Back to Journals » Neuropsychiatric Disease and Treatment » Volume 16
Commensal Microbiota Regulation of Metabolic Networks During Olfactory Dysfunction in Mice
Authors Wang H, Liu L, Rao X, Chai T, Zeng B, Zhang X, Yu Y, Zhou C, Pu J, Zhou W, Li W, Zhang H, Wei H, Xie P
Received 30 October 2019
Accepted for publication 1 March 2020
Published 19 March 2020 Volume 2020:16 Pages 761—769
DOI https://doi.org/10.2147/NDT.S236541
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jun Chen
Haiyang Wang,1,2,* Lanxiang Liu,1,3,* Xuechen Rao,1,2,* Tingjia Chai,1,* Benhua Zeng,4 Xiaotong Zhang,1 Ying Yu,1 Chanjuan Zhou,1 Juncai Pu,1,3 Wei Zhou,1 Wenxia Li,4 Hanping Zhang,1,3 Hong Wei,4 Peng Xie1–3
1NHC Key Laboratory of Diagnosis and Treatment on Brain Functional Diseases, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, People’s Republic of China; 2College of Biomedical Engineering, Chongqing Medical University, Chongqing 400016, People’s Republic of China; 3Department of Neurology, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, People’s Republic of China; 4Department of Laboratory Animal Science, College of Basic Medical Sciences, Third Military Medical University, Chongqing 400038, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng Xie
NHC Key Laboratory of Diagnosis and Treatment on Brain Functional Diseases, The First Affiliated Hospital of Chongqing Medical University, 1 Youyi Road, Yuzhong District, Chongqing 400016, People’s Republic of China
Tel +86-23-68485490
Email [email protected]
Hong Wei
Department of Laboratory Animal Science, College of Basic Medical Sciences, Third Military Medical University, Chongqing 400038, People’s Republic of China
Tel +86-23-68752051
Email [email protected]
Introduction: Recently, an increasing number of studies have focused on commensal microbiota. These microorganisms have been suggested to impact human health and disease. However, only a small amount of data exists to support the assessment of the influences that commensal microbiota exert on olfactory function.
Methods: We used a buried food pellet test (BFPT) to investigate and compare olfactory functions in adult, male, germ-free (GF) and specific-pathogen-free (SPF) mice, then examined and compared the metabolomic profiles for olfactory bulbs (OBs) isolated from GF and SPF mice to uncover the mechanisms associated with olfactory dysfunction.
Results: We found that the absence of commensal microbiota was able to influence olfactory function and the metabolic signatures of OBs, with 38 metabolites presenting significant differences between the two groups. These metabolites were primarily associated with disturbances in glycolysis, the tricarboxylic acid (TCA) cycle, amino acid metabolism, and purine catabolism. Finally, the commensal microbiota regulation of metabolic networks during olfactory dysfunction was identified, based on an integrated analysis of metabolite, protein, and mRNA levels.
Conclusion: This study demonstrated that the absence of commensal microbiota may impair olfactory function and disrupt metabolic networks. These findings provide a new entry-point for understanding olfactory-associated disorders and their potential underlying mechanisms.
Keywords: gut microbiota, germ-free, olfactory bulb, metabolomic, gas chromatography-mass spectrometry
Introduction
Olfaction, or sense of smell, serves various important functions during everyday life for mammals, mediating functions such as the detection of environmental hazards, food preferences, spatial orientation, social interactions, and emotional behaviors.1 The olfactory bulb (OB), located in the rostral extension of the forebrain, is an important relay station for the olfactory system.2 It receives sensory inputs from one source (axons that project from the olfactory receptor neurons of the olfactory epithelium) and relays these signals through two outputs (axons that project from mitral and tufted cells). Reduced OB volumes have been associated with olfactory dysfunction in patients with major depressive and post-traumatic stress disorders.3–5 Moreover, olfactory dysfunction has also been demonstrated during many neurodegenerative diseases,6 particularly Parkinson’s disease7 and Alzheimer’s disease.8 Thus, the important role played by the OB in the olfactory system makes it a preferred area of research when examining the mechanisms of olfactory-associated disorders.
Reductions in the relative sensitivity and specificity of the olfactory system have also been identified in patients with mental disorders. Our previous studies consistently demonstrated that decreased neurogenesis and enhanced apoptosis in the OB were associated with depression.9,10 In addition, previous proteomic profiling analyses revealed that neurogenesis and energy metabolism disturbances may be involved in stress-induced mechanisms of OB dysfunction.11 Interestingly, OB dysfunction, accompanied by disruptions in the cAMP response element-binding protein (CREB) signaling pathway, was identified during gut microbiome remolding in depressive mice.12 Emerging evidence has indicated that microbes can influence the host’s olfactory signaling systems by producing metabolites that function as odorants.13 Commensal microbiota is necessary for the appropriate regulation of normal olfactory epithelium development14 and may play potential roles in animal physiology and behavior, through olfaction alterations.
Germ-free (GF) mice, which are devoid of any bacterial contamination, can act as an important model when studying the effects of commensal microbiota on host physiology and behavior.15 We have successfully used GF mice to study the molecular underpinnings of the effects of commensal microbiota on the hosts’ brain and behavioral functions.16–20 Moreover, a previous study using GF mice found that microbiota can modulate the physiology of the olfactory epithelium, resulting in a thinner cilia layer and the reduced expression of genes related to the olfactory transduction pathway.21 Thus, we hypothesized that commensal microbiota dysbiosis may influence the biochemistry of the OB and affect olfaction through host-microbe interactions.
Therefore, we performed a buried food pellet test (BFPT) with GF mice and compared the results with those for specific-pathogen-free (SPF) mice to determine whether the absence of commensal microbiota could influence olfactory function. Subsequently, a gas chromatography-mass spectrometry (GC-MS) metabolomics approach, coupled with multivariate statistical methods, was employed to explore changes in the biochemical features of the OB in GF mice compared with those in SPF mice. Moreover, we performed an integrated analysis of our previous proteomic profiles and mRNA microarray results and the metabolic changes assessed in this study for further insights into the molecular underpinnings of OB dysfunction that may be influenced by the presence of commensal microbiota.
Materials and Methods
Animals
Adult, male, GF and SPF Kunming mice (age: 8 weeks; weight: 35–40 g) were obtained from the Experimental Animal Research Center at Third Military Medical University (Chongqing, China). GF mice were maintained under GF conditions, in vinyl isolators, until the beginning of these experiments. We verified the germ-free statuses of all GF mice by testing the feces and skin, according to the Chinese Laboratory Animal-Microbiological standards (GB 14922.2–2011), by PCR analysis, using a universal primer for the V3 region of the bacterial 16S rRNA gene.22,23 SPF mice were housed in a barrier facility, which has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International for housing SPF mice (Laboratory Animal Facility of Tsinghua University). All animals were fed with γ-ray-irradiated food and water and maintained under standard conditions (12-h light/dark cycle, with lights on from 7:30 to 19:30; humidity: 55 ± 5%; temperature: 21–22°C). Experimental procedures were approved by the Institutional Animal Care and Use Committee of Third Military Medical and performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978).
Behavioral Tests
The BFPT was employed, with modification.24–26 A Purina mouse chow pellet (GB 14924.3–2010) was used in place of an exogenously scented pellet, to eliminate the pre-training necessary for mice to learn an exogenous food odor. GF and SPF mice (n = 8 per group) were food-deprived for 16 h. A single mouse was placed at the center of the test cage (45 cm × 24 cm × 20 cm) and allowed to recover a 1-g buried food pellet. The latency time, defined as the time between placing the mouse into the cage and the mouse grasping the food pellet with its forepaws or teeth, was recorded. Animals that did not find the food pellet within 10 min were removed from the test cage and placed back into the home cage. This trial was repeated three times with 1-h intervals. The food pellet was placed into a randomly chosen corner for each trial. The latency values were averaged among the three trials. In addition, one hour later, the latency time to reach a visible food pellet was recorded for each mouse, as a control, and this control trial was only performed once for each mouse. The behavioral data were analyzed using a nonparametric Mann–Whitney U-test because of the small sample size, and a p-value of less than 0.05 was considered to be significant.
Sample Collection and Pretreatment
The GF and SPF mice (n = 10 per group) were anesthetized with chloral hydrate. Following decapitation, the OBs were rapidly separated on an ice-cold plate, snap-frozen in liquid nitrogen, and stored at −80°C, until analysis. The GC-MS approach was described previously.27,28 Briefly, the OB samples were homogenized, using 400 μL water-methanol-chloroform (2:5:2, v/v/v), and centrifuged at 14,000 rpm, for 15 min, at 4°C. A dulcitol solution was used as an internal standard. Then, 350 μL of supernatant was collected. The residue was resuspended in 100 μL methanol, followed by vortexing and centrifugation at 14,000 rpm for 15 min, at 4°C, after which, 100 μL of supernatant was extracted and mixed with the first supernatant. A 400-μL aliquot of the mixed supernatant was evaporated to dryness under a stream of nitrogen gas, and derivatized with 30 μL methoxamine hydrochloride (20 mg/mL pyridine), for 90 min, at 37°C, with continuous shaking. Then, 30 μL of N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA), containing 1% trimethylchlorosilane (TMCS) was added to the mixture and allowed to react for 1 h, at 70°C.
GC-MS Analysis
A 1-μL volume of each derivative sample transferred by splitless injection into an Agilent 7980 GC system (Agilent Technologies Inc., USA). A helium carrier gas, at a flow rate of 1 mL/min, was used to separate the mixture on an HP-5 MS fused silica capillary column (30 m × 0.25 mm × 0.25 μm, Agilent, USA). The injector temperature was set to 280°C, and the column temperature was maintained at 80°C for 2 min and then maintained at 300°C for 6 min. The column effluent was transferred into the ion source of an Agilent 5975 mass selective detector (Agilent Technologies). The temperature of the ion source and the MS quadrupole were set to 230°C and 150°C, respectively. Masses were acquired from 50 to 600 m/z.
GC-MS Data Analysis
The GC-MS metabolite profiles were processed, after conversion into the NetCDF file format, using TagFinder.35. Following deconvolution, alignment, and data reduction, a list of mass and retention time pairs, with the corresponding intensities for all detected peaks from each data file, was obtained. The normalized dataset, including peak indices (RT-m/z pairs), sample names (observations), and normalized peak area percentages, was introduced into SIMCA-P 12.0 (Umetrics, Umeå, Sweden) for multivariate statistical analysis. An unsupervised principal component analysis (PCA) was performed to obtain an overview of the variations among individuals. Partial least squares discriminant analysis (PLS-DA) and pair-wise orthogonal projections to latent structures discriminant analysis (OPLS–DA) were performed to visually discriminate between GF and SPF mice. Differentially expressed metabolites were identified according to a threshold for the variable influence on projection values (VIP > 1) derived from the OPLS–DA model and a false discovery rate (FDR < 0.05), which was adjusted by the Benjamini–Hochberg procedure.
Bioinformatics Analysis
To further understand the biological functions associated with metabolic molecules that presented significant alterations, we performed enrichment pathway analysis on all differently expressed metabolites, using the online MetaboAnalyst software.29 We then visualized the results using an enrichment plot. Both deficiencies and alterations in the composition of commensal microbiota represent types of dysbiosis. Thus, we integrated the present metabolomics data with previous proteomic profiles, derived from mice undergoing gut microbiota remolding,12 and mRNA microarrays from GF mice,20 facilitating further insights into the molecular underpinnings of olfactory dysfunction that may be influenced by commensal microbiota.
Results
Olfactory Function Analysis
Olfactory function was evaluated by observing behavioral changes during the BFPT. Based on the average latency time across three trials, GF mice took longer to find the buried pellet than SPF mice (Z = −2.521, p = 0.012, Figure 1A). However, no difference was observed for the latency time to reach a visible pellet between GF and SPF mice (Z = −0.525, p = 0.6, Figure 1B). These results indicated that although both GF and SPF mice demonstrated an equivalent desire for the food pellet, the absence of commensal microbiota resulted in impaired olfactory function in GF mice compared with that in SPF mice.
OB Metabolite Signature in GF Mice
Typical GC-MS total ion current chromatograms were performed for both GF and SPF mice. In total, 326 metabolites, which were identified in at least 80% of all samples in each group, were characterized. From the PCA score plots (R2X = 0.685, Figure 2A), the SPF samples were clustered tightly, suggesting the detection of only small changes in metabolite levels within the SPF group. PLS-DA was performed to explore the metabolic differences between the GF and SPF groups, and the resulting score plot demonstrated significant discrimination between the two groups (R2Y=0.994, Q2=0.944, Figure 2B). Moreover, OPLS-DA was also performed to obtain more precise information regarding the identified metabolites in the GF and SPF groups. The OPLS-DA score plot also demonstrated significant discrimination between the two groups (R2Y=0.970, Q2=0.882, Figure 2C). Based on the thresholds described above (VIP > 1, FDR < 0.05), a total of 38 differential metabolites were identified between the GF and SPF groups (Table 1). Compared with the SPF group, 23 metabolites were upregulated in GF mice. In contrast, 15 metabolites were downregulated in the GF group relative to the SPF group.
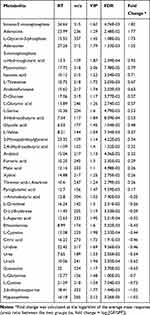 |
Table 1 Differentially Expressed Metabolites Identified in the Olfactory Bulb Between GF and SPF Mice |
Functional Enrichment Analysis
According to the functional enrichment analysis (Figure 3A), many metabolites were involved in the urea cycle (ie, adenosine-5-monophosphate, fumaric acid, L-glutamic acid, L-glutamine, L-aspartic acid, L-ornithine, and urea) and purine metabolism (ie, adenosine-5-monophosphate, adenosine, guanosine, hypoxanthine, inosine-5ʹ-monophosphate, 2,6-dihydroxypurine, fumaric acid, L-glutamic acid, L-glutamine, and L-aspartic acid). Among these metabolites, hypoxanthine and 2,6-dihydroxypurine (xanthine), which are the end-products of purine metabolism, were downregulated in GF mice compared with SPF mice, suggesting that the absence of commensal microbiota may disrupt purine metabolism. To our knowledge, the urea cycle primarily occurs in the liver; thus, the L-ornithine and urea that were identified in the OB may be byproducts of other metabolic pathways. In addition, pathway analysis for the differentially expressed metabolites revealed that arginine and proline metabolism, alanine, aspartate, and glutamate metabolism, and purine metabolism were the primary perturbed pathways (Figure 3B).
 |
Figure 3 The function enrichment (A) and pathway (B) analyses for differential metabolites, using MetaboAnalyst software. |
Construction of Multi-Omics-Based Networks
Based on the identified metabolic changes and the integrated analysis that included previously reported protein and mRNA alterations, we constructed multi-omics-based networks that may be regulated by commensal microbiota dysbiosis (Figure 4). In the tricarboxylic acid (TCA) cycle network, mRNAs encoding two rate-limiting enzymes, pyruvate dehydrogenase (PDH), and isocitrate dehydrogenase (IDH), were downregulated, suggesting that TCA cycle activity may be reduced in the absence of commensal microbiota. Interestingly, mRNAs encoding succinate-CoA ligase (SUCL), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH) were also downregulated; however, the corresponding proteins were upregulated, a paradox that may be attributable to different subtypes of commensal microbiota dysbiosis. Moreover, the inhibitory neurotransmitter, γ-Aminobutyric acid (GABA), was downregulated, likely due to the decreased expression of glutamate decarboxylase (GAD). Additionally, in the purine metabolism network, the mRNAs encoding adenylosuccinate synthetase (ADSS), adenylosuccinate lyase (ADSL), 5ʹ-nucleotidase (5ʹ-NT), and purine-nucleoside phosphorylase (PNP) were downregulated, and the mRNA encoding xanthine dehydrogenase (XDH), which is the rate-limiting enzyme for purine metabolism, was upregulated, suggesting a disturbance in purine metabolism dynamics in the absence of commensal microbiota.
Discussion
The OB is anatomically and functionally related to the hippocampus and amygdala.30 Anatomically, the OB is located only three synapses away from the hippocampus and two synapses away from the amygdala. In the olfactory pathway, olfactory receptor neurons recognize odor molecules and transfer these olfactory signal to the OB, which then amplifies the signals and transfers them to the primary olfactory cortex (piriform cortex), hippocampus, and amygdala, which have been identified as the important brain regions in the olfactory system. Previous studies have reported that the hippocampus and amygdala were significantly influenced by commensal microbiota.16,31,32 In addition, our previous study showed that commensal microbiota may influence the host by modulating the host’s metabolism.16 Expanding upon previous findings, in this study, we found that the absence of commensal microbiota can influence olfactory function and the metabolic signatures of the OB in male mice. We did not use female mice in this study because female hormone cycles may influence olfactory function.33 Moreover, by integrating these metabolic changes with previously reported alterations in associated proteins and mRNAs, we provided a new entry-point for understanding the molecular mechanisms underlying impaired olfactory function caused by commensal microbiota dysbiosis.
In the present study, compared with SPF mice, 11 metabolites involved in glycolysis and the TCA cycle (ie, L-glycerol-3-phosphate, α-hydroxyglutaric acid, itaconic acid, D-glucose, 3-hydroxybutyric acid, arabitol, 2,4-dihydroxybutyric acid, xylitol, fumaric acid, malic acid, and citric acid) were found to be altered in GF mice, and most of these metabolites were upregulated in GF mice compared with SPF mice, except for citric acid, which is a key intermediate metabolite of the TCA cycle. The PDH complex catalyzes the pivotal and irreversible oxidative decarboxylation of pyruvate to form acetyl-CoA, which leads to the consumption of glucose in the energy-generating pathways.34 Interestingly, the mRNA encoding PDH was downregulated in GF mice compared with SPF mice. Moreover, the mRNA encoding IDH, a multi-subunit enzyme located in the mitochondrial matrix that is thought to play a central role during aerobic energy production in the TCA cycle,35 was consistently downregulated in GF mice compared with SPF mice. Taken together, these results revealed that glycolysis was enhanced, whereas the TCA cycle was inhibited, in GF mice compared with SPF mice, suggesting deficiencies in the OB energy supply in the absence of commensal microbiota.
Moreover, a panel of amino acids (ie, L-glutamic acid, L-glutamine, L-aspartic acid, GABA, L-threonine, L-serine, L-valine, L-ornithine L-cysteine, and L-cystine) were found to be significantly disturbed in GF mice relative to SPF mice, and some of these perturbed amino acids act as neurotransmitters (ie, L-glutamic acid, L-glutamine, L-aspartic acid, and GABA). Here, the level of primary excitatory neurotransmitters, such as L-glutamic acid, was upregulated in the OB of GF mice compared with SPF mice. However, L-glutamine was simultaneously downregulated in GF mice compared with SPF mice, implying the inhibition of the transformation of glutamate into glutamine. By contrast, the level of inhibitory neurotransmitters, such as GABA, was downregulated in GF mice compared with SPF mice, likely due to the decreased expression of the GAD enzyme. Previous studies have shown that microbiota can modulate excitatory and inhibitory neurotransmitter levels.36–38 This imbalance between excitatory and inhibitory neurotransmitters may result in excitotoxicity,39 which could lead to decreased OB neurogenesis and account for impaired olfactory function. Importantly, the OB efferents are primarily glutamatergic,40 and olfactory dysfunction would likely disrupt the physiology of OB, which may subsequently contribute to excitatory-inhibitory imbalances in their target regions. In addition, L-ornithine was downregulated, accompanied by the downregulation of the mRNAs encoding Spermidine/spermine N1-acetyltransferase (SAT) and spermine oxidase (SMOX),41 supporting a disturbance in polyamine catabolism during olfactory dysfunction.
Purinergic catabolism plays an important role in the mitochondrial antioxidant defense.42 In the present study, upstream metabolites of purinergic catabolism (ie, inosine-5′-monophosphate, adenosine, and adenosine-5-monophosphate) were upregulated in GF mice relative to the SPF mice. In contrast, the downstream metabolites of purinergic catabolism (ie, uridine, uracil, guanosine, 2,6-dihydroxypurine, and hypoxanthine) were significantly downregulated in GF mice. Moreover, the mRNAs encoding ADSS, ADSL, 5ʹ-NT, and PNP, the enzymes that catalyze purine catabolism, were downregulated in GF mice, whereas the mRNA encoding XDH was upregulated, changes which may be associated with the immune dysfunction.43,44 Previous studies have also indicated that commensal microbiota may influence purine metabolism by altering the expression of XDH.45,46 These findings indicated that the absence of commensal microbiota resulted in purinergic catabolism disturbance, which may be attributed to increased oxidative stress damage. Interestingly, we found that the levels of two amino acids, L-cysteine and L-cystine, which function to fight against oxidative stress damage,47 were downregulated in GF mice compared to SPF mice, further supporting the high-oxidative-stress status of GF mice.
Importantly, this study has some limitations. First, although the sample size used in this study was calculated to provide sufficient statistical power, the sample size was relatively small and remains a possible limitation; thus, further studies using large sample sizes are required. Second, only the GC-MS approach was used to identify metabolic changes in the OB; thus, the combination of multiple metabolomics techniques should be considered in future metabolic studies. Third, the primary findings in the present study were not experimentally verified because the multi-omics integrated analysis was able to provide very high confidence in these results. Fourth, only the OB was examined in this study, due to its pivotal role in the olfactory system. However, other brain regions should be included in future analyses to obtain a comprehensive map of the mechanisms through which commensal microbiota may mediate changes in olfactory function.
Conclusion
Here, we provided evidence that the absence of commensal microbiota significantly impaired olfactory function and that the molecular mechanisms underlying these changes were primarily associated with disturbances in glycolysis, the TCA cycle, amino acid metabolism, and purine catabolism. These findings highlight the potential causal role played by commensal microbiota on olfactory-associated disorders and their potential underlying mechanisms.
Abbreviations
5ʹ-NT, 5ʹ-nucleotidase; ADSL, adenylosuccinate lyase; ADSS, adenylosuccinate synthetase; BFPT, buried food pellet test; FDR, false discovery rate; GABA, γ-Aminobutyric acid; GAD, glutamate decarboxylase; GC-MS, gas chromatography–mass spectrometry; GF, germ-free; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; OB, olfactory bulb; OPLS–DA, pair-wise orthogonal projections to latent structures discriminant analysis; PDH, pyruvate dehydrogenase; PLS-DA, partial least squares discriminant analysis; PNP, purine-nucleoside phosphorylase; SAT, Spermidine/spermine N1-acetyl transferase; SDH, succinate dehydrogenase; SMOX, spermine oxidase; SPF, specific pathogen free; SUCL, succinate-CoA ligase; TCA, tricarboxylic acid; VIP, variable influence on projection values; XDH, xanthine dehydrogenase.
Acknowledgments
This study was supported by the National Key R&D Program of China (Grant no. 2017YFA0505700), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (Grant no. 2019PT320002300) and the Natural Science Foundation Project of China (Grant no. 81820108015).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhou G, Lane G, Cooper SL, Kahnt T, Zelano C. Characterizing functional pathways of the human olfactory system. Elife. 2019;8.
2. Mori K, Takahashi YK, Igarashi KM, Yamaguchi M. Maps of odorant molecular features in the Mammalian olfactory bulb. Physiol Rev. 2006;86(2):409–433. doi:10.1152/physrev.00021.2005
3. Buschhuter D, Smitka M, Puschmann S, et al. Correlation between olfactory bulb volume and olfactory function. Neuroimage. 2008;42(2):498–502. doi:10.1016/j.neuroimage.2008.05.004
4. Mueller A, Rodewald A, Reden J, Gerber J, von Kummer R, Hummel T. Reduced olfactory bulb volume in post-traumatic and post-infectious olfactory dysfunction. Neuroreport. 2005;16(5):475–478. doi:10.1097/00001756-200504040-00011
5. Negoias S, Croy I, Gerber J, et al. Reduced olfactory bulb volume and olfactory sensitivity in patients with acute major depression. Neuroscience. 2010;169(1):415–421. doi:10.1016/j.neuroscience.2010.05.012
6. Attems J, Walker L, Jellinger KA. Olfactory bulb involvement in neurodegenerative diseases. Acta Neuropathol. 2014;127(4):459–475. doi:10.1007/s00401-014-1261-7
7. Doty RL. Olfactory dysfunction in Parkinson disease. Nat Rev Neurol. 2012;8(6):329–339. doi:10.1038/nrneurol.2012.80
8. Murphy C. Olfactory and other sensory impairments in Alzheimer disease. Nat Rev Neurol. 2019;15(1):11–24. doi:10.1038/s41582-018-0097-5
9. Yang D, Liu X, Zhang R, et al. Increased apoptosis and different regulation of pro-apoptosis protein bax and anti-apoptosis protein bcl-2 in the olfactory bulb of a rat model of depression. Neurosci Lett. 2011;504(1):18–22. doi:10.1016/j.neulet.2011.08.046
10. Yang D, Li Q, Fang L, et al. Reduced neurogenesis and pre-synaptic dysfunction in the olfactory bulb of a rat model of depression. Neuroscience. 2011;192:609–618. doi:10.1016/j.neuroscience.2011.06.043
11. Cheng K, Li J, Yang D, et al. 2D-gel based proteomics unravels neurogenesis and energetic metabolism dysfunction of the olfactory bulb in CUMS rat model. Behav Brain Res. 2016;313:302–309. doi:10.1016/j.bbr.2016.05.026
12. Huang C, Yang X, Zeng B, et al. Proteomic analysis of olfactory bulb suggests CACNA1E as a promoter of CREB signaling in microbiota-induced depression. J Proteomics. 2019;194:132–147. doi:10.1016/j.jprot.2018.11.023
13. Carthey AJR, Gillings MR, Blumstein DT. The extended genotype: microbially mediated olfactory communication. Trends Ecol Evol. 2018;33(11):885–894. doi:10.1016/j.tree.2018.08.010
14. Koskinen K, Reichert JL, Hoier S, et al. The nasal microbiome mirrors and potentially shapes olfactory function. Sci Rep. 2018;8(1):1296. doi:10.1038/s41598-018-19438-3
15. Grover M, Kashyap PC. Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol Motil. 2014;26(6):745–748. doi:10.1111/nmo.2014.26.issue-6
16. Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21(6):786–796. doi:10.1038/mp.2016.44
17. Zeng L, Zeng B, Wang H, et al. Microbiota modulates behavior and protein kinase C mediated cAMP response element-binding protein signaling. Sci Rep. 2016;6(1):29998. doi:10.1038/srep29998
18. Luo Y, Zeng B, Zeng L, et al. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl Psychiatry. 2018;8(1):187. doi:10.1038/s41398-018-0240-5
19. Huo R, Zeng B, Zeng L, et al. Microbiota modulate anxiety-like behavior and endocrine abnormalities in hypothalamic-pituitary-adrenal axis. Front Cell Infect Microbiol. 2017;7:489. doi:10.3389/fcimb.2017.00489
20. Chen JJ, Zeng BH, Li WW, et al. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav Brain Res. 2017;322(Pt A):34–41. doi:10.1016/j.bbr.2017.01.021
21. François A, Grebert D, Rhimi M, et al. Olfactory epithelium changes in germfree mice. Sci Rep. 2016;6:24687. doi:10.1038/srep24687
22. Zeng B, Li G, Yuan J, Li W, Tang H, Wei H. Effects of age and strain on the microbiota colonization in an infant human flora-associated mouse model. Curr Microbiol. 2013;67(3):313–321. doi:10.1007/s00284-013-0360-3
23. Yuan J, Zeng B, Niu R, et al. The development and stability of the genus Bacteriodes from human gut microbiota in HFA mice model. Curr Microbiol. 2011;62(4):1107–1112. doi:10.1007/s00284-010-9833-9
24. Bianchi P, Bettini S, Guidi S, et al. Age-related impairment of olfactory bulb neurogenesis in the Ts65Dn mouse model of Down syndrome. Exp Neurol. 2014;251:1–11. doi:10.1016/j.expneurol.2013.10.018
25. Gao X, Li N, Zhang J. SB203580, a p38MAPK inhibitor, attenuates olfactory dysfunction by inhibiting OSN apoptosis in AR mice (activation and involvement of the p38 mitogen-activated protein kinase in olfactory sensory neuronal apoptosis of OVA-induced allergic rhinitis). Brain Behav. 2019;9(6):e01295. doi:10.1002/brb3.1295
26. Nathan BP, Yost J, Litherland MT, Struble RG, Switzer PV. Olfactory function in apoE knockout mice. Behav Brain Res. 2004;150(1–2):1–7. doi:10.1016/S0166-4328(03)00219-5
27. Liu L, Zhou X, Zhang Y, et al. The identification of metabolic disturbances in the prefrontal cortex of the chronic restraint stress rat model of depression. Behav Brain Res. 2016;305:148–156. doi:10.1016/j.bbr.2016.03.005
28. Liu L, Zhou X, Zhang Y, et al. Hippocampal metabolic differences implicate distinctions between physical and psychological stress in four rat models of depression. Transl Psychiatry. 2018;8(1):4. doi:10.1038/s41398-017-0018-1
29. Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–w494. doi:10.1093/nar/gky310
30. Smitka M, Puschmann S, Buschhueter D, et al. Is there a correlation between hippocampus and amygdala volume and olfactory function in healthy subjects? Neuroimage. 2012;59(2):1052–1057. doi:10.1016/j.neuroimage.2011.09.024
31. Stilling RM, Ryan FJ, Hoban AE, et al. Microbes & neurodevelopment–absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 2015;50:209–220. doi:10.1016/j.bbi.2015.07.009
32. Pearson-leary J, Zhao C, Bittinger K, et al. The gut microbiome regulates the increases in depressive-type behaviors and in inflammatory processes in the ventral hippocampus of stress vulnerable rats. Mol Psychiatry. 2019. doi:10.1038/s41380-019-0380-x
33. Watanabe K, Umezu K, Kurahashi T. Human olfactory contrast changes during the menstrual cycle. Jpn J Physiol. 2002;52(4):353–359. doi:10.2170/jjphysiol.52.353
34. Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4(14):3224–3233. doi:10.1096/fsb2.v4.14
35. Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102(13):932–941. doi:10.1093/jnci/djq187
36. Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28(8):1221–1238. doi:10.1210/me.2014-1108
37. Moloney RD, Desbonnet L, Clarke G, Dinan TG, Cryan JF. The microbiome: stress, health and disease. Mamm Genome. 2014;25(1–2):49–74. doi:10.1007/s00335-013-9488-5
38. Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693(Pt B):128–133. doi:10.1016/j.brainres.2018.03.015
39. Wajner M, Kölker S, Souza DO, Hoffmann GF, de Mello CF. Modulation of glutamatergic and GABAergic neurotransmission in glutaryl-CoA dehydrogenase deficiency. J Inherit Metab Dis. 2004;27(6):825–828. doi:10.1023/B:BOLI.0000045765.37043.fb
40. Skelin I, Sato H, Diksic M. Olfactory bulbectomy reduces cerebral glucose utilization: 2-[14C]deoxyglucose autoradiographic study. Brain Res Bull. 2008;76(5):485–492. doi:10.1016/j.brainresbull.2008.01.020
41. Gross JA, Turecki G. Suicide and the polyamine system. CNS Neurol Disord Drug Targets. 2013;12(7):980–988. doi:10.2174/18715273113129990095
42. Yao JK, Dougherty GG, Reddy RD, Matson WR, Kaddurah-daouk R, Keshavan MS. Associations between purine metabolites and monoamine neurotransmitters in first-episode psychosis. Front Cell Neurosci. 2013;7:90. doi:10.3389/fncel.2013.00090
43. Grunebaum E, Zhang J, Roifman CM. Novel mutations and hot-spots in patients with purine nucleoside phosphorylase deficiency. Nucleosides Nucleotides Nucleic Acids. 2004;23(8–9):1411–1415. doi:10.1081/NCN-200027647
44. Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles for ecto-5ʹ-nucleotidase (CD73). Purinergic Signal. 2006;2(2):351–360. doi:10.1007/s11302-005-5302-5
45. Crane JK. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Gut Microbes. 2013;4(5):388–391. doi:10.4161/gmic.25584
46. Guo Z, Zhang J, Wang Z, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. 2016;6:20602. doi:10.1038/srep20602
47. Rodrigues SD, Batista GB, Ingberman M, Pecoits-filho R, Nakao LS. Plasma cysteine/cystine reduction potential correlates with plasma creatinine levels in chronic kidney disease. Blood Purif. 2012;34(3–4):231–237. doi:10.1159/000342627
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



