Back to Journals » Journal of Inflammation Research » Volume 15
Comparative Transcriptome Analysis of Gingival Immune-Mediated Inflammation in Peri-Implantitis and Periodontitis Within the Same Host Environment
Authors Yuan S, Wang C, Jiang W, Wei Y, Li Q, Song Z, Li S, Sun F, Liu Z, Wang Y, Hu W
Received 22 February 2022
Accepted for publication 13 May 2022
Published 25 May 2022 Volume 2022:15 Pages 3119—3133
DOI https://doi.org/10.2147/JIR.S363538
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adam D Bachstetter
Shasha Yuan,1 Cui Wang,1 Wenting Jiang,1 Yiping Wei,1 Qingqing Li,2,3 Zhanming Song,2,3 Siqi Li,1 Fei Sun,1 Zhongtian Liu,2,3 Ying Wang,2,3 Wenjie Hu1,4
1Department of Periodontology, National Clinical Research Center for Oral Diseases, National Engineering Laboratory for Digital and Material Technology of Stomatology, Beijing Key Laboratory of Digital Stomatology, Peking University School and Hospital of Stomatology, Beijing, People’s Republic of China; 2Department of Immunology, School of Basic Medical Sciences, and NHC Key Laboratory of Medical Immunology, Peking University, Beijing, People’s Republic of China; 3Center for Human Disease Genomics, Peking University, Beijing, People’s Republic of China; 4NHC Research Center of Engineering and Technology for Computerized Dentistry, Beijing, People’s Republic of China
Correspondence: Ying Wang, Department of Immunology, School of Basic Medical Sciences, and NHC Key Laboratory of Medical Immunology, Peking University, No. 38, College Road, Haidian District, Beijing, People’s Republic of China, Tel +86 10 8280115, Email [email protected] Wenjie Hu, Department of Periodontology, National Clinical Research Center for Oral Diseases, National Engineering Laboratory for Digital and Material Technology of Stomatology, Beijing Key Laboratory of Digital Stomatology, Peking University School and Hospital of Stomatology, No. 22, Zhongguancun South Street, Haidian District, Beijing, People’s Republic of China, Tel +86 10 82195374, Email [email protected]
Objective: This investigation aimed to determine whether and to what extent there are transcriptional differences between periodontitis and peri-implantitis in the same susceptible host.
Background: As an immune-mediated inflammatory disease resulting in aggressive bone resorption around dental implants, peri-implantitis constitutes a major threat to dental implants’ long-term success. Compared to periodontitis, its etiological molecular mechanism remains elusive. Currently, there are few investigations on these two diseases at the transcriptional level within the same basal environment.
Methods: Ligature-induced peri-implantitis and periodontitis were generated in the same mice. Gingival tissues of healthy, periodontitis, and peri-implantitis sites from the same oral cavity were collected and used for RNA sequencing. Differentially expressed genes (DEGs) were screened between periodontitis/peri-implantitis and healthy sites. Enrichment analysis of DEGs was performed. The comprehensive immune landscape was annotated by seq-ImmuCC. Hub genes from peri-implantitis-specific DEGs were filtered using the STRING database and Cytoscape. Validation of the selected hub genes was performed on the GEO106090 dataset (gingival tissues from six periodontitis patients, six peri-implantitis patients, and six healthy controls).
Results: The results indicated that peri-implantitis and periodontitis exhibited significantly distinct transcriptional signatures, with the complement and coagulation cascade pathways and osteoclast differentiation predominating in peri-implantitis mucosa. Compared with periodontitis, peri-implantitis sites exhibited elevated macrophage proportions and relatively enriched macrophage activation and bone loss. Hub genes were selected, and IL1B, CCL3, and CLEC4E were significantly highly expressed in human peri-implantitis mucosa.
Conclusion: The study suggests that the interplay between macrophages and bone resorption seems to be more robust than in periodontitis. IL1B, CCL3, and CLEC4E may be considered promising therapeutic targets for peri-implantitis. These critical biological processes and identified genes may contribute to the etiology of peri-implantitis, which is unique from periodontitis. This work may make way for deeper exploration and contribute significantly to the treatment and prevention of peri-implantitis.
Keywords: peri-implantitis, periodontitis, transcriptome analysis, immune profiling, bioinformatics analysis
Introduction
Peri-implantitis is an immune-mediated inflammatory condition that manifests as soft tissue inflammation and accelerated bone resorption surrounding dental implants, which compromises the long-term success and even ultimately the loss of implants.1,2 With approximately a quarter of all patients experiencing peri-implantitis and the growing number of dental implants placed per year, peri-implantitis has sparked growing concern among researchers globally.3,4
Similar to periodontitis regarding clinical and radiographic signs in some ways, peri-implantitis is also treated with mechanical and antimicrobial treatment approaches.5 Unfortunately, the current treatment of peri-implantitis yields highly variable and unpredictable outcomes, with some interventions exhibiting a recurrence rate of up to 100% after 12 months.6–8 The lack of efficacy of conventional treatment approaches suggests that peri-implantitis may be different from periodontitis in terms of pathogenesis to some extent, which is also supported by evidence from recent open-ended investigations.9
Past studies focused on the different molecular mechanisms in peri-implantitis and periodontitis have revealed some important differences from the histopathological and immunological aspects. Histopathological analysis revealed that inflammatory regions are more prominent in peri-implantitis, with higher proportions of infiltrating leukocytes.10–12 Immunological studies regarding the proinflammatory cytokines of peri-implant crevicular fluid reported that IL-6, IL-1β, and TNF-α can be used as biomarkers to distinguish peri-implantitis from periodontitis.13–15 It should be noted that the histochemical staining/immunoassay used in the abovementioned research could only identify a very limited range of known markers, which is inadequate to obtain entire thorough molecular profiles in the course of peri-implantitis. Given that innate immunity systems respond to stimuli through sophisticated and coordinated gene expression programs that include the transcription of various genes, the use of RNA sequencing (RNA-seq) technology in conjunction with unbiased bioinformatics analysis undoubtedly offers a robust approach to deconstructing the molecular profiles of peri-implantitis.
Published studies found that periodontitis and peri-implantitis display distinctive signatures of transcriptional gene expression.16–18 However, different host environments were included in those studies to assess the differences between the two analogous diseases with the impact of individual variation overlooked. Individual variation in the immune system response appears to be preexisting, and it is likely that these differences contribute to differences in susceptibility to inflammation,19,20 which makes it more challenging to analyze the fundamental pathogenesis of peri-implantitis. These issues warrant further investigation of the two analogous diseases observed within the same oral cavity. In fact, dental implants are regularly placed in patients with a history of periodontitis, and long-term studies indicate that the coexistence of peri-implantitis and periodontitis is becoming more common clinically.21,22 There is a need to compare their characteristics in one susceptible host with these two diseases.
Therefore, the study’s principal goal was to delineate the distinctions between peri-implantitis and periodontitis by employing genome-wide mRNA expression profiling in the same mice by establishing ligature-induced experimental peri-implantitis and periodontitis, thus providing novel information on the differences between the two diseases. The second aim was to screen signatures that are unique to peri-implantitis through deeper bioinformatics analysis using genes expressed exclusively in peri-implantitis. The results reported in the present study could lay the foundation for understanding the pathogenesis of peri-implantitis and provide potential therapeutics for treating this disease.
Materials and Methods
Mouse Models
Five-week-old male wild-type C57BL/6J mice were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice used during the research were maintained in pathogen-free facilities at Peking University’s Health Science Center and were given an ad libitum regular diet. All experiments were conducted in accordance with relevant provisions for the use of animals following the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. The RNASeqPower package in R was used to calculate the sample sizes and determine the statistical power for our data.23 All animal care and experiments were conducted under the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Peking University Health Science Center Ethics Committee (Ethics number: LA2021495).
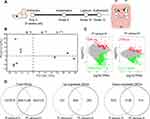
The complete procedures used were identical to those previously described with certain alterations (Figure 1A).24 In short, all mice were given general anesthesia, and the right maxillary first molars were extracted. After six weeks, sequential 1.0-mm drill holes in extraction sockets were prepared with a micro hand drill (0.3 mm in diameter; D. P. Machining Inc., La Verne, CA, USA). The screw-shaped titanium implant (1 mm in length and 0.5 mm in diameter; D. P. Machining Inc., La Verne, CA, USA) was screwed into the drill holes clockwise until torque could be achieved. After four weeks of osseointegration, peri-implantitis and periodontitis were experimentally established. On the right side of the maxilla, a 5–0 silk ligature was placed subgingivally around the implants and corresponding left maxillary molars (Supplementary Figure 1). All mice were sacrificed two weeks after ligature placement using CO2 inhalation, and soft tissues were collected. The successful establishment of the models was confirmed by scanning alveolar bone using microCT (Inveon MM Gantry-STD 3121, Siemens, Germany), The X-ray beam was set at 60 kVp and 220 μA. All maxillae were scanned in the sagittal position at a voxel resolution of 18 μm (Supplementary Figure 2).
Soft Tissue Collection and RNA Extraction
The soft tissues were cut in an imaginary line 1 mm around the implants and molars, and the soft tissues were peeled off and stored at −80℃ (Figure 1A). Soft tissues surrounding ligatured implants and corresponding ligatured molars were regarded as peri-implantitis sites (PI) and periodontitis sites (P), respectively, while tissues around the third molars were collected as healthy controls (H). Blinding was established by marking each tissue with letters and using the same letter for each group. Except for the investigators who established the animal models and collected tissues, the investigators responsible for extracting RNA and performing the analysis were kept blinded to the group assignments.
TRIzol® reagent (Invitrogen, Rockville, MD, USA) was used to extract total RNA from soft tissues (n = 3 mice) according to the manufacturer’s instructions. The quality of the RNA was then evaluated using an Agilent 2100 Bioanalyzer (Santa Clara, CA) and quantified using an ND-2000 system (NanoDrop Technologies, USA). Each sample had an RNA integrity value ≥ 7.5, optical density 260/280 ≥ 1.8, and optical density 260/230 ≥ 2.0.
Transcriptome Analysis
The Illumina TruSeqTM RNA Sample Preparation Kit (San Diego, CA) was used to create the RNA-seq transcriptome library using 1 μg of total RNA. On an Illumina platform, sequencing was performed (NovaSeq 6000 sequencer, 150 paired-end reads). Each sample contained 41–45 million clean reads and Q30 > 94% (Supplementary Table 1). The clean reads were separately aligned to the mouse reference genome (GRCm38.p6, http://asia.ensembl.org/Mus_musculus/Info/Index). The Majorbio I-Sanger Cloud Platform was used to upload and process the sequencing data (www.i-sanger.com). The RNA-seq data obtained by this research are available in the publicly accessible Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE186882.
Identification of Differentially Expressed Genes
DEseq2 (Ver. 1.2) was used to identify differentially expressed genes (DEGs), and genes with q value (adjusted p value) ≤ 0.001 and |fold change| ≥ 2 were considered significantly different in expression. Pairwise comparisons were made between the healthy gingival controls and two disease phenotypes (P versus H, PI versus H).
Bioinformatics Analysis
A Venn diagram and principal component analysis were performed using R software. We used Gene Ontology (GO; www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to identify the biological functions of the DEGs with a Bonferroni-corrected P value ≤ 0.001.
Gene set enrichment analysis (GSEA) was applied to validate whether the particular signature was considerably enriched at the top or bottom of the predefined gene set. We downloaded GSEA software (Ver. 4, software.broadinstitute.org/gsea/index.jsp/) and predesigned the gene sets of interest, including the “macrophage activation” and “bone loss” gene sets (Supplementary Table 2). The list of genes with differential expression between peri-implantitis/periodontitis and healthy controls was input into the software with 1000 permutations, and gene sets were regarded as highly enriched when the false discovery rate (FDR) q-value was < 0.05.
In addition to obtaining the comprehensive landscape of the immune microenvironment and identifying the distinct signatures of immune cell proportions in both phenotypes, seq-ImmuCC (http://218.4.234.74:3200/immune/) was applied to normalize and process the raw RNA-seq read counts.25 It can determine the relative percentage of main immune cell types by using the deconvolution model, which relies on a presumed linear connection between mixed expression profiles in tissue samples and isolated cell type expression profiles to estimate the comparative fraction of main immune cell types.
Core Gene Selection and Reanalysis
The STRING online database (https://string-db.org/) was utilized to forecast the protein–protein interaction (PPI) network, which could help to clarify the distinct mechanism of the initiation and course within peri-implantitis.26 The PPI network of DEGs exclusively expressed in peri-implantitis was evaluated, and interactions with an overall score > 0.7 were considered statistically significant. For ranking nodes in a PPI network, cytoHubba, a plug-in for Cytoscape, was used to perform topological analysis methods.27 Four centrality indexes, including the maximal clique centrality (MCC), the maximum neighborhood component (MNC), the edge percolated component (EPC), and the degree, were chosen to filter the top 20 genes. A total of seven overlapping hub genes among these top genes were screened according to these four methods.
GEO serves as a public repository of high-throughput functional genomics expression data. Here, the GSE106090 dataset from GEO was acquired to validate the expression levels of seven hub genes associated with peri-implantitis. GSE106090 consisted of 18 human soft tissue samples and was divided into three groups, namely, the peri-implantitis, periodontitis, and healthy control groups, with six samples in each group. Core genes were identified when P < 0.05 and fold change > 1.5. Three core genes were identified.
A summary of the three core genes interacting networks was created using GeneMANIA (http://genemania.org/) to concurrently identify genes with functionalities comparable to those of the target genes and anticipate gene functions simultaneously.28 The visualization of functions was achieved by the ClueGO plug-in for Cytoscape.
Statistical Analysis
The statistically analyzed data are presented as the mean ± standard error of the mean (SEM). Statistical analysis was performed using GraphPad (GraphPad Software Inc.). Bioinformatics analysis was performed using R 3.6. The statistical analysis of RNA-seq data is described above.
Results
Transcriptome Profiling
Significant DEGs were identified with a significance threshold of |fold change| ≥ 2 and q value ≤ 0.001. Identification of DEGs revealed different gene expression modulations of periodontitis and peri-implantitis in the same basal background. Figure 1C illustrates that 539 genes were upregulated and 1808 genes were downregulated when periodontitis was compared with the healthy control group (P versus H comparison). Comparing peri-implantitis patients with healthy controls (PI versus H comparison) revealed 755 upregulated genes and 1312 downregulated genes. Detailed information on the significant DEGs is provided in Supplementary Tables 3 and 4.
Principal component analysis was performed to determine if the expression profiles obtained for groups represented a distinct molecular signature suggestive of peri-implantitis/periodontitis, as illustrated in Figure 1B. Principal component 1 (PC1), which accounted for 54.16% of the variance, distinctly separated the periodontitis/peri-implantitis groups from the healthy control group. PC2, which accounted for 12.02% of the variance, separated the peri-implantitis group from the periodontitis group. The results showed a clear separation of the three groups in the present study.
Given the similar inflammatory phenotypes of periodontitis and peri-implantitis, we evaluated the overlapping DEGs between the two entities. Venn diagrams revealed several overlapping and exclusive DEGs in each disease group versus the healthy control group. A total of 402 upregulated and 1138 downregulated DEGs in both P versus H and PI versus H were involved during disease progression (Figure 1D). The results suggested that the gene modulation of gingival inflammation around two distinct structures of teeth and implants was similar to a large degree but still had certain separate properties.
RNA-seq generates much more information than can be used for deeper explorations. To identify key pathologic mechanisms of peri-implantitis compared with periodontitis, the overlapping and exclusive DEGs annotated by the GO and KEGG databases were examined.
Investigation of the DEGs and Pathways Shared in Both Diseases
To determine the mechanisms in response to ligature in peri-implantitis and periodontitis, the overlapping DEGs (402 upregulated and 1138 downregulated DEGs) were selected for GO and KEGG enrichment analyses. As illustrated in Figure 2A, the findings suggested that the shared DEGs in the two entities were predominantly enriched in pathways that were mostly associated with the inflammatory process. In particular, these genes were enriched in hematopoietic cell lineage, immune cell activation, cytokine–cytokine receptor interaction, and chemokine signaling pathway, which were highly identical to the clinical processes of periodontitis and peri-implantitis.
Investigation of the Exclusive DEGs and Pathways in Peri-Implantitis and Periodontitis
To determine the distinct pathologic mechanisms in both analogous diseases, we further explored the exclusive DEGs within the two entities. The Venn diagram of both entities’ upregulated and downregulated DEGs revealed that 137 and 353 upregulated DEGs and 670 and 174 downregulated DEGs were specifically expressed in the peri-implantitis and periodontitis groups, respectively, compared to the healthy control group (Figure 1D). More specific upregulated DEGs (353) were identified in the PI versus H comparison, while the number of specific downregulated DEGs (670) was drastically reduced in the P versus H comparison. To identify the function of these exclusive DEGs, these specific upregulated and downregulated DEGs were analyzed by GO and KEGG data enrichment analyses separately (Figure 2B).
In the GO analysis, the exclusive DEGs in the P versus H comparison were mainly enriched in pathways involved in cell adhesion and differentiation; in comparison, the exclusive DEGs in the PI versus H comparison were mainly concentrated in the response to stimulus (detection of external biotic stimulus, cellular response to molecule of bacterial origin), abnormal immune cell activation (neutrophil migration, granulocyte migration, positive regulation of chemotaxis), regulation of reactive oxygen species (ROS) (positive regulation of ROS metabolic process, regulation of ROS metabolic process), and regulation of bone mineralization.
The KEGG analysis indicated that the specific upregulated DEGs in PI versus H and downregulated DEGs in P versus H were enriched in multiple pathways, while no pathway was enriched with the specific downregulated DEGs in PI versus H and upregulated DEGs in P versus H. The exclusively upregulated DEGs of the PI versus H comparison were mainly enriched in pathways associated with immune-mediated inflammation activation and imbalance (complement and coagulation cascades, lysosome, and TNF signaling pathway) and cell proliferation, adhesion, and migration (cytokine–cytokine receptor interaction, osteoclast differentiation, glycosaminoglycan biosynthesis, and C-type lectin receptor signaling pathway). In contrast, these pathways were not enriched in the P versus H comparison. The exclusively downregulated DEGs in the P versus H comparison were mainly enriched in pathways involved in hematopoietic cell lineage, B-cell receptor signaling and natural killer cell–mediated cytotoxicity.
Given that enhanced immune-mediated inflammatory signatures were observed in the GO and KEGG analyses of the PI versus H comparison, further exploration was focused on inflammation caused by abnormal immune cell activation.
Macrophage Compositions are Promoted in Peri-Implantitis Sites
Seq-ImmuCC was then applied to generate the relative abundance of the major immune subset population. As illustrated in Figure 3A, plotting the 10 kinds of mainly expressed immune cells showed that monocytes and macrophages were activated in different proportions in both diseases. The recruitment of monocytes was higher in the P group, while the activation of macrophages was much stronger in the PI group. In addition, other cell types did not change among different sites.
Given the close link between macrophages and bone remodeling, we further predesigned two gene sets to examine changes in macrophages and bone remodeling through GSEA, designated “macrophage activation” and “bone loss”. The two gene sets were significantly enriched in soft tissues in the PI versus H comparison, while both showed no significant difference in the P versus H comparison (Figure 3B). These data demonstrated that the activation of macrophages and bone resorption in the peri-implantitis site were considerably enhanced.
Identification of Core Genes in Inflammation Signaling
DEGs may not be indicative of their importance to the phenotype on their own, but they may instead reflect the indirect interaction with a gene network. Thus, to explore the possible target core genes in peri-implantitis, the STRING database was used to anticipate potential associations among the upregulated and downregulated DEGs specifically expressed in the PI versus H comparison. The PPI network consisting of 170 nodes and 188 edges is shown in Figure 4. Furthermore, the top 20 genes were obtained by using four algorithms (MCC, MNC, degree, and EPC) in cytoHubba. Seven overlapping hub genes were selected for further analysis, namely, Il1b, Tlr2, Ccl3, Ccl4, Irg1, Clec4e, and Cd80 (Table 1).
 |
Table 1 Comparison of Hub Genes for Specifically Expressed Genes in PI versus H Ranked in the cytoHubba Plugin of Cytoscape |
Reanalysis of the Seven Selected Hub Genes
GSE106090, containing information on human gingival samples, was used to further validate the expression of these seven selected hub genes from RNA-seq (Figure 5). Irg1 is more commonly known as Acod1 (aconitate decarboxylase 1) in human databases. GSE106090 verified that the expression of six of the seven selected genes was significantly higher in both diseases than in healthy gingival samples (H). Moreover, soft tissues derived from peri-implantitis (PI) exhibited higher expression levels of IL1B, CCL3, and CLEC4E than periodontitis (P) tissues, implying that these three genes could be key regulators in peri-implantitis. To explore the biological processes of IL1B, CCL3 and CLEC4E, the GeneMANIA algorithm was employed to construct gene association networks among these core genes and their 20 related genes (Figure 6A). In addition, the exploration of their biological functions is shown in Figure 6B, indicating that the core genes were mainly involved in the chemotaxis and migration of immune cells, including cytokine-mediated signaling pathways, cytokine binding, and immune receptor activity.
Discussion
The pathophysiology of peri-implantitis is not entirely understood, and numerous obstacles and uncertainties hinder further exploration, particularly in comparison with periodontitis. RNA-seq technology offers apparent advantages in regard to elucidating the potential mechanisms underlying complicated diseases.29 There are many challenges and complexities in peri-implantitis pathology. RNA-seq technology allows an accurate comparison of the pathology of these diseases due to the high resolution and high data throughput. To the best of our knowledge, this research presents the first RNA-seq experiment incorporating experimental peri-implantitis and periodontitis in the same host environment to compare these two diseases to date. Our study aimed to uncover key pathways and molecules linked with peri-implantitis and potential therapeutic targets for peri-implantitis at the molecular level.
The current study identified 1540 shared DEGs in pathologic events of periodontitis and peri-implantitis. In addition, 527 DEGs (353 upregulated and 174 downregulated DEGs) were specifically expressed in peri-implantitis, and 807 DEGs (137 upregulated and 670 downregulated DEGs) were specifically expressed in periodontitis. The results revealed that the gene modulation of gingival inflammation around teeth and implants was largely similar but had certain distinct characteristics. These distinct molecular signatures emphasize that these two disease entities should be viewed as distinct disease events.
Accordingly, the following pathway analysis suggested that the 1540 shared DEGs of peri-implantitis/periodontitis were mainly enriched in pathways associated with cell activities such as proliferation, adhesion, migration, differentiation, and apoptosis. These results showed a high degree of consistency with prior research, indicating that regenerative processes such as proliferative and apoptotic processes, along with defensive and immunological pathways, could play a crucial part in inflammation.16,18 It is important to note that the similarities of pathways within peri-implantitis and periodontitis appear to be greater than the previous study’s overlap, further explaining why peri-implantitis and periodontitis are often summarized into one group.16,18
Interestingly, among the enrichment analyses of the exclusive DEGs, the pathways involved in the response to stimulus, immune-mediated inflammation activation, regulation of ROS, and regulation of bone remodeling represented the most prominent pathways in peri-implantitis. These pathways were significantly upregulated in peri-implantitis, but no change or opposite regulation patterns were observed in the periodontitis setting, which suggested that these pathways may be the key mechanism of peri-implantitis. The host response to stimulation seems to be more robust in peri-implantitis soft tissues. Indeed, the degree of immune cell invasion appeared to be stronger in peri-implantitis, whereas lesions within the connective tissue compartment were well restrained in periodontitis.30–32 This could be explained by the anatomical variations between periodontal and peri-implant soft tissues. Due to the decreased cellularity and vascularity of peri-implant connective tissue, implants may be more vulnerable to pathogenic stimuli and disease initiation. Furthermore, prior research has revealed that soft tissues around peri-implantitis have a more prominent inflammatory microenvironment, which includes macrophage activation and migration, as well as the production of proinflammatory and bone remodeling-related cytokines and oxidative stress. Inflammatory microenvironment-mediated oxidative stress is characterized by the overproduction of ROS, which triggers severe inflammation and extended tissue destruction.33 Previous studies have indicated the vital role of ROS in peri-implantitis and periodontitis. Interestingly, GO analysis of the exclusive DEGs revealed the positive regulation of ROS as the prominent signaling pathway that was upregulated in peri-implantitis compared to periodontitis, which suggests higher ROS activity in peri-implantitis.34 In particular, increasing evidence has demonstrated that scavenging ROS can inhibit osteoclastogenesis and that alveolar bone resorption is suppressed by ROS scavengers through local injection,35–38 confirming that ROS participate in bone remodeling by enhancing osteoclast capacity. In agreement, the osteoclast differentiation pathway was relatively upregulated in peri-implantitis, which might explain the accelerated rate of bone resorption in clinical peri-implantitis lesions. Even when both disease entities begin at the same time, more bone loss occurs around implants than around natural teeth, implying a faster progression in peri-implantitis.39,40
Conventional immunohistochemistry/immunofluorescence analysis is bound to have difficulty annotating the global immune landscape due to its technical sensitivity and the limited number of mature immune markers for staining. Thus, we used the RNA-seq-based approach seq-ImmuCC to deduce the precise annotation of the comprehensive immune landscape. The data showed that the immune landscape of peri-implantitis tends to show more macrophages and fewer monocytes than that of periodontitis. Consistent with our data, histopathological studies of human biopsy material from periodontitis and peri-implantitis indicated that peri-implantitis lesions were much wider and featured significantly more macrophages in terms of area proportions, numbers, and densities than periodontitis lesions.10–12 Compelling evidence indicates that the interplay between alveolar bone and macrophages in the microenvironment might be a key process in distinguishing peri-implantitis from periodontitis.41 A recent study examined how bone stromal cells regulate immune cell populations by performing single-cell RNA-seq of mouse alveolar bone and found that macrophages are the largest subpopulation that interacts with bone stromal cells, thereby regulating the remodeling of alveolar bone.42 Previous research has shown that titanium implants or byproducts, as foreign bodies, can activate macrophages, boosting the secretion of proinflammatory cytokines. Some of these cytokines could aid in prolonging the lifespan of leucocytes at the site of inflammation, affecting peri-implant bone immunity.9,43,44 As an example, IL-1β, G-CSF and tumor necrosis factor are released by macrophages and can enhance bone resorption. The comprehensive immune landscape profile and GSEA detected the robust activation of macrophages and bone loss. These findings suggest that peri-implantitis may elicit a more enhanced host response and extensive infiltration of macrophages and osteoclastogenesis than periodontitis within the same susceptible host environment.
After further analysis and validation of the exclusive DEGs of peri-implantitis, three core genes, IL1B, CCL3 and CLEC4E, were filtered out, which could be the key regulatory factors in peri-implantitis. Upon reviewing the published literature, these core genes were discovered to have a wide range of functions in immune cell chemotaxis and migration. IL1B, as a classical proinflammatory cytokine-coding gene, is one of the most extensively studied immunologic indicators for peri-implantitis. Mounting evidence has supported that IL1B could be a potential biomarker of peri-implantitis owing to its close relationship with the development of peri-implantitis.14,45–47 The production of IL1B may stimulate osteoclast differentiation via a RANKL-independent mechanism in activated macrophages. Thus, proinflammatory cytokines are effective stimulators of osteoclast-mediated bone resorption in inflammatory bone disorders, possibly explaining the increased bone resorption observed in peri-implantitis. As evidenced above, it has been proposed that IL1B might be an effective therapeutic target in peri-implantitis. Likewise, in patient samples, it was verified that CCL3 expression was considerably higher in peri-implantitis. CCL3 is a gene that encodes a chemotactic (stimulation of cell migration) protein that plays a critical function in inflammation and enhanced osteoclast activation.48,49 The increased CCL3 protein level is related to greater severity in chronic periodontitis.50 Although the specific role of CCL3 in peri-implantitis remains unknown,51,52 together, these findings may explain the enhanced bone loss activity in peri-implantitis. What excited the authors was ClEC4E, coding the macrophage-inducible C-type lectin in macrophages, which is a critical transmembrane pattern recognition receptor capable of sensing a diverse array of sugar-containing ligands produced by pathogens.53 The majority of investigations have concentrated exclusively on the roles of Toll-like receptors in macrophage pathogen detection during periodontal infections. Recent investigations, however, have indicated that ClEC4E is a key receptor for sensing and modulating macrophage responses to the major periodontal pathogen Tannerella forsythia, which is also emerging as a pathogen in peri-implantitis.46,54 This is consistent with the current result that macrophages are dominant in peri-implantitis. Considering that the core genes identified reflect characteristics that distinguish peri-implantitis from periodontitis, they may provide a possible therapeutic target for peri-implantitis. Understandably, given that our findings are based on transcriptome data, rigorous validation of individual protein levels and experiments examining their putative function are needed to elucidate the clinical significance of our findings. In summary, soft tissues around implants likely trigger more enhanced host immune responses, such as dominant infiltration of macrophages, to promote osteoclastogenesis than those in periodontitis. Further studies in clinical cohorts and animal models are needed to delineate the interplay between macrophages and bone loss in the pathogenesis of peri-implantitis. Moreover, the placement of implants may alter the gene expression profile in soft tissue to some extent. An animal model combining healthy teeth, healthy implants, periodontitis and peri-implantitis may be introduced to perform deeper analysis. Importantly, our results suggest that IL1B, CCL3, and CLEC4E could serve as promising therapeutic targets for peri-implantitis. If further intervention studies corroborate the vital role and therapeutic effect of these core genes, they may contribute to the prevention and treatment of peri-implantitis.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The article contains information about the repositories and their accession numbers.
Ethics Statement
Animal care and all animal experiments were reviewed and approved by the Ethics Committee of Peking University Health Science Center (Beijing, China).
Acknowledgments
The authors would like to express their gratitude to Majorbio Technology Co., Ltd. for providing exceptional technical help during the sequencing process.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82173647), Peking University Clinical Scientist Program (BMU2019LCKXJ010), and the Beijing Municipal Natural Science Foundation (7214273).
Disclosure
The authors declare no conflicts of interest with respect to this current study.
References
1. Berglundh T, Armitage G, Araujo MG, et al. Peri-implant diseases and conditions: consensus report of workgroup 4 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Clin Periodontol. 2018;45(Suppl 20):S286–S291. doi:10.1111/jcpe.12957
2. Schwarz F, Derks J, Monje A, Wang HL. Peri-implantitis. J Clin Periodontol. 2018;45(Suppl 20):S246–S266. doi:10.1111/jcpe.12954
3. Derks J, Tomasi C. Peri-implant health and disease. A systematic review of current epidemiology. J Clin Periodontol. 2015;42(Suppl 16):S158–S171. doi:10.1111/jcpe.12334
4. Tarnow DP. Increasing prevalence of peri-implantitis: how will we manage? J Dent Res. 2016;95(1):7–8. doi:10.1177/0022034515616557
5. Heitz-Mayfield LJ, Lang NP. Comparative biology of chronic and aggressive periodontitis vs. peri-implantitis. Periodontol 2000. 2010;53:167–181. doi:10.1111/j.1600-0757.2010.00348.x
6. Daugela P, Cicciù M, Saulacic N. Surgical regenerative treatments for peri-implantitis: meta-analysis of recent findings in a systematic literature review. J Oral Maxillofac Surg. 2016;7(3):e15. doi:10.5037/jomr.2016.7315
7. Derks J, Schaller D, Håkansson J, Wennström JL, Tomasi C, Berglundh T. Effectiveness of implant therapy analyzed in a Swedish population: prevalence of peri-implantitis. J Dent Res. 2016;95(1):43–49. doi:10.1177/0022034515608832
8. Roccuzzo M, Layton DM, Roccuzzo A, Heitz-Mayfield LJ. Clinical outcomes of peri-implantitis treatment and supportive care: a systematic review. Clin Oral Implants Res. 2018;29(Suppl 16):331–350. doi:10.1111/clr.13287
9. Kotsakis GA, Olmedo DG. Peri-implantitis is not periodontitis: scientific discoveries shed light on microbiome-biomaterial interactions that may determine disease phenotype. Periodontol 2000. 2021;86(1):231–240. doi:10.1111/prd.12372
10. Carcuac O, Berglundh T. Composition of human peri-implantitis and periodontitis lesions. J Dent Res. 2014;93(11):1083–1088. doi:10.1177/0022034514551754
11. Dionigi C, Larsson L, Carcuac O, Berglundh T. Cellular expression of DNA damage/repair and reactive oxygen/nitrogen species in human periodontitis and peri-implantitis lesions. J Clin Periodontol. 2020;47(12):1466–1475. doi:10.1111/jcpe.13370
12. Fretwurst T, Garaicoa‐Pazmino C, Nelson K, et al. Characterization of macrophages infiltrating peri‐implantitis lesions. Clin Oral Implants Res. 2020;31(3):274–281. doi:10.1111/clr.13568
13. Duarte PM, de Lorenzo Abreu L, Vilela A, Feres M, Giro G, Miranda TS. Protein and mRNA detection of classic cytokines in corresponding samples of serum, gingival tissue and gingival crevicular fluid from subjects with periodontitis. J Periodontal Res. 2019;54(2):174–179. doi:10.1111/jre.12617
14. Ghassib I, Chen Z, Zhu J, Wang HL. Use of IL-1 beta, IL-6, TNF-alpha, and MMP-8 biomarkers to distinguish peri-implant diseases: a systematic review and meta-analysis. Clin Implant Dent Relat Res. 2019;21(1):190–207. doi:10.1111/cid.12694
15. Renvert S, Widen C, Persson GR. Cytokine expression in peri-implant crevicular fluid in relation to bacterial presence. J Clin Periodontol. 2015;42(7):697–702. doi:10.1111/jcpe.12422
16. Becker ST, Beck-Broichsitter BE, Graetz C, Dorfer CE, Wiltfang J, Hasler R. Peri-implantitis versus periodontitis: functional differences indicated by transcriptome profiling. Clin Implant Dent Relat Res. 2014;16(3):401–411. doi:10.1111/cid.12001
17. Liu Y, Liu Q, Li Z, et al. Long non-coding RNA and mRNA expression profiles in peri-implantitis vs periodontitis. J Periodontal Res. 2020;55(3):342–353. doi:10.1111/jre.12718
18. Zhou H, Chen D, Xie G, Li J, Tang J, Tang L. LncRNA-mediated ceRNA network was identified as a crucial determinant of differential effects in periodontitis and periimplantitis by high-throughput sequencing. Clin Implant Dent Relat Res. 2020;22(3):424–450. doi:10.1111/cid.12911
19. Fairfax BP, Humburg P, Makino S, et al. Innate Immune Activity Conditions the Effect of Regulatory Variants upon Monocyte Gene Expression. Science. 2014;343(6175):1246949. doi:10.1126/science.1246949
20. Lee MN, Ye C, Villani A-C, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343(6175):1246980. doi:10.1126/science.1246980
21. Meyle J, Gersok G, Boedeker R-H, Gonzales JR. Long-term analysis of osseointegrated implants in non-smoker patients with a previous history of periodontitis. J Clin Periodontol. 2014;41(5):504–512. doi:10.1111/jcpe.12237
22. Smith MM, Knight ET, Al-Harthi L, Leichter JW. Chronic periodontitis and implant dentistry. Periodontol 2000. 2017;74(1):63–73. doi:10.1111/prd.12190
23. Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher J-P. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20(12):970–978. doi:10.1089/cmb.2012.0283
24. Hiyari S, Wong RL, Yaghsezian A, et al. Ligature-induced peri-implantitis and periodontitis in mice. J Clin Periodontol. 2018;45(1):89–99. doi:10.1111/jcpe.12817
25. Chen Z, Quan L, Huang A, et al. seq-ImmuCC: cell-centric view of tissue transcriptome measuring cellular compositions of immune microenvironment from mouse RNA-Seq data. Front Immunol. 2018;9:1286. doi:10.3389/fimmu.2018.01286
26. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi:10.1093/nar/gku1003
27. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11. doi:10.1186/1752-0509-8-s4-s11
28. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi:10.1093/nar/gkq537
29. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi:10.1101/gr.079558.108
30. Belibasakis GN. Microbiological and immuno-pathological aspects of peri-implant diseases. Arch Oral Biol. 2014;59(1):66–72. doi:10.1016/j.archoralbio.2013.09.013
31. Fretwurst T, Muller J, Larsson L, et al. Immunohistological composition of peri-implantitis affected tissue around ceramic implants-A pilot study. J Periodontol. 2021;92(4):571–579. doi:10.1002/JPER.20-0169
32. Degidi M, Artese L, Piattelli A, et al. Histological and immunohistochemical evaluation of the peri-implant soft tissues around machined and acid-etched titanium healing abutments: a prospective randomised study. Clin Oral Investig. 2012;16(3):857–866. doi:10.1007/s00784-011-0574-3
33. Chapple IL, Matthews JB. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontol 2000. 2007;43:160–232. doi:10.1111/j.1600-0757.2006.00178.x
34. Mijiritsky E, Ferroni L, Gardin C, et al. Presence of ROS in inflammatory environment of peri-implantitis tissue: in vitro and in vivo human evidence. J Clin Med. 2019;9(1):38. doi:10.3390/jcm9010038
35. Kanzaki H, Wada S, Narimiya T, et al. Pathways that regulate ROS scavenging enzymes, and their role in defense against tissue destruction in periodontitis. Front Physiol. 2017;8:351. doi:10.3389/fphys.2017.00351
36. Saita M, Kaneko J, Sato T, et al. Novel antioxidative nanotherapeutics in a rat periodontitis model: reactive oxygen species scavenging by redox injectable gel suppresses alveolar bone resorption. Biomaterials. 2016;76:292–301. doi:10.1016/j.biomaterials.2015.10.077
37. Li J, Deng C, Liang W, et al. Mn-containing bioceramics inhibit osteoclastogenesis and promote osteoporotic bone regeneration via scavenging ROS. Bioact Mater. 2021;6(11):3839–3850. doi:10.1016/j.bioactmat.2021.03.039
38. Zhang YT, Hu C, Zhang SX, et al. Euphoesulatin A prevents osteoclast differentiation and bone loss via inhibiting RANKL-induced ROS production and NF-κB and MAPK signal pathways. Bioorg Chem. 2021;119:105511. doi:10.1016/j.bioorg.2021.105511
39. Charalampakis G, Abrahamsson I, Carcuac O, Dahlén G, Berglundh T. Microbiota in experimental periodontitis and peri-implantitis in dogs. Clin Oral Implants Res. 2014;25(9):1094–1098. doi:10.1111/clr.12235
40. Tzach-Nahman R, Mizraji G, Shapira L, Nussbaum G, Wilensky A. Oral infection with Porphyromonas gingivalis induces peri-implantitis in a murine model: evaluation of bone loss and the local inflammatory response. J Clin Periodontol. 2017;44(7):739–748. doi:10.1111/jcpe.12735
41. Sima C, Glogauer M. Macrophage subsets and osteoimmunology: tuning of the immunological recognition and effector systems that maintain alveolar bone. Periodontol 2000. 2013;63(1):80–101. doi:10.1111/prd.12032
42. Lin W, Li Q, Zhang D, et al. Mapping the immune microenvironment for mandibular alveolar bone homeostasis at single-cell resolution. Bone Res. 2021;9(1):17. doi:10.1038/s41413-021-00141-5
43. Caicedo MS, Desai R, McAllister K, Reddy A, Jacobs JJ, Hallab NJ. Soluble and particulate Co-Cr-Mo alloy implant metals activate the inflammasome danger signaling pathway in human macrophages: a novel mechanism for implant debris reactivity. J Orthop Res. 2009;27(7):847–854. doi:10.1002/jor.20826
44. Wang X, Li Y, Feng Y, Cheng H, Li D. Macrophage polarization in aseptic bone resorption around dental implants induced by Ti particles in a murine model. J Periodontal Res. 2019;54(4):329–338. doi:10.1111/jre.12633
45. Duarte PM, Serrao CR, Miranda TS, et al. Could cytokine levels in the peri-implant crevicular fluid be used to distinguish between healthy implants and implants with peri-implantitis? A systematic review. J Periodontal Res. 2016;51(6):689–698. doi:10.1111/jre.12354
46. Faot F, Nascimento GG, Bielemann AM, Campão TD, Leite FR, Quirynen M. Can peri-implant crevicular fluid assist in the diagnosis of peri-implantitis? A systematic review and meta-analysis. J Periodontol. 2015;86(5):631–645. doi:10.1902/jop.2015.140603
47. Hentenaar DFM, De Waal YCM, Vissink A, et al. Biomarker levels in peri-implant crevicular fluid of healthy implants, untreated and non-surgically treated implants with peri-implantitis. J Clin Periodontol. 2021;48(4):590–601. doi:10.1111/jcpe.13423
48. Brylka LJ, Schinke T. Chemokines in physiological and pathological bone remodeling. Front Immunol. 2019;10:2182. doi:10.3389/fimmu.2019.02182
49. Taddei SR, Queiroz-Junior CM, Moura AP, et al. The effect of CCL3 and CCR1 in bone remodeling induced by mechanical loading during orthodontic tooth movement in mice. Bone. 2013;52(1):259–267. doi:10.1016/j.bone.2012.09.036
50. Souto GR, Queiroz CM
51. Bhavsar I, Miller CS, Ebersole JL, Dawson DR
52. Marques Filho JS, Gobara J
53. Patin EC, Orr SJ, Schaible UE. Macrophage inducible C-type lectin as a multifunctional player in immunity. Front Immunol. 2017;8:861. doi:10.3389/fimmu.2017.00861
54. Chinthamani S, Settem RP, Honma K, Kay JG, Sharma A, Yilmaz Ö. Macrophage inducible C-type lectin (Mincle) recognizes glycosylated surface (S)-layer of the periodontal pathogen Tannerella forsythia. PLoS One. 2017;12(3):e0173394. doi:10.1371/journal.pone.0173394
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.