterial expressing unmutated MTB Ddn.
 |
Table 2 MIC of Ddn Mutants in M. smegmatis Against PA-824 |
Discussion
PA-824 is a promising drug candidate for the treatment of tuberculosis (TB). In line with results of previous reports, this study shows that PA-824 has antibacterial activity against MTB.4,23,24 Moreover, additional in vitro susceptibility testing in this work indicates that PA-824 susceptibility across NTM species is highly diverse. On the one hand, M. avium, M. intracellulare, M. abscessus and M. fortuitum are naturally resistant to PA-824, due to their lack of a functional reductase that would deactivate PA-824; on the other hand, PA-824 exhibited moderate in vitro efficacy against M. kansasii, with MIC values of approximately half of isolates below the maximal attainable PA-824 serum concentration in healthy human subjects.25 Thus, PA-824 could serve as a salvage therapy for patients with M. kansasii infections. It is interesting to speculate about the underlying reason for NTM species diversity with regard to PA-824 sensitivity. Although the precise cause of this diversity remains unclear, comparative genome analysis of MTB Ddn and Ddn homologous sequences of NTMs has revealed high homology among slowly growing MTB and M. kansasii.26 Meanwhile, results of this work involving phylogenetic analysis of MTB Ddn sequences and homologous proteins in NTM species indicate that MTB Ddn is more similar to Ddn homologue of M. kansasii than to Ddn homologous proteins of other NTMs. Therefore, taking species similarity into account may facilitate prediction of drug susceptibilities of new agents across NTM species.
Ddn is a switch for MTB against PA-824, which can directly activate PA-824 and play a bactericidal role.7 Comparative analysis of MTB Ddn and NTM species homologous Ddn sequences in this work demonstrated that Y65, V75, A76, Q125, Y133 and V148 are potential Ddn active site residues involved in PA-824 conversion. Our experiments using M. smegmatis modeling confirmed that M. smegmatis transformation with expression plasmids encoding mutants with substitutions of residues Y65L, A76V or Y133F were resistant to PA-824, since mutation of these sites may dramatically affect Ddn enzymatic activity. In alignment with our observations, a recent study demonstrated that amino acid substitutions at sites Y65, A76 or Y133 led to the loss of Ddn enzymatic activity in vitro. Regarding Y133, that Ddn residue is thought to play a critical role in the anchoring of the deazaflavin ring of the coenzyme of PA-824 within the active site, while both Y65 and A76 are likely to play roles in stabilizing the F420 complex. Interestingly, Susan and colleagues found that substituting an A for a G at codon 76 resulted in Ddn inactivation, as determined using a biochemical assay.12 In the present study, M. smegmatis expressing MTB Ddn A76V was resistant to PA-824, echoing previous findings demonstrating A76 to be an important Ddn active site residue needed to catalyze reductive activation of PA-824. Nevertheless, additional studies are urgently needed to confirm our observations in PA-824-resistant MTB isolates.
Interestingly, mutants with V75L, Q125K or V148I substitutions remained sensitive to PA-824, suggesting that these mutations had little effect on Ddn enzyme activity. A structural study revealed that Ddn segments containing residues 75–88 and 122–148 exerted less influence on overall enzyme function when combined with F420, reflecting that residues within these segments are conformationally flexible and probably less functionally relevant.12 However, in that study valine residues encoded by codons 75 and 148 were replaced by leucine and isoleucine, respectively. In view of the fact that these three amino acids are classified as nonpolar amino acids, the substitution of one nonpolar amino acid for another nonpolar amino acid may not have much impact on Ddn protein structure, thereby resulting in little loss of enzymatic activity.
There were several obvious limitations of this study. First, mutations conferring PA-824 resistance were not validated in clinical MTB isolates, mainly because PA-824 has not been used in clinical practice in China. Second, M. smegmatis, rather than MTB, was used as a model to study the underlying molecular mechanism for PA-824 susceptibility in vitro. Although these two species share similar catalytic mechanisms for achieving reductive activation of PA-824, the disparity in their genetic backgrounds may negatively impact the evaluation of anti-TB compounds, while results using MTB would likely have greater relevance to human disease. Third, PA-824 exhibits great efficacy against intracellular MTB, while the activity of mutants was not evaluated in this study. Despite these limitations, our results extend our knowledge of the molecular mechanisms for PA-824 resistance using comprehensive comparative analysis in mycobacteria.
In conclusion, our data demonstrate that PA-824 shows excellent and moderate in vitro activities for inhibiting the growth of MTB and M. kansasii, respectively. However, M. avium, M. intracellulare, M. abscessus and M. fortuitum were naturally resistant to PA-824, suggesting that replacements of MTB amino acid residues Y65, A76 or Y133 with residues found in NTMs leads to high-level PA-824 resistance. Further studies are urgently needed to confirm these results by analyzing MTB isolates that are resistant to PA-824.
Acknowledgments
This study was supported by the Tongzhou District Science and Technology Committee (KJ2019CX016), the Beijing Talents Foundation (2017000021223ZK39), the Beijing Municipal Administration of Hospitals’ Youth Programme (QML20171601). We would like to thank the staff from National Clinical Laboratory on Tuberculosis for their technical assistance.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pasipanodya JG, Gumbo T. A new evolutionary and pharmacokinetic–pharmacodynamic scenario for rapid emergence of resistance to single and multiple anti-tuberculosis drugs. Curr Opin Pharmacol. 2011;11(5):457–463. doi:10.1016/j.coph.2011.07.001
2. World Health Organization(WHO). Global tuberculosis report 2019. WHO/CDS/TB/2019.20. Geneva, Switzerland: World Health Organization; 2019.
3. Stover CK, Warrener P, VanDevanter DR, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405(6789):962–966. doi:10.1038/35016103
4. Wen S, Jing W, Zhang T, et al. Comparison of in vitro activity of the nitroimidazoles delamanid and pretomanid against multidrug-resistant and extensively drug-resistant tuberculosis. Eur J Clin Microbiol Infect Dis. 2019;38(7):1293–1296. doi:10.1007/s10096-019-03551-w
5. Keam SJ. Pretomanid: first approval. Drugs. 2019.
6. Manjunatha U, Boshoff HI, Barry CE. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun Integr Biol. 2009;2(3):215–218. doi:10.4161/cib.2.3.7926
7. Singh R, Manjunatha U, Boshoff HI, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322(5906):1392–1395. doi:10.1126/science.1164571
8. Choi KP, Kendrick N, Daniels L. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F(420) and FO biosynthesis. J Bacteriol. 2002;184(9):2420–2428. doi:10.1128/JB.184.9.2420-2428.2002
9. Choi KP, Bair TB, Bae YM, Daniels L. Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG. J Bacteriol. 2001;183(24):7058–7066. doi:10.1128/JB.183.24.7058-7066.2001
10. Manjunatha UH, Lahiri R, Randhawa B, Dowd CS, Krahenbuhl JL, Barry CR. Mycobacterium leprae is naturally resistant to PA-824. Antimicrob Agents Chemother. 2006;50(10):3350–3354. doi:10.1128/AAC.00488-06
11. Haver HL, Adeline C, Pramila G, et al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(9):5316–5323. doi:10.1128/AAC.00308-15
12. Cellitti SE, Shaffer J, Jones DH, et al. Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824. Structure. 2012;20(1):101–112. doi:10.1016/j.str.2011.11.001
13. Pang Y, Tan Y, Chen J, et al. Diversity of nontuberculous mycobacteria in eastern and southern China: a cross-sectional study. Eur Respir J. 2017;49:3.
14. Naraoka MOMOI, Fukasawa GOTO. Isolation and identification of a naturally occurring 7,8-didemethyl-8-hydroxy-5-deazariboflavin derivative from Mycobacterium avium. BBA - Gen Subj. 1984;797(3):377–380. doi:10.1016/0304-4165(84)90260-5
15. Purwantini E, Daniels L. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1996;178(10):2861–2866. doi:10.1128/JB.178.10.2861-2866.1996
16. Purwantini E, Gillis TP, Daniels L. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol Lett. 1997;146(1):129–134. doi:10.1111/j.1574-6968.1997.tb10182.x
17. Zhang Z, Pang Y, Wang Y, Cohen C, Zhao Y, Liu C. Differences in risk factors and drug susceptibility between Mycobacterium avium and Mycobacterium intracellulare lung diseases in China. Int J Antimicrob Ag. 2015;45(5):491–495. doi:10.1016/j.ijantimicag.2015.01.012
18. Zhang Z, Pang Y, Wang Y, Liu C, Zhao Y. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int J Antimicrob Ag. 2014;43(3):231–235. doi:10.1016/j.ijantimicag.2013.12.007
19. Clinical and Laboratory Standards Institute. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes; Approved Standard, 2nd Ed; CLSI Document M24–A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2011.
20. Xiao J, Jia H, Pan L, et al. Application of the CRISPRi system to repress sepF expression in Mycobacterium smegmatis. Infect Genet Evol. 2019;72:183–190. doi:10.1016/j.meegid.2018.06.033
21. Wang J, Li BX, Ge PP, et al. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat Immunol. 2015;16(3):237–245. doi:10.1038/ni.3096
22. Rengarajan J, Murphy E, Park A, et al. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A. 2008;105(1):264–269. doi:10.1073/pnas.0710601105
23. Lenaerts AJ, Veronica G, Marietta KS, et al. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother. 2005;49(6):2294–2301. doi:10.1128/AAC.49.6.2294-2301.2005
24. Soni I, De Groote MA, Dasgupta A, Chopra S. Challenges facing the drug discovery pipeline for non-tuberculous mycobacteria. J Med Microbiol. 2016;65(1):1–8. doi:10.1099/jmm.0.000198
25. Ginsberg AM, Laurenzi MW, Rouse DJ, Whitney KD, Spigelman MK. Safety, tolerability, and pharmacokinetics of PA-824 in healthy subjects. Antimicrob Agents Chemother. 2009;53(9):3720–3725. doi:10.1128/AAC.00106-09
26. Wee WY, Dutta A, Choo SW. Comparative genome analyses of mycobacteria give better insights into their evolution. PLoS One. 2017;12(3):e0172831. doi:10.1371/journal.pone.0172831
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.