Back to Journals » International Journal of Nanomedicine » Volume 16
Comparison of Curative Effect of Human Umbilical Cord-Derived Mesenchymal Stem Cells and Their Small Extracellular Vesicles in Treating Osteoarthritis
Authors Tang S, Chen P, Zhang H, Weng H, Fang Z, Chen C, Peng G, Gao H, Hu K, Chen J, Chen L, Chen X
Received 1 September 2021
Accepted for publication 4 November 2021
Published 16 December 2021 Volume 2021:16 Pages 8185—8202
DOI https://doi.org/10.2147/IJN.S336062
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Farooq A. Shiekh
Shijie Tang,1– 4,* Penghong Chen,1– 4,* Haoruo Zhang,1– 4 Haiyan Weng,1– 4 Zhuoqun Fang,1– 4 Caixiang Chen,1– 4 Guohao Peng,1– 4 Hangqi Gao,1– 4 Kailun Hu,1– 4 Jinghua Chen,5 Liangwan Chen,3,6 Xiaosong Chen1– 3
1Department of Plastic Surgery, Fujian Medical University Union Hospital, Fuzhou, 350001, People’s Republic of China; 2Department of Plastic Surgery and Regenerative Medicine Institute, Fujian Medical University, Fuzhou, 350001, People’s Republic of China; 3Engineering Research Center of Tissue and Organ Regeneration, Fujian Province University, Fuzhou, 350001, People’s Republic of China; 4Oncology Institution, Fujian Medical University, Fuzhou, 350004, People’s Republic of China; 5Department of Pharmaceutical Analysis, the School of Pharmacy, Fujian Medical University, Fuzhou, 350100, People’s Republic of China; 6Department of Cardiac Surgery, Fujian Medical University Union Hospital, Fuzhou, 350001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaosong Chen; Liangwan Chen Email [email protected]; [email protected]
Introduction: Human umbilical cord-derived mesenchymal stem cells (hUC-MSCs) and their small extracellular vesicles (hUC-MSC-sEVs) have shown attractive prospects applying in regenerative medicine. This study aimed to compare the therapeutic effects of two agents on osteoarthritis (OA) and investigate underlying mechanism using proteomics.
Methods: In vitro, the proliferation and migration abilities of chondrocytes treated with hUC-MSCs or hUC-MSC-sEVs were detected by Cell Counting Kit-8 assay and scratch wound assay. In vivo, hUC-MSCs (a single dose of 5 × 105) or hUC-MSC-sEVs (30 μg/time) were injected into the knee joints of anterior cruciate ligament transection-induced OA model. Hematoxylin and eosin, Safranin O/Fast Green staining were used to observe cartilage degeneration. The levels of cartilage matrix metabolic molecules (Collagen II, MMP13 and ADAMTS5) and macrophage polarization markers (CD14, IL-1β, IL-10 and CD206) were assessed by immunohistochemistry. Finally, proteomics analysis was performed to characterize the proteinaceous contents of two agents.
Results: In vitro data showed that hUC-MSC-sEVs were taken up by chondrocytes. A total of 15 μg/mL of sEVs show the greatest proliferative and migratory capacities among all groups. In the animal study, hUC-MSCs and hUC-MSC-sEVs alleviated cartilage damage. This effect was mediated via maintaining cartilage homeostasis, as was confirmed by upregulation of the COL II and downregulation of the MMP13 and ADAMTS5. Moreover, the M1 macrophage markers (CD14) were significantly reduced, while the M2 macrophage markers (CD206 and IL-10) were increased in the hUC-MSCs and hUC-MSC-sEVs relative to the untreated group. Mechanistically, we found that many proteins connected to cartilage repair were more abundant in sEVs. Notably, compared to hUC-MSCs, the upregulated proteins in sEVs were mostly involved in the regulation of immune effector process, extracellular matrix organization, PI3K-AKT signaling pathways, and Rap1 signaling pathway.
Conclusion: Our study indicated that hUC-MSC-sEVs protect cartilage from damage and many cartilage repair-related proteins are probably involved in the restoration process. These data suggest the promising potential of hUC-MSC-sEVs as a therapeutic agent for OA.
Keywords: human umbilical cord-derived mesenchymal stem cells, small extracellular vesicles, osteoarthritis, proteomics, cell-free therapy
Introduction
Knee osteoarthritis (OA), a chronic degenerative joint disease and the most common form of arthritis, is a disease of the synovial joints that is characterized by cartilage degradation and bony overgrowth in the form of osteophytes and subchondral thickening,1 tends to occur in middle-aged and elderly female individuals.2,3 In OA, there are well-described progressive destructive changes in the articular cartilage, which parallel characteristic changes in the underlying bone.4 Its complex pathogenesis and articular cartilage structure, without a vascular, neural, or lymphatic network, challenges traditional treatment modalities for OA. Numerous attempts have been made to prevent the progression of knee OA, but appropriate and effective strategies are still being explored.
Mesenchymal stem cells (MSCs) have shown promise in treating OA, thanks to their anti-inflammatory, regenerative and immunomodulatory properties. It is generally accepted that stem cells derived from different tissues play a fundamental role in regenerating cartilage and ameliorating the symptoms of OA, including bone marrow-derived MSCs,5 adipose-derived MSCs,6 amniotic fluid-derived MSCs,7 and human umbilical cord-derived MSCs (hUC-MSCs).8 Among the various sources of MSCs, the human umbilical cord among the most widely used in preclinical and clinical trials due to their richness and availability, noninvasive collection method, and strong proliferative ability in vitro.9 Although investigations have provided preliminary confirmation of the feasibility and safety of MSCs, the need to overcome many obstacles of MSC transplantation remains a major long-term challenge, including ethical issues, the possibility of immune rejection,10 and uncontrollable proliferation in vivo,11 among other problems. Consequently, it is imperative to develop alternative strategies to eliminate complications in cell transplantation.
The therapeutic effects of MSCs on cartilage damage have recently been attributed to the paracrine mechanism, particularly the exosomes, one of the small extracellular vesicles (sEVs).12 sEVs are bilipid and nano-sized (<200 nm) membrane vesicles that transfer various proteins, nucleic acids, lipids, and other bioactive substances.13 In recent years, approaches using MSC-sEVs are beginning to emerge,14 that have the remarkable potential to regenerate damaged cartilage and restore biological function. A growing number of researchers have attributed the roles and therapeutic potential of MSC-sEVs in OA to their participation in many biological processes, including immunomodulation and cartilage microenvironment homeostasis.15–17 This biological function is closely related to the identity of sEVs as a medium of intercellular communication, whereby they deliver genetic information through the transfer of protein cargoes.18 For example, exosomes derived from primary chondrocytes are rich in various mitochondrial proteins, which functionally abolish mitochondrial dysfunction and participate in immune regulation in OA,19 indicating that proteins play an important role in functional execution. However, whether hUC-MSC-derived sEVs (hUC-MSCs-sEVs) ameliorate OA by transferring proteins, and to what extent they can promote the repair and regeneration of cartilage remains unclear.
In this study, we compared the therapeutic effects of hUC-MSCs and sEVs derived from hUC-MSCs on cartilage damage. In in vitro experiments, we used CCK-8 and scratching-assay to explore whether both therapies could promote chondrocyte proliferation and migration. In in vivo experiments, we assessed whether hUC-MSCs-sEVs could replicate the regenerative effects of hUC-MSCs. hUC-MSCs or hUC-MSCs-sEVs were injected into the knee joint of anterior cruciate ligament rupture (ACLT)-induced OA rats and collected the cartilage and synovium tissues. The microstructure and morphology of the tissues were observed on hematoxylin and eosin (H&E) and Safranin O/Fast Green staining specimen. Collagen II (COLII), MMP13, and ADAMTS5 expression were examined by immunohistochemistry, and macrophage polarization phenotype was analyzed by CD14, IL-1β, IL-10, and CD206. Finally, we used liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics to explore the potential therapeutic mechanisms of the two therapeutic agents. We speculated that sEVs obtained from hUC-MSCs may be a more fantastic therapeutic strategy for future clinical OA.
Materials and Methods
Ethics Statement
All animal experimental procedures were approved by the institutional Animal Ethics Committee of Fujian Medical University, Fuzhou, Fujian, China (FJMU IACUC 2020–0052).
Human Umbilical Cord Mesenchymal Stem Cells Culture
Passage 3–5 (P3–P5) hUC-MSCs (Qilu Cell Therapy Technology Co., Ltd., China) were purchased and maintained in Mesenchymal Stem Cell Basal Medium (without FBS) (MSCYF01-500, YINFENG BIOLOGICAL, China) supplemented with Mesenchymal Stem Cell Supplement (MSCYF02-20, YINFENG BIOLOGICAL, China) and penicillin/streptomycin (P-S) (100 U/mL) (HyClone) at 37°C in a humid air with 5% CO2.
Identification of Human Umbilical Cord Mesenchymal Stem Cells
For the phenotype characterization of hUC-MSCs, the mesenchymal markers, CD105, CD73, and CD90, and the hematopoietic cell markers CD34, CD45, and HLA-DR were detected with a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA). For trilineage differentiation assay, the adipogenic induction medium, osteogenic induction medium and chondrogenic induction medium (all from ScienCell, USA) were used to induce differentiation and assessed by oil red staining for lipid droplets (in adipogenesis), Safranin O staining for proteoglycans (in chondrogenesis), and alizarin red s stains calcium in bone (in osteogenesis).
Preparation and Identification of Human Umbilical Cord Mesenchymal Stem Cells-sEVs
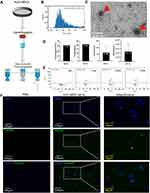
hUC-MSC-conditioned medium was collected after 48 h culture. The medium was centrifuged at 300 × g for 10 min at 4°C to remove dead cells and large apoptotic bodies. Subsequently, the supernatant was filtered using a 0.22 μm filter to remove cell debris, and the supernatant was transferred to a 100 KD ultrafiltration tube (UFC9100; Amicon® Ultra Ireland) to obtain 3 mL concentrated medium. sEVs were purified by size-exclusion chromatography (SEC), as previously described.20 Briefly, the CL-2B column (Echo9103A-30 mL; ECHO BIOTECH, China) was washed with 60 mL sterile PBS to elute any residual ethyl alcohol. The concentrated sample was loaded on the column and fractions 6 to 10 (5 mL) were collected (Figure 1A). Finally, these fractions were concentrated with a 100 KD ultrafiltration tube and stored at −80°C for further experiments.
The morphology of the sEVs was observed using transmission electron microscopy (TEM). The particle size distribution and concentration of the sEVs were measured using nFCM (N30E Nanoflow Analyzer; NanoFCM Inc., Xiamen, China). The surface biosignature proteins of the sEVs were detected by nano flow cytometry. Antibodies, including FITC mouse anti-human CD63 (556019, BD, Franklin Lake, New Jersey, USA), FITC mouse anti-human CD9 (555371, BD, Franklin Lake, New Jersey, USA), FITC mouse anti-human CD81 (551108, BD, Franklin Lake, New Jersey, USA), and FITC mouse IgG1 (400108, BioLegend, San Diego, USA)
Uptake of sEVs by Chondrocytes
Purified sEVs were labeled with PKH67 membrane dye following to the manufacturer’s instructions (Sigma Aldrich, USA). The labeled sEVs were collected by additional ultracentrifugation and resuspended in PBS. Subsequently, PKH67-labeled sEVs were co-cultured with chondrocytes (CP-R092, Procell Life Sci & Tech Co. Ltd., Wuhan, China) for 12 h, and the internalization of sEVs was evaluated using fluorescence microscopy (Leica DMi8 S, Germany).
Cell Proliferation Assay
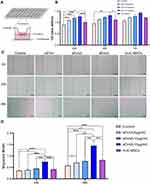
The effect of hUC-MSCs and hUC-MSC-sEVs on the proliferation of chondrocytes was evaluated by a Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) following the manufacturer’s specifications. Briefly, the chondrocytes were stimulated with 1 ng/mL interleukin (IL)-1β (CM002-1000HP, Chamot Biotechnology, Shanghai, China) for 24 h, and then IL-1β-treated cells (2000 cells per well) were seeded in 96-well plates, sEVs1 (5 μg/mL), sEVs2 (10 μg/mL), sEVs3 (15 μ/mL), or hUC-MSCs were added into each well at 37°C for 24, 48, or 72 h. A 96-well transwell system (PSHT004R1 Merck Millipore, Germany) was utilized to co-cultivate two cell types (Figure 2A), and the number of co-culture MSCs was chosen on the basis of the total sEVs in the 15 μg/mL group. We found that 5 × 105 MSCs secreted about 30 μg protein. Then, a total of 10 μL CCK-8 was added into each well and co-incubated for 3 h. The absorbance was evaluated in 450 nm using a microplate spectrophotometer (Thermo Scientific).
Cell Scratch Wound Assay
The scratch wound assay was used to analyze the effects of hUC-MSCs and hUC-MSC-sEVs on the migration of chondrocytes. Briefly, 5 × 105 chondrocytes were plated in a 24-well plate and incubated at 37°C for 8 h. The adherent cell layer was carefully scratched using a p200 pipette tip. Next, the cell fragments were washed three times with PBS. Then, sEVs (sEVs1–3), hUC-MSCs, or an equal volume of PBS were added. The hUC-MSCs group was performed using a 24-well transwell system (TCS016024, Biofil, Guangzhou, China). Chondrocytes were photographed at 0, 24, and 48 h after wounding. The change in the width of the scratched areas was measured using Image J software (NIH, USA).
Anterior Cruciate Ligament Transection Osteoarthritis Rat Model
Male Sprague-Dawley (SD) rats (8 weeks old) were randomized into four groups: normal group (n = 6), treatment with hUC-MSCs (n = 6), treatment with hUC-MSC-sEVs (n = 6), and control (OA model treated with PBS) (n = 6). The OA model was induced by anterior cruciate ligament transection (ACLT), as previously described.21 The postoperative treatment lasted for 4 weeks. Rats in the hUC-MSC-sEVs group were given 200 μL hUC-MSC-sEVs (30 μg) suspended in PBS by intra-articular injection once a week for 4 weeks. The hUC-MSCs group was treated with a single dose of 200 μL hUC-MSCs (5 × 105), while the rats in the control group were injected with 200 μL PBS. All rats were euthanized, and the joints and synovial tissues were collected for further evaluation at 9 weeks post-surgery.
Histological Analyse and Immunohistochemistry
Knee joint samples were fixed in 10% paraformaldehyde for 24 h and then were treated with Ethylene Diamine Tetraacetic Acid (EDTA) until decalcification was complete. The decalcified specimens were dehydrated, embedded in paraffin, and sectioned at a 4 μm thickness. Next, the slices were processed for hematoxylin and eosin (H&E) and Safranin O/Fast Green staining. For immunohistochemical (IHC) staining, the sections were separately incubated with type 2 collagen (COL II) (1:300, GB11027, Servicebio), MMP13 (1:200, GB11247, Servicebio), ADAMTS5 (1:100, DF13268, Affbiotech), CD14 (1:1000, GB11254, Servicebio), CD206 (1:500, GB13438, Servicebio), IL-10 (1:500, GB11108, Servicebio) and IL-1β (1:800, GB11113, Servicebio), followed by incubation with horseradish peroxidase-conjugated secondary antibody (1:200, GB23303, Servicebio). Finally, the staining color was developed using the DAB Detection Kit (Servicebio).
The Osteoarthritis Research Society International (OARSI) score was applied to evaluate cartilage degradation (Supplementary Table 1).22 H&E and Safranin O/Fast Green staining were observed in a low-magnification field. The IHC staining was measured using the %Area parameter or the percentage of pixels in the image or selection highlighted in red using Image→Adjust→Threshold. Each parameter was graded and independently measured by two observers.
LC-MS/MS of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs
Each sample included three specimens from the same group. For hUC-MSCs and hUC-MSC-sEV, an SDT (4%SDS, 100 mM Tris-HCl, 1 mM DTT, pH 7.6) buffer was used for sample lysis and protein extraction. The amount of protein was quantified with the BCA Protein Assay Kit (Bio-Rad, USA). The digest peptides of each sample were then desalted and concentrated. To evaluate the effects of sample extraction, 20 µg of protein for each sample was mixed with 5X loading buffer, respectively, and boiled for 5 min. The proteins were separated on 12.5% SDS-PAGE gel (constant current 14 mA, 90 min). The protein bands were visualized by Coomassie Blue R-250 staining.
The LC-MS/MS analyses were performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Thermo Fisher Scientific) for 120 min. The peptides were loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18) connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min, controlled by IntelliFlow technology. The mass spectrometer was operated in positive ion mode. MS data were acquired using a data-dependent top-10 method, dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. The automatic gain control (AGC) target was set to 3e6 and maximum inject time to 10 ms. Dynamic exclusion duration was 40.0 s. Survey scans were acquired at a resolution of 70,000 at m/z 200, and the resolution for HCD spectra was set to 17,500 at m/z 200, and the isolation width was 2 m/z. The normalized collision energy was 30 eV, and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The MS raw data for each sample were combined and searched using MaxQuant 1.5.3.17 software for identification and quantitation analyses. The parameters and instructions are shown in Supplementary Table 2.
Bioinformatics Analyses
The raw MS data were processed using the MaxQuant software (v.1.5.3.17) for identification and quantitation analyses. Hierarchical clustering analyses were performed in Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and Java Treeview (http://jtreeview.sourceforge.net) using the average-linkage clustering method. Volcanic maps of the differentially expressed genes were generated with R package ggplots2. The upregulated proteins were used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses by the clusterProfiler R package. The protein–protein interaction (PPI) information of the differentially expressed proteins was constructed in the STRING database (http://string-db.org/) and visualized in Cytoscape (http://www.cytoscape.org/, version 3.2.1).
Statistical Analyses
All experiments were performed in triplicate, and the data are expressed as means ± SD. The one-way ANOVA and nonparametric tests were performed using IBM SPSS Statistics Version 21. Values of P < 0.05 or less were considered statistically significant.
Results
Characterization of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs
hUC-MSCs exhibited a typical fibroblast morphology in the bright field images (Supplementary Figure 1A). The results of trilineage differentiation confirmed that hUC-MSCs had multilineage differentiation potential into adipocytes, osteocytes, and chondrocytes (Supplementary Figure 1B–D). Moreover, the phenotype characterization of hUC-MSCs was analyzed by flow cytometry, and it was found that the majority of hUC-MSCs expressed CD105, CD73, CD90 and are negative for CD34, CD45 and HLA-DR (Supplementary Figure 1E).
Morphological analyses of hUC-MSC-sEVs with TEM clearly revealed that saucer-like structures, with a diameter of 70–90 nm (Figure 1C). Data from nanoflow cytometry (nFCM) demonstrated that the mean diameter of the sEVs was 80.48 nm (Figure 1B). Furthermore, the percentages of CD9, CD63, or CD81 positive sEVs were 38.8%, 42.1%, and 33.5%, respectively (Figure 1E). These data indicated the successful purification of sEVs from the supernatant of hUC-MSCs.
The yield of hUC-MSC-sEVs was further assessed (n = 3). According to particle quantitative analyses, the mean particle concentration was 2.301 × 107 ± 1.774 × 107 particles per mL CM and 61.089 ± 49.966 particles per cell. The protein yield of hUC-MSC-sEVs was 20.920 ± 1.226 μg per mL CM and 31.394 ± 1.417 μg per 5 × 105 cells (Figure 1D).
Effects of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs on Chondrocyte Proliferation and Migration
We evaluated the effects of two therapeutic agents on chondrocytes. First, fluorescence microscope images clearly showed that PKH-67-labelled sEVs were distributed around the nuclei after co-culturing with chondrocytes (Figure 1F), suggesting that hUC-MSC-sEVs were successfully internalized by chondrocytes and may modulate the biological processes of chondrocytes.
Subsequently, CCK-8 assay was used to evaluate the proliferation of chondrocytes. Both treatments showed greater proliferation potential compared to the control group at 24, 48, and 72 h. The sEVs treatment groups, including sEVs1 (5 μg/mL), sEVs2 (10 μg/mL), sEVs3 (15 μg/mL), showed significantly stimulated chondrocyte proliferation in a dose-dependent manner, particularly at a concentration of 15 μg/mL (in 24h and 48h group, compared to the control group; Figure 2B).
Likewise, scratch wound assays demonstrated that hUC-MSC-sEVs dose-dependently enhanced the migration of the chondrocytes (Figure 2C), whereas the effects of hUC-MSCs treatment is significantly weaker than that of the sEVs3 group (24 and 48h, Figure 2D). These data further showed that sEVs were more efficient to increase motility of chondrocytes than hUC-MSCs.
Effect of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs on the Repair of Cartilage Damage in ACLT-Induced OA Rats
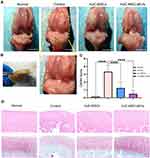
An ACLT-triggered rat OA model offers an excellent option for evaluating the protective effects of the two treatments on articular cartilage. We delivered MSCs or sEVs by intra-articular injection into articular cavity at 4 weeks postoperatively (Figure 3B). At week 9 after OA induction, knee joint specimens were collected for further analyses. Their gross appearance is shown in Figure 3A. The control group exhibited a markedly rough articular surface and local erosion compared with the normal group, suggesting the successful establishment of the OA model. By contrast, we observed that the cartilage was generally repaired in the hUC-MSC and hUC-MSC-sEV groups to some extent. Compared to treatment with hUC-MSCs, the articular surface was smoother and more polished in the hUC-MSC-sEV group.
H&E and Safranin O/Fast Green staining revealed that the surface layer of cartilage was smooth, the hierarchical structure was clear, the staining was positive and uniform in the normal group. The cartilage injuries in the control group were the most serious of those in all groups; they exhibited fractured cartilage, thickened subchondral bone, abnormal distribution of chondrocytes, and loss of proteoglycan (Figure 3D), and the OARSI score was 3.33 ± 0.82 (Figure 3C). By contrast, when the hUC-MSCs and hUC-MSC-sEVs were injected, cartilage was well reconstructed, which was characterized by a regular surface, restored cartilage thickness, near-normal morphology chondrocytes, and Safranin O/Fast Green staining of proteoglycans showed intense red and even distribution in the articular cartilage. Consistently, according to OARSI grading, hUC-MSC-sEV treatment significantly reduced the OARSI scores relative to those of the control group (0.50 ± 0.45 vs 3.33 ± 0.82, P < 0.0001) and hUC-MSCs group (1.25 ± 0.76 vs 3.33 ± 0.82, P < 0.0001) (Figure 3C), indicating that both the treatments effectively reduced cartilage damage during the OA progression in rats.
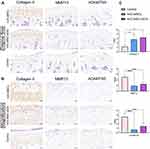
To further explore the effects of two treatments on cartilage matrix in vivo, immunohistochemical staining was performed to clarify the expression of COL II, MMP13, and ADAMTS5. The results showed that relative to the control group, the decrease in COL II (cartilage matrix synthetic protein) expression in matrix could be significantly reversed in the hUC-MSCs and hUC-MSC-sEVs group (Figure 4A and B). Similarly, the percentage of MMP13+ cells and ADAMTS5+ cells in hUC-MSC-sEVs-treated group was significantly lower compared to that in the control group, 2.5437 ± 0.8316 vs 5.0167 ± 0.8315, P < 0.05; 1.4532 ± 0.6383 vs 3.5250 ± 4.2, P < 0.05, respectively. In addition, the hUC-MSC treatment group also displayed a significant reduction of matrix-degrading proteins with a lower percentage of MMP13 (1.4532 ± 0.6384 vs 5.0167 ± 0.8315, P < 0.0001) and ADAMTS5 (0.6704 ± 0.3107 vs 3.5250 ± 0.6992, P < 0.0001). Relative to the hUC-MSC group, we found that the levels of COL II, MMP13, and ADAMTS5 were higher in hUC-MSC-sEVs group, although the differences were not statistically significant (Figure 4C).
Taken together, these data suggest that two therapeutic agents are involved in articular cartilage homeostasis through the control of matrix anabolism and catabolism in chondrocytes. Importantly, the capacity of promoting the cartilage matrix synthetic protein production of the hUC-MSC-sEVs was superior to that in their parent cells.
Role of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs in Reprogramming Macrophages in Synovium
There is increasing evidence that the abnormal activation of macrophages in synovium drives the progression of OA,23 so the extent of macrophage infiltration in the synovial tissues was evaluated using immunohistochemical staining. The results showed a dramatic increase in CD14-positive staining (M1-like macrophage marker) and the expression of CD206, IL-10 (M2-like macrophage markers) were lower in OA rats compared to the two treatment groups (Figure 5A and B). Furthermore, the percentage of IL-1β-positive areas, a M1 macrophage-associated pro-inflammatory cytokine, also significantly increased in the control group. Consistent with the results from H&E and Safranin O/Fast Green staining, hUC-MSCs and hUC-MSC-sEVs showed the potential to rescue the above phenotypes. As shown in Figure 5C, with the hUC-MSC-sEVs and hUC-MSCs treatments, the staining levels of M2 markers (CD206 and IL-10) and M1 marker (CD14) were consistently enhanced and reduced, respectively, suggesting a successful modulation of M2/M1 macrophage polarization status leading to a healthy environment. Comparably, there were no statistical differences between the hUC-MSC-sEVs group and hUC-MSCs-treated group.
Biological Mechanisms of Human Umbilical Cord Mesenchymal Stem Cells and Human Umbilical Cord Mesenchymal Stem Cells-sEVs in Cartilage Repair
To further explore the molecular mechanism and elucidate differences in the protein content of hUC-MSCs and hUC-MSC-sEVs, LC-MS/MS was applied for proteomic analyses in biological triplicates. A total of 457,585 spectrum, 50,947 peptides, and 6279 proteins were detected (Figure 6A). In the protein expression profiles of hUC-MSCs and hUC-MSC-sEVs, we detected 6204 and 4478 proteins, respectively (Figure 6B). Next, we were interested in comparing the level of LFQ intensity of a class of chondrogenesis‐related proteins in hUC-MSC-sEVs and hUC-MSCs (Supplementary Table 3). The data showed that the majority of these chondrogenesis‐related proteins are highly enriched in hUC-MSC-sEV, such as Alpha2-macroglobulin (A2M)24 (P < 0.001), TGFB125 (P < 0.01), Versican (VCAN)26 (P < 0.001), Runt-related transcription factor 1 (RUNX1)27 (P > 0.05), epidermal growth factor receptor (EGFR)28 (P < 0.001) and Biglycan (BGN)29 (P < 0.005), while only ADAM930 (P < 0.005) and RUNX2 (P > 0.05)31 are mainly in hUC-MSCs (Figure 6C).
A Venn diagram was used to identify with-group protein identification overlap (Supplementary Figure 2A and B), and Figure 7A shows that 4403 proteins were shared between two groups, with 1801 unique proteins in hUC-MSCs and 75 unique proteins in hUC-MSC-sEVs, suggesting that hUC-MSCs secrete more proteins and may perform a wider range of functions. Differentially expressed proteins are shown in Figure 7B through hierarchical clustering analyses (fold change > 2.0, p < 0.05), indicating the diversity of the proteins between hUC-MSCs and hUC-MSC-sEVs. The volcano plot and histogram (Figure 7C and D and Supplementary Table 4) identified 333 upregulated and 1407 downregulated proteins in the hUC-MSC-sEV group relative to their parent cell hUC-MSCs (fold change > 2.0 or < 0.5, p < 0.05). Upregulated proteins including ITGA4, ORM2, CD82, and AMFR, and so forth. Meanwhile downregulated proteins including LAMC1, RPS8, PABPC1, and CAD, and so forth.
Subsequently, the upregulated proteins were subject to GO and KEGG enrichment analyses. The GO analyses indicated that the molecular function category identified these proteins as being mainly enriched in integrin binding, glycosaminoglycan binding, and extracellular matrix structural constituent (Figure 7F). The GO cellular component (CC) pathways included collagen-containing extracellular matrix, secretory granule membrane, and vesicle lumen (Figure 7G). The analyses of biological process (BP) revealed that proteins that are involved in the regulation of multiple biological processes related to cartilage repair, such as “regulation of immune effector process”, “extracellular matrix organization”, and “extracellular structure organization” terms were enriched (Figure 7E). KEGG pathway analyses demonstrated that these proteins were obviously enriched in “PI3K-AKT signaling pathways”, “Rap1 signaling pathway”, “focal adhesion”, and “ECM-receptor interaction” (Figure 7H). Importantly, these biological functions and pathways are crucial for chondrogenesis, inflammatory regulation, and macrophage polarization. In addition, we constructed a PPI network with 3491 nodes and 212,704 interactions (Supplementary Figure 2C), which indicated highly aggregated proteins may perform biological functions through synergies (Supplementary Table 5).
Our proteomics studies indicated that hUC-MSCs and hUC-MSC-sEVs have distinct differences in protein composition and abundance, and these differential proteins were closely related to fundamental biological processes.
Discussion
In this study, we first compared the effects of hUC-MSC-sEVs and hUC-MSCs on the treatment of OA. In both in vivo and in vitro experiments, our results revealed that the MSC-sEVs mimic the biological functions of parent cells, as evidenced by effectively and safely promoting cartilage regeneration. In addition, this study demonstrated that chondrogenesis-associated proteins of hUC-MSC-sEVs are promising candidate molecules for sEV-based cartilage repair.
OA is the most prevalent debilitating condition worldwide, causing a significant decline in patients’ quality of life and their well-being.32 Although the interesting therapeutic properties of MSCs, such as their regenerative capability and immunomodulatory effect, make them a novel candidate for regenerative medicine, security is a primary concern when developing an MSC-based therapy. Increasing evidence suggests that the great therapeutic contribution of MSCs is the result of its paracrine actions,12,33 and exosomes (one of sEVs) are a key paracrine regulator involved in MSCs-mediated tissue repair.34 This clue has enlightened us regarding the use of sEVs as a new class of therapeutic option to substitute for direct cellular therapy in OA. The ideal therapeutic efficacy of MSC-sEVs has been widely reported. The authors evaluated the effectiveness of exosomes derived from synovial membrane MSCs (SMMSC-Exos) and induced pluripotent stem cell-derived MSCs (iMSC-Exos) in a collagenase-induced OA mouse and demonstrated that iMSC-Exos possesses a superior effect on cartilage repair by enhancing chondrocyte migration and proliferation.35 In another study, the DMM-induced OA mice were injected embryonic MSCs (ESC-MSCs) and their exosomes into articular cavity, although this comparison was set in different periods and with different frequency of injection, the ESC-MSC-exosomes group exhibited their potential in cartilage protection.36 A more recent publication demonstrated that osteochondral regeneration can be achieved by pretreatment with human umbilical cord Wharton’s jelly MSC-exosomes.16 However, no studies have explored the differences between hUC-MSC-sEVs and hUC-MSCs and possible therapeutic mechanisms underlying cartilage repair from the perspective of proteomics.
Researches show that chondrocytes are hypo-replicative during homeostasis, while they do maintain the potential to proliferate in some settings. For example, chondrocytes proliferate in the form of ‘clusters’ during the early stages of OA, which is commonly viewed as an attempt to repair damaged matrix.1,37,38 In addition, chondrocytes have been shown to facilitate intercellular communication through the production of extracellular vesicles. In consequence, the proliferation and migration of chondrocytes are important factors for the repair of osteoarthritis. In concordance with those studies, our in vitro data suggested that both hUC-MSCs and their sEVs can markedly stimulate cellular activities in an inflammatory environment compared the control group, including quantity and mobility of chondrocytes. Notably, the effects of hUC-MSC-sEVs at a concentration of 15 μg/mL on chondrocyte migration were best among all other groups. This thus indicates that hUC-MSC-sEVs may be more potency and efficacy to a certain extent than hUC-MSCs. In addition, as shown in Figure 1F, hUC-MSC-sEVs were successfully taken up by chondrocytes, thus directly affected the target cells. This action may be related to the Caveolin-1 involved signaling pathway.39
IL-1β is one of the key factors inducing inflammatory and ECM destruction OA,40 evidenced by inhibiting extracellular matrix synthesis protein expression, such as aggrecan and collagen type II, promoting matrix-degrading protein (MMP13 and ADAMTS5).41,42 Imbalance of catabolism and metabolism of cartilage is a major link in the progression of osteoarthritis.38,43,44 Thus, on the basis of in vitro experiments, we further explored the regenerative effects of the two treatments in an ACLT-induced OA model. Repeated local injections of sEVs are required to maintain the effects of sEVs-based therapy (one per week),45,46 while cell-based therapy often requires only a single administration of MSCs to be effective, because the implanted cells may continue to secrete their secretoma for at least 3 weeks.7,47 This paradigm forms the basis of our injection plans in the current study. The results confirm that both treatments strongly inhibit IL-1β expression and IL-1β-induced upregulation of MMP13 and ADAMTS5. Our results are in keeping with those of previous studies.17,48 Another significant feature is the high content of COL II after pretreatment with hUC-MSC-sEVs and hUC-MSCs, demonstrating good repair effect. The adverse effects of synovitis on the pathological process of OA are also a focus of our research. The features of synovial inflammation include increased synovial cellularity and macrophage accumulation, leading to severe clinical symptoms.49,50 Vigorous research has confirmed that different macrophage phenotypes are key mediators of cartilage homeostasis,51 including pro-inflammatory M1-like macrophages and anti-inflammatory M2-like macrophages. Recent studies have reported that exosomes from bone marrow MSCs promoted a phenotypic transformation of macrophages from M1 to M2.52 In this study, we found that hUC-MSC-sEVs inhibited the infiltration of synovial M1-like macrophages, reducing the expression of CD14 and pro-inflammatory cytokines, IL-1β. In addition, the expression of CD206 and IL-10 was enhanced, indicating that sEVs possess effective therapeutic properties in inhibiting the inflammatory response and subsequent amelioration of OA similar to MSCs. Importantly, our findings also agree with previous results.52 Thus, a phenotypic switch of macrophages therapeutically guided by sEVs may be a promising approach for clinical practice.
sEVs contain numerous molecules, such as proteins, lipids, long noncoding RNAs and microRNAs, which are potential candidates for disease diagnosis and treatment.14 Considerable attention was paid to non-coding RNAs in EVs.15,53,54 However, little investigations has been done on the proteome of MSC-derived sEVs. Because proteins directly reflect the pattern of cellular behavior, their role cannot be neglected. Here, we analyzed differences in the expression of proteins between hUC-MSC-sEVs and hUC-MSCs. We found that the gene expression patterns in hUC-MSCs are quite different from those in hUC-MSC-sEVs. This result is consistent with previous reports based on proteomic analyses of human urine-derived stem cells and their exosomes.55 Although hUC-MSCs carry more proteins than their sEVs, they share a number of common proteins, indicating that sEVs have a functional role in human biology similar to MSCs, and may be a good substitute for therapeutic MSCs. Next, we selected a series of proteins related to ECM organization and chondrogenesis from among the shared proteins and then identified different abundances between the two groups. Surprisingly, these proteins are expressed at a significantly higher level in sEVs. For example, A2M, a protease inhibitor can produce a strong inhibitory effect on a variety of cartilage-degradation-related factors,56 including ADAMTS4, ADAMTS5, and MMP13.57 Moreover, a close association between EGFR signaling pathway and chondrocyte metabolism has been reported.28,58 A similar conclusion was reached by Chen et al,59 from a study using antibody array technology. We found that proteoglycan 4, a critical cytoprotective glycoprotein,60 is absent from MSCs. Importantly, we also found that the upregulated proteins were highly enriched in the regulation of immune effector process, extracellular matrix organization, PI3K-AKT signaling pathways, Rap1 signaling pathway, and ECM-receptor interaction. Thus, based on these bioinformatics analyses and our experimental results, the important conclusion was drawn: sEVs derived from MSCs may be more efficient than their parent cells in the treatment of OA.
On the one hand, we must acknowledge that as a cell-derived nanotherapeutic agent, sEVs have numerous advantages over intact stem cells: first, sEVs’ acellular status poses less risk than cellular transplantation, evading immune rejection and oncogenicity and providing high stability. Second, the small size with a lipid bilayer membrane structure makes them easier to store and produce. In addition, sEVs demonstrate an innate ability to home in on to tissue injury sites,61,62 and native nanoparticles can be engineered to optimize their targeting ability, therapeutic potency and drug loading capacity. On the other hand, the disadvantages of sEVs therapy cannot be ignored, such as the fact that multiple injections produce additional pain, the isolation methods have high cost, and sEVs have low enrichment efficiency.63 Objectively, these are the main issues to be overcome in the future.
Although this effects of hUC-MSCs-derived sEVs on preventing OA progression are as potent as those of hUC-MSCs, our study had certain inevitable limitations. The development of any new therapeutic agent requires the exploration of the optimal dosage. Meanwhile, the dose-response assessment of EVs is highly recommended by the MISEV2018 guidelines.13 Our work logically found a proportional relationship between the observed outcome and the dose of sEVs administered; we identified 15 μg/mL as the standard dose. However, there was a huge difference in the choice of the dose of EVs. A total of 5 μg/mL MSC exosomes were used to alleviate temporomandibular joint osteoarthritis,64 while doses as high as 150 μg/mL65 were used in the OA model. This phenomenon results from the inherent heterogeneity of sEVs and the lack of separation standard.66,67 Thus, we need to determine the optimal dosage of therapeutic sEVs on the basis of specific conditions or diseases for promoting clinical translation. In addition, the safety and effectiveness concerns of sEVs administration must be methodically addressed in large animals. Finally, additional work is necessary to determine the active ingredients in protein profiles of sEVs and their mode of action. These limitations will be addressed in the future work.
Conclusions
hUC-MSCs and their derivatives (such as sEVs) have cut a striking figure in the field of promoting OA repair, and our study complements the exploration of the differences between hUC-MSC-sEVs and hUC-MSCs from a proteomic perspective and possible therapeutic mechanisms for cartilage repair. Our study shows that both hUC-MSCs and their sEVs significantly stimulated chondrocyte activity and matrix remodeling processes in an inflammatory environment, and that sEVs had effective therapeutic properties similar to MSCs in suppressing inflammatory responses and subsequently ameliorated OA. And on top of that, hUC-MSC-sEVs may be more effectively than hUC-MSCs to some extent. Consequently, in future studies, we hope to refine the dose-response assessment and determine the active ingredients in protein profiles of sEVs and their mode of action, with a view to providing a new, complete and promising treatment option for OA.
Abbreviations
KOA, knee osteoarthritis; MSCs, mesenchymal stem cells; hUC-MSCs, human umbilical cord-derived mesenchymal stem cells; sEVs, small extracellular vesicles; hUC-MSCs-sEVs, human umbilical cord MSCs derived sEVs; LC-MS/MS, liquid chromatography-tandem mass spectrometry; ECM, extracellular matrix; CM, conditioned medium; SEC, size-exclusion chromatography; TEM, transmission electron microscopy; CCK-8, Cell Counting Kit-8; IL-1β, interleukin −1β; ACLT, Anterior cruciate ligament transection; EDTA, ethylene diamine tetraacetic acid; H&E, Hematoxylin and eosin; IHC, immunohistochemical; COL II, Type 2 collagen; OARSI, Osteoarthritis Research Society International; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction; A2M, alpha2-macroglobulin; VCAN, Versican; RUNX1, runt-related transcription factor 1; EGFR, epidermal growth factor receptor; BGN, biglycan; MF, molecular function; CC, cellular component; BP, biological process; SMMSC-Exos, synovial membrane MSCs-derived exosomes; iMSC-Exos, exosomes secreted by the induced pluripotent stem cell-derived MSCs; ESC-MSCs, embryonic mesenchymal stem cells; PRG4, proteoglycan 4.
Data Sharing Statement
All the data and materials were presented in the main paper.
Ethics approval
All animal protocols were implemented under the Animal Ethical Committee of Fujian Medical University’s (PR China) supervision and approval, which conforms to the guide for the National Institutes of Health to use laboratory animals (Permit Number: FJMU IACUC 2020-0052).
Consent for publication
All authors have read and consent to publish this article.
Acknowledgments
We thank the Public Technology Service Center of Fujian Medical University (PR China), Oncology Institution of Fujian Medical University and Central Laboratory of Fujian Medical University Union Hospital (PR China) for providing technical support as well as experimental platforms.
Funding
This study was financed by National Natural Science Foundation of China (grant No. 81971855), Joint funding Project of Science and Technology Innovation in Fujian Province (grant No. 2017Y9101), Special Financial Funds of Fujian Province (grant Nos. 2018B054 and 2020CZ016) and Industrial Technology Joint Innovation Project of Fujian Provincial Development and Reform Commission (grant No. 2011601).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Coryell PR, Diekman BO, Loeser RF. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat Rev Rheumatol. 2021;17(1):47–57. doi:10.1038/s41584-020-00533-7
2. Katz JN, Arant KR, Loeser RF. Diagnosis and treatment of hip and knee osteoarthritis: a review. JAMA. 2021;325(6):568–578. doi:10.1001/jama.2020.22171
3. Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet. 2015;386(9991):376–387. doi:10.1016/S0140-6736(14)60802-3
4. Findlay DM, Kuliwaba JS. Bone-cartilage crosstalk: a conversation for understanding osteoarthritis. Bone Res. 2016;4:16028. doi:10.1038/boneres.2016.28
5. Gao J, Zhang G, Xu K, et al. Bone marrow mesenchymal stem cells improve bone erosion in collagen-induced arthritis by inhibiting osteoclasia-related factors and differentiating into chondrocytes. Stem Cell Res Ther. 2020;11(1):171. doi:10.1186/s13287-020-01684-w
6. Brindo da Cruz IC, Velosa APP, Carrasco S, et al. Post-Adipose-Derived Stem Cells (ADSC) Stimulated by Collagen Type V (Col V) Mitigate the Progression of Osteoarthritic Rabbit Articular Cartilage. Front Cell Dev Biol. 2021;9:606890. doi:10.3389/fcell.2021.606890
7. Zavatti M, Beretti F, Casciaro F, Bertucci E, Maraldi T. Comparison of the therapeutic effect of amniotic fluid stem cells and their exosomes on monoiodoacetate-induced animal model of osteoarthritis. Biofactors. 2020;46(1):106–117. doi:10.1002/biof.1576
8. Matas J, Orrego M, Amenabar D, et al. Umbilical Cord-Derived Mesenchymal Stromal Cells (MSCs) for Knee Osteoarthritis: repeated MSC Dosing Is Superior to a Single MSC Dose and to Hyaluronic Acid in a Controlled Randomized Phase I/II Trial. Stem Cells Transl Med. 2019;8(3):215–224. doi:10.1002/sctm.18-0053
9. Yang W, Zhang J, Xu B, et al. HucMSC-derived exosomes mitigate the age-related retardation of fertility in female mice. Mol Ther. 2020;28(4):1200–1213. doi:10.1016/j.ymthe.2020.02.003
10. Shi Y, Hu G, Su J, et al. Mesenchymal stem cells: a new strategy for immunosuppression and tissue repair. Cell Res. 2010;20(5):510–518. doi:10.1038/cr.2010.44
11. Leferink PS, Breeuwsma N, Bugiani M, van der Knaap MS, Heine VM. Affected astrocytes in the spinal cord of the leukodystrophy vanishing white matter. Glia. 2018;66(4):862–873. doi:10.1002/glia.23289
12. Lai RC, Yeo RW, Lim SK. Mesenchymal stem cell exosomes. Semin Cell Dev Biol. 2015;40:82–88. doi:10.1016/j.semcdb.2015.03.001
13. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi:10.1080/20013078.2018.1535750
14. Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal Stem Cell-derived Extracellular Vesicles: toward Cell-free Therapeutic Applications. Mol Ther. 2015;23(5):812–823. doi:10.1038/mt.2015.44
15. Hu H, Dong L, Bu Z, et al. miR-23a-3p-abundant small extracellular vesicles released from Gelma/nanoclay hydrogel for cartilage regeneration. J Extracell Vesicles. 2020;9(1):1778883. doi:10.1080/20013078.2020.1778883
16. Jiang S, Tian G, Yang Z, et al. Enhancement of acellular cartilage matrix scaffold by Wharton’s jelly mesenchymal stem cell-derived exosomes to promote osteochondral regeneration. Bioact Mater. 2021;6(9):2711–2728. doi:10.1016/j.bioactmat.2021.01.031
17. Woo CH, Kim HK, Jung GY, et al. Small extracellular vesicles from human adipose-derived stem cells attenuate cartilage degeneration. J Extracell Vesicles. 2020;9(1):1735249. doi:10.1080/20013078.2020.1735249
18. Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. doi:10.1038/nrm3335
19. Zheng L, Wang Y, Qiu P, et al. Primary chondrocyte exosomes mediate osteoarthritis progression by regulating mitochondrion and immune reactivity. Nanomedicine. 2019;14(24):3193–3212. doi:10.2217/nnm-2018-0498
20. Takov K, Yellon DM, Davidson SM. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J Extracell Vesicles. 2019;8(1):1560809. doi:10.1080/20013078.2018.1560809
21. Lorenz J, Grassel S. Experimental osteoarthritis models in mice. Methods Mol Biol. 2014;1194:401–419.
22. Pritzker KP, Gay S, Jimenez SA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14(1):13–29. doi:10.1016/j.joca.2005.07.014
23. Manferdini C, Paolella F, Gabusi E, et al. From osteoarthritic synovium to synovial-derived cells characterization: synovial macrophages are key effector cells. Arthritis Res Ther. 2016;18:83. doi:10.1186/s13075-016-0983-4
24. Zhang Y, Wei X, Browning S, Scuderi G, Hanna LS, Wei L. Targeted designed variants of alpha-2-macroglobulin (A2M) attenuate cartilage degeneration in a rat model of osteoarthritis induced by anterior cruciate ligament transection. Arthritis Res Ther. 2017;19(1):175. doi:10.1186/s13075-017-1363-4
25. Lee MC, Ha CW, Elmallah RK, et al. A placebo-controlled randomised trial to assess the effect of TGF-ss1-expressing chondrocytes in patients with arthritis of the knee. Bone Joint J. 2015;97-B(7):924–932. doi:10.1302/0301-620X.97B7.35852
26. Choocheep K, Hatano S, Takagi H, et al. Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J Biol Chem. 2010;285(27):21114–21125. doi:10.1074/jbc.M109.096479
27. Zhao X, Meng F, Hu S, et al. The Synovium Attenuates Cartilage Degeneration in KOA through Activation of the Smad2/3-Runx1 Cascade and Chondrogenesis-related miRNAs. Mol Ther Nucleic Acids. 2020;22:832–845. doi:10.1016/j.omtn.2020.10.004
28. Long DL, Ulici V, Chubinskaya S, Loeser RF. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) is increased in osteoarthritis and regulates chondrocyte catabolic and anabolic activities. Osteoarthritis Cartilage. 2015;23(9):1523–1531. doi:10.1016/j.joca.2015.04.019
29. Embree MC, Kilts TM, Ono M, et al. Biglycan and fibromodulin have essential roles in regulating chondrogenesis and extracellular matrix turnover in temporomandibular joint osteoarthritis. Am J Pathol. 2010;176(2):812–826. doi:10.2353/ajpath.2010.090450
30. Djouad F, Delorme B, Maurice M, et al. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res Ther. 2007;9(2):R33. doi:10.1186/ar2153
31. Lee KS, Kim HJ, Li QL, et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783–8792. doi:10.1128/MCB.20.23.8783-8792.2000
32. Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi:10.1016/S0140-6736(19)30417-9
33. Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi:10.1146/annurev-physiol-021014-071641
34. Toh WS, Lai RC, Hui JHP, Lim SK. MSC exosome as a cell-free MSC therapy for cartilage regeneration: implications for osteoarthritis treatment. Semin Cell Dev Biol. 2017;67:56–64. doi:10.1016/j.semcdb.2016.11.008
35. Zhu Y, Wang Y, Zhao B, et al. Comparison of exosomes secreted by induced pluripotent stem cell-derived mesenchymal stem cells and synovial membrane-derived mesenchymal stem cells for the treatment of osteoarthritis. Stem Cell Res Ther. 2017;8(1):64. doi:10.1186/s13287-017-0510-9
36. Wang Y, Yu D, Liu Z, et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther. 2017;8(1):189. doi:10.1186/s13287-017-0632-0
37. Lotz MK, Otsuki S, Grogan SP, Sah R, Terkeltaub R, D’lima D. Cartilage cell clusters. Arthritis Rheum. 2010;62(8):2206–2218. doi:10.1002/art.27528
38. Chen G, Liu T, Yu B, Wang B, Peng Q. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle. 2020;19(13):1696–1705. doi:10.1080/15384101.2020.1772545
39. Zhai M, Zhu Y, Yang M, Mao C. Human Mesenchymal Stem Cell Derived Exosomes Enhance Cell-Free Bone Regeneration by Altering Their miRNAs Profiles. Adv Sci. 2020;7(19):2001334. doi:10.1002/advs.202001334
40. Goldring MB. Osteoarthritis and cartilage: the role of cytokines. Curr Rheumatol Rep. 2000;2(6):459–465. doi:10.1007/s11926-000-0021-y
41. Mengshol JA, Vincenti MP, Coon CI, Barchowsky A, Brinckerhoff CE. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor kappaB: differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 2000;43(4):801–811. doi:10.1002/1529-0131(200004)43:4<801::AID-ANR10>3.0.CO;2-4
42. Verma P, Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J Cell Biochem. 2011;112(12):3507–3514. doi:10.1002/jcb.23298
43. Zhu C, Shen K, Zhou W, Wu H, Lu Y. Exosome-mediated circ_0001846 participates in IL-1β-induced chondrocyte cell damage by miR-149-5p-dependent regulation of WNT5B. Clin Immunol. 2021;232:108856. doi:10.1016/j.clim.2021.108856
44. Zhen G, Wen C, Jia X, et al. Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19(6):704–712. doi:10.1038/nm.3143
45. Liu X, Yang Y, Li Y, et al. Integration of stem cell-derived exosomes with in situ hydrogel glue as a promising tissue patch for articular cartilage regeneration. Nanoscale. 2017;9(13):4430–4438. doi:10.1039/C7NR00352H
46. Zhang S, Chu WC, Lai RC, Lim SK, Hui JH, Toh WS. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage. 2016;24(12):2135–2140. doi:10.1016/j.joca.2016.06.022
47. Jing XH, Yang L, Duan XJ, et al. In vivo MR imaging tracking of magnetic iron oxide nanoparticle labeled, engineered, autologous bone marrow mesenchymal stem cells following intra-articular injection. Joint Bone Spine. 2008;75(4):432–438. doi:10.1016/j.jbspin.2007.09.013
48. Wu J, Kuang L, Chen C, et al. miR-100-5p-abundant exosomes derived from infrapatellar fat pad MSCs protect articular cartilage and ameliorate gait abnormalities via inhibition of mTOR in osteoarthritis. Biomaterials. 2019;206:87–100. doi:10.1016/j.biomaterials.2019.03.022
49. Sellam J, Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol. 2010;6(11):625–635. doi:10.1038/nrrheum.2010.159
50. Dijkstra CD, Dopp EA, Joling P, Kraal G. The heterogeneity of mononuclear phagocytes in lymphoid organs: distinct macrophage subpopulations in the rat recognized by monoclonal antibodies ED1, ED2 and ED3. Immunology. 1985;54(3):589–599.
51. Orlowsky EW, Kraus VB. The role of innate immunity in osteoarthritis: when our first line of defense goes on the offensive. J Rheumatol. 2015;42(3):363–371. doi:10.3899/jrheum.140382
52. Zhang J, Rong Y, Luo C, Cui W. Bone marrow mesenchymal stem cell-derived exosomes prevent osteoarthritis by regulating synovial macrophage polarization. Aging. 2020;12(24):25138–25152. doi:10.18632/aging.104110
53. Kotani A, Ito M, Kudo K. Non-coding RNAs and lipids mediate the function of extracellular vesicles in cancer cross-talk. Semin Cancer Biol. 2021;74:121.
54. Yan L, Liu G, Wu X. The umbilical cord mesenchymal stem cell-derived exosomal lncRNA H19 improves osteochondral activity through miR-29b-3p/FoxO3 axis. Clin Transl Med. 2021;11(1):e255. doi:10.1002/ctm2.255
55. Chen CY, Rao SS, Ren L, et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics. 2018;8(6):1607–1623. doi:10.7150/thno.22958
56. Wang S, Wei X, Zhou J, et al. Identification of alpha2-macroglobulin as a master inhibitor of cartilage-degrading factors that attenuates the progression of posttraumatic osteoarthritis. Arthritis Rheumatol. 2014;66(7):1843–1853. doi:10.1002/art.38576
57. Tortorella MD, Arner EC, Hills R, et al. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J Biol Chem. 2004;279(17):17554–17561. doi:10.1074/jbc.M313041200
58. Zhang X, Zhu J, Liu F, et al. Reduced EGFR signaling enhances cartilage destruction in a mouse osteoarthritis model. Bone Res. 2014;2:14015. doi:10.1038/boneres.2014.15
59. Chen L, Xiang B, Wang X, Xiang C. Exosomes derived from human menstrual blood-derived stem cells alleviate fulminant hepatic failure. Stem Cell Res Ther. 2017;8(1):9. doi:10.1186/s13287-016-0453-6
60. Alquraini A, Jamal M, Zhang L, Schmidt T, Jay GD, Elsaid KA. The autocrine role of proteoglycan-4 (PRG4) in modulating osteoarthritic synoviocyte proliferation and expression of matrix degrading enzymes. Arthritis Res Ther. 2017;19(1):89. doi:10.1186/s13075-017-1301-5
61. Pironti G, Strachan RT, Abraham D, et al. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation. 2015;131(24):2120–2130. doi:10.1161/CIRCULATIONAHA.115.015687
62. de Couto G, Gallet R, Cambier L, et al. Exosomal MicroRNA Transfer Into Macrophages Mediates Cellular Postconditioning. Circulation. 2017;136(2):200–214. doi:10.1161/CIRCULATIONAHA.116.024590
63. Kesimer M, Gupta R. Physical characterization and profiling of airway epithelial derived exosomes using light scattering. Methods. 2015;87:59–63. doi:10.1016/j.ymeth.2015.03.013
64. Zhang S, Teo KYW, Chuah SJ, Lai RC, Lim SK, Toh WS. MSC exosomes alleviate temporomandibular joint osteoarthritis by attenuating inflammation and restoring matrix homeostasis. Biomaterials. 2019;200:35–47. doi:10.1016/j.biomaterials.2019.02.006
65. Rong Y, Zhang J, Jiang D, et al. Hypoxic pretreatment of small extracellular vesicles mediates cartilage repair in osteoarthritis by delivering miR-216a-5p. Acta Biomater. 2021;122:325–342. doi:10.1016/j.actbio.2020.12.034
66. Liang Y, Lehrich BM, Zheng S, Lu M. Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J Extracell Vesicles. 2021;10(7):e12090. doi:10.1002/jev2.12090
67. Huang G, Lin G, Zhu Y, Duan W, Jin D. Emerging technologies for profiling extracellular vesicle heterogeneity. Lab Chip. 2020;20(14):2423–2437. doi:10.1039/D0LC00431F
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.