Back to Journals » OncoTargets and Therapy » Volume 11
Combined detection of IL-6 and IL-8 is beneficial to the diagnosis of early stage esophageal squamous cell cancer: a preliminary study based on the screening of serum markers using protein chips
Authors Tong Q, Wang XL, Li SB, Yang GL, Jin S, Gao ZY, Liu XB
Received 16 April 2018
Accepted for publication 13 June 2018
Published 12 September 2018 Volume 2018:11 Pages 5777—5787
DOI https://doi.org/10.2147/OTT.S171242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Qiang Tong,1 Xiao-long Wang,2 Sheng-bao Li,1 Gong-li Yang,1 Shu Jin,1 Zi-ye Gao,3 Xiao-bo Liu1
1Department of Gastroenterology, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, 442000, People’s Republic of China; 2Department of Gastroenterology, Dongfeng General Hospital, Hubei University of Medicine, Shiyan, Hubei, 442000, People’s Republic of China; 3Department of Oncology, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, 442000, People’s Republic of China
Background: The diagnosis rate of early stage esophageal squamous cell carcinoma (ESCC) is low due to the lack of specific tumor markers. Seeking for these markers is beneficial to improve the early diagnosis rate and the prognosis of patients. This study profiles the differentially expressed proteins of early stage ESCC patients via the AAH-BLG-507 protein chip, which further consolidates the clinical evidence of ESCC diagnosis.
Materials and methods: In this study, 20 serum samples were collected from Taihe Hospital between August 2016 and June 2017. Ten of them carried ESCC, while the rest were healthy controls. To profile the proteins’ expression level, the AAH-BLG-507 protein chip was used, and both highly expressed and lowly expressed proteins were fished out. Meanwhile, their biological roles were examined by using Gene Ontology (GO) database and String database, and they were further verified by ELISA.
Results: Results showed that the expression levels of AXL, ARTN, Ang2, BDNF, BMP7, cripto-1, CCL28, E-selectin, IL-6, IL-8 and SHH in the serum of early ESCC were significantly upregulated (P<0.05), particularly IL-6 and IL-8. The expression levels of TSP1 and MMP-8 were markedly downregulated (P<0.05). Analysis showed that these proteins were mainly involved in angiogenesis, signal transduction, cell proliferation and migration, indicating the close relationship with the development of ESCC.
Conclusion: It suggested that IL-6 and IL-8 proteins could be considered as the markers for ESCC diagnosis.
Keywords: early stage esophageal cancer, squamous cell carcinoma, protein chip, tumor marker, bioinformatics analysis
Introduction
About 450,000 cases of esophageal cancer (EC) are diagnosed each year globally, and among whom, >400,000 die from EC.1 EC can be divided into esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC).2 People’s Republic of China is one of the countries showing a high incidence of EC, accounting for 50% cases worldwide.3,4 The 5-year survival rate of early EC is >90%.5 However, >70% has turned to advanced stage when diagnosed.6,7 Studies show that adjuvant chemotherapy, radiotherapy, and endoscopic treatments elongate the 5-year survival rate marginally,8 significantly decreasing to 30%–40%.5
The early stage diagnosis of EC is essential in prolonging the survival time of EC patients. However, due to the lack of appropriate screening technologies, most of them are at very late stage, leading to losing the best treatment period and causing poor prognosis. Although gastroscopy and pathological examination are effective, the aggressive, expensive and low-acceptance rate markedly hinders their applicability. Hence, seeking for high-sensitive and specific biomarkers can be beneficial for early stage EC diagnosis. It has been reported that the diagnosis sensitivity of squamous cell carcinoma antigen (SCC-Ag) is 8%–37% for EC and 32%–45% for cytokeratin 19 fragment (Cyfra 21-1), while their combinatorial diagnosis sensitivity is 50%.9,10 Unfortunately, Cyfra 21-1 and SCC-Ag can only apply to diagnose the stage II patients but not the early stage.11 Cheng et al12 had found that phenylalanine, 4-hydroxyphenyllactic acid, 3,4-two hydroxyphenylalanine and 3,4-two hydroxyphenylacetic acid can be used as diagnostic biomarkers for ESCC. Jin et al13 demonstrated that the combined detection of three serum metabolites valine, γ-aminobutyric acid and pyrrole-2-carboxylic acid has a high diagnostic value in ESCC metastasis. The calculation of canine urinary ammonia, 5-hydroxytryptophan, 5-hydroxyindole-3-acetic acid, 5-hydroxytryptamine and the ratio of their tryptophan may be screened from healthy people for ESCC and metastatic ESCC patients.14 To date, there is no specific marker for ESCC.
Proteome has been promising in quantifying and qualifying cancer cells from normal cells.15,16 It is because the groups of proteins and their expression levels altered in the serum,17 which further reflects the progress of the tumor to a certain extent. Hence, detecting and distinguishing the differentially expressed proteins in tissues or serum can be beneficial to diagnose EC.18–20
Obtaining the whole proteome of serum/tissues is straightforward, such as bi-directional gel electrophoresis, mass spectrometric identification and protein chip. Among them, protein chip is a high-throughput, multi-targeted and high-sensitive technology for protein detection. This technique has been successfully used to identify the key factors of some diseases, screen the markers of cancers and determine the drug targets.21–26 It has been reported that the AAH-BLG-507 protein chip has the capacity to simultaneously detect 507 human proteins including angiogenic factors, inflammatory factors, growth factors, cell adhesion molecules, soluble receptors and chemokines.
To our knowledge, few studies exist diagnosing the early stage ESCC via proteomics approach. In this study, we adopted the AAH-BLG-507 protein chip provided by Ray Biotech (Norcross, GA, USA) to screen the proteins contained in the serum of ESCC samples and those from the normal ones. We aimed at discovering protein markers for the diagnosis of early stage ESCC.
Materials and methods
Sample preparation
Ten cases of patients, including five males and five females, were randomly selected from the patients suffering from early esophageal cancer (the tumor group) from August 2016 to June 2017 from Taihe Hospital. The normal samples (the control group) were collected from the biometrics center of the same hospital during the same period. The average age of the cancer group and the normal group was 57.90±2.10 and 58.90±2.60 years, respectively. Statistical analysis showed no significant difference between the two groups (Table 1).
  | Table 1 General clinical data of two groups |
All ESCC samples have been confirmed by pathological examination after endoscopic mucosal dissection (ESD) or surgery; TNM staging was Tis or T1N0M0 (UICC2002). Criteria of exclusion: the impact caused by other tumors, autoimmune diseases and acute or chronic infections was excluded, such as liver cancer, kidney cancer and endocrine diseases.
The healthy control group’s selection criteria were as follows: no history of malignancy or family history of cancer, no significant organ dysfunction and matched age, gender and residence with those of ESCC patients. The exclusion criteria were as follows: systemic or local acute and chronic infections and liver, kidney, endocrine metabolism and autoimmune disease history, combined with other organ diseases or malignant tumor.
The study has been approved by the ethics committee of Taihe Hospital. All patients included in this study provided written informed consent.
Software and database
Software and database are given in Table 2.
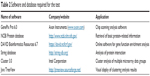  | Table 2 Software and database required for the test |
Methods
Collection of serum samples
A total of 3 mL of fasting venous blood was drawn in the morning. After sitting at room temperature for 1 hour, it was centrifuged for 15 minutes at 8,000 r/min under 4°C. The supernatant was dispensed into eppendorf tubes at 100 μL per tube. The tubes were then numbered and stored in a −80°C freezer.
Determination of protein expression level
According to the standard operating procedures of AAH-BLG-507 protein chip kit (RayBiotech), 20 μL of the serum was taken from each tube and undergone six steps to obtain the protein expression level. These steps were dialysis, biotin labeling, secondary dialysis, chip closure, sample hybridization and washing and centrifugation. Later, the expression levels were obtained by using the fluorescence scanning (GenePix 4000B, Axon Instruments, Inc, Foster City, CA, USA) followed by the built-in data transformation tool. The values were presented as mean values rounded to the nearest integer.
Bioinformatics analysis
Analysis based on Gene Ontology (GO)
The differentially expressed proteins were loaded into the DAVID 6.7 online server to annotate their functionality under the maximum false discovery rate of 0.05. The involved biological processes and signaling pathways of these proteins were also obtained. The detailed experimental parameters were set as follows – “analysis type”: “functional annotation”, “data type”: “gene list” and “output data”: “functional annotation chart”. Functional annotations of differentially expressed proteins were used to reveal the biological processes involved.
Protein network construction
String database was used to analyze the interactions of the differentially expressed proteins. Parameters were set as follows: “analysis of category selection”: “multiple names”, “species”: “Homo sapiens” and “reliability”: “high confidence (0.7)”. The software and databases used for the study are given in Table 2.
Cross-reference validation
Reference retrieval
PubMed, Embase and Cochrane library were considered for retrieving EC-related publications before December 2017. The key words used for retrieval were: “differences in protein”, “esophageal cancer” and “esophageal carcinoma”.
Inclusion criteria
The study was performed on serum, cancer or cancer cell lines, and the pathology was confirmed as esophageal cancer.
Exclusive criteria
Duplicate results or repeated publication, animal research, summary, systematic reviews, letters from readers, etc. were excluded from the study.
Validation by ELISA
A total of 30 cases were randomly selected from the early stage cancer group and the control group according to the previous criteria. These samples were tested by the instructions of the ELISA kit (Abcam, Cambridge, UK). The OD of each sample was read at the wavelength of 450 nm. The standard curve was calculated, and the corresponding concentration at each OD was calculated. Then, the levels of IL-6 and IL-8 of the serum were determined.
Statistical analyses
The t-test module from the SPSS 19.0 (IBM Corporation, Armonk, NY, USA) was used to analyze the statistical significance of the protein expression levels. A differential expression was considered as statistically significant if the unpaired t-test P-value was no larger than 0.05.
Results
Protein chip test results
The protein chip scanning results are shown in Figures 1A and B and 2. The results indicate that 13 proteins were differentially expressed, including AXL, artemin (ARTN), angiopoietin-2 (Ang2), brain-derived neurotrophic factor (BDNF), bone morphogenetic protein7 (BMP7), cripto-1 (CR-1/teratocarcinoma-derived growth factor 1 (TDGF-1)), CCL28, E-selectin, interleukin-6 (IL-6), interleukin-8 (IL-8), sonic Hedgehog (SHH), matrix metalloproteinase 8 (MMP-8) and thrombospondin-1 (TSP1). The protein expression levels of MMP-8 and TSP1 were downregulated, while those of the rest of proteins were upregulated (Table 3). There was no significant difference in the expression of various proteins in esophageal precancerous lesions (P>0.05, fold change<1.50, data not shown).
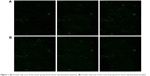  | Figure 1 (A) Protein chip scan of the tumor group (from three representative patients). (B) Protein chip scan of the control group (from three representative people). |
  | Figure 2 The differential protein expression of the cancer group and the control group. |
  | Table 3 Summary of the differentially expressed proteins between the two groups |
Bioinformatics analysis results
GO analysis results
According to the GO analysis, the 13 differentially expressed proteins were involved in seven biological processes, including cell communication, cell proliferation, signal transduction, angiogenesis, cell migration, reaction to external stimuli and phosphorylation (Table 4).
  | Table 4 GO analysis results |
KEGG pathway analysis
The differential expressed proteins were loaded into the online analysis software of String. The results showed that there were direct or indirect interactions among 12 proteins except TDGF-1 (CR-1; Figure 3). KEGG pathway analysis revealed that these proteins were involved in three signaling pathways: TGFβ signaling pathway, chemokine signaling pathway and cytokine–cytokine receptor interaction.
ELISA validation
The result of ELISA analysis showed that the concentration of IL-6 in the early cancer group was significantly higher than that in the control group (18.13±2.54 ng/mL vs 7.87±1.25 ng/mL, P<0.01). Similarly, the concentration of IL-8 in the serum of the early cancer group was significantly higher than that in healthy controls (84.92±3.06 ng/mL vs 55.83±8.01 ng/mL, P<0.01; Table 5). The results were in line with those obtained from the protein chip analysis (Figure 4A and B).
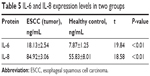  | Table 5 IL-6 and IL-8 expression levels in two groups |
  | Figure 4 The expression of IL-6 (A) and IL-8 (B) protein was verified by ELISA. |
Discussion
In this study, the L series protein chip AAH-BLG-507 provided by RayBiotech was used to compute the expression levels of 507 target proteins collected from the serum of ten randomly selected patients having early stage ESCC and ten normal samples. Based on the screening criteria of P-value<0.05 and fold change>1.50, 13 proteins were identified showing significant differential expression levels. Among them, eleven were upregulated, which were AXL, ARTN, Ang2, BDNF, BMP7, CR-1, CCL28, E-selectin, IL-6, IL-8 and SHH. The rest two TSP1 and MMP-8 were downregulated. The same resident population was selected as the testing object in this study. Besides, the sampling conditions, sample processing and detection technology were the same, eliminating the influence of other factors. Therefore, the test results were considered as authentic and credible. Database was searched to retrieve all the information of esophageal cancer to assess the validity of the identified differential in proteins. Following is the details of the identified differentially expressed proteins that are associated with ESCC.
AXL
AXL is a receptor tyrosine kinase belonging to the TAM receptor family. It promotes the growth of xenografts of various human cancers. In addition, AXL expression can activate AKT, ERK and NF-κB pathways to induce cancer cells to target drug resistance.27 AXL was up-regulated in malignant tumors, such as, lung cancer, prostate cancer, breast cancer and pancreatic cancer. Thus, it can be used as a therapeutic target.28
AXL overexpression occurs in >50% of EAC.29 Hsieh et al30 found that AXL expression was enhanced in 80% of ESCC and was significantly associated with EC progression (P<0.001), distant metastasis (P<0.05) and AXL-positive patients with increased risk of death (P<0.028). Paccez et al31 confirmed that AXL was expressed in EC cell lines and cancer tissues, especially in advanced-stage patients. Blocking AXL expression can inhibit the survival, proliferation, migration and invasion of cancer cells and tumor growth of tumor tissue in vivo. This indicates that AXL shows a clear role in ESCC tumor and has potential therapeutic significance.
ARTN
ARTN belongs to the glial cell-derived neurotrophic factor (GDNF) family of ligands (GFL), together with GDNF, neurturin and persephin. ARTN plays an important role in tumor growth, migration, adhesion and invasion. Its expression is increased in a variety of human tumors.32,33 Li et al34 detected the expression of ARTN in EC cell lines KYSE-150, KYSE-510, EC9706 and TE13 and tissues and corresponding paracancerous tissues using Western blot. The results showed that the expression of ARTN in cancer tissues was high in paracancerous tissues; they were differentially expressed in various cell lines (KYSE-150>KYSE-510, TE13>EC9706). In Chen et al,35 the expression of ARTN in ESCC was significantly higher than that in normal tissues (P<0.05). Immunohistochemical results showed that the positive expression rates of ARTN protein in normal esophageal mucosa, high-grade intraepithelial neoplasia, carcinoma in situ and ESCC were 0% (0/25), 23.5% (6/17) and 80.7% (94/114), respectively. The difference was statistically significant (P<0.01).
Ang2
Loges et al36 detected 54 cases of primary EC (ECA:ESCC=19:35) with a positive serum Ang2 positive rate of 100%. Compared with patients with cancer AEG in esophageal gastric junction, Ang2 expression was higher in ESCC patients.37 Zhou et al38 examined 91 cases of normal tissues, 44 cases of esophagitis, 85 cases of esophageal hyperplasia, 13 cases of early ESCC and 28 patients with advanced ESCC Ang2 expression level in serum. The results showed that the expression of Ang2 in the early ESCC group was significantly higher than that in other groups (P=0.009). The diagnostic sensitivity of early and advanced-stage ESCC was 23.1% and 78.6%, respectively, suggesting that serum Ang2 levels were involved with ESCC occurrence and development. However, unfortunately Ang2 cannot be used for screening of early ESCC.
BDNF
BDNF can promote the survival and differentiation of the central and peripheral nervous system cells. It has been found in the heart, lungs, platelets, lymphocytes and lacrimal glands.39 BDNF polymorphisms are associated with an increased esophageal sensitivity to experimental electrical stimulation, and BDNF genotypes may be the useful biomarkers of electrical sensitivity in healthy human esophageal tubes.40 BDNF can significantly promote the proliferation and migration of esophageal cancer cell line ECa9706.41 Xu et al42 reported that the expression of nerve growth factor (NGF) and BDNF was upregulated in esophageal cancer cell line EC109.
BMP7
BMP is a signal molecule secreted by the growth factor TGFβ superfamily. BMP7 was expressed in the cytoplasm of esophageal carcinoma cells. The positive rate was 61.7%. Compared with BMP7 negative ESCC patients, BMP7 positive patients had deeper tumor progression (P<0.001), later stage (P<0.005), more severe venous invasion (P<0.005) and worse prognosis (P<0.0005). Multivariate analysis showed that BMP7 was one of the independent prognostic factors of ESCC (P<0.05), which might be a good prognostic indicator for ESCC patients.43
CCL28
The genetic profiles of ECA tumors after neoadjuvant chemoradiotherapy that demonstrated a pathologic complete response were significantly different from those of typical no-or-incomplete response. Gene CCL28 was overexpressed in pathologic complete response patients, and it could be a potential predictor of treatment response.44 Lysyl oxidase-like 2 (LOXL2), a member of the lysyl oxidase family, plays an important role in the biosynthesis and tumor development of extracellular matrix protein; wild-type LOXL2 (LOXL2Δ72) significantly promotes esophageal squamous cells by upregulating CCL28 cancer ESCC cell migration.45
CR-1
CR-1 is a glycoprotein linked to glycosylphosphatidylinositol (GPI) on the cell surface. It belongs to the epidermal growth factor EGF-CFC protein family. CR-1 maintains pluripotency of embryonic stem cells and is critical for early embryonic development. It is expressed in cancer stem cell (CSC) populations and promotes epithelial–mesenchymal transition (EMT), which significantly enhances tumor cell migration, invasion and angiogenesis.46 The expression of CR-1 in esophagus tissues of Luo et al47 was significantly higher than that in normal esophageal mucosa tissues (86.7% vs 16.7%, respectively, P<0.05). Lymph node metastasis and distant metastasis were significantly higher than those in negative ones (P<0.05). In ESCC cells, inhibit CR-1 gene expression can decrease the stem cells and EMT, tumorigenicity and thus affect the body and the body’s ability to transfer. The expression of CR-1 in ESCC tissues was positively correlated with TNM stage, depth of invasion and lymph node metastasis. CR-1 is an independent prognostic indicator of ESCC and a potential therapeutic target for ESCC.48
MMP-8
MMP-8 belongs to the collagenase subfamily of MMPs, also known as collagenase-2 or neutrophil collagenase. It is predominantly a neutrophil product but is also expressed in fibroblasts, endothelial cells, keratinocytes, epithelial cells, chondrocytes, macrophages and plasma cells.49 In tongue squamous cell carcinoma, the expression of MMP-8 was positively correlated with the increase in the survival rate.50 The level of MMP-8 in breast cancer was positively correlated with lymph node metastasis and negatively correlated with the risk of distant metastasis. It suggested that MMP-8 protein levels were in contrary between blood and tissue. When blood proteins were isolated in tissue, the level of MMP-8 in the blood was low and the level of blood was higher after secretion, and MMP-8 in breast cancers may metastasize to lymph nodes, which showed a protective effect.51 Wang et al52 detected the expression of MMP-8 protein in 15 cases of ESCC and paired distant cancer tissues, showing an increasing trend from T1 to T4 stage and no significant difference between different lymph node metastasis groups.
E-selectin
E-selectin is associated with blood metastasis in tumor patients. Preoperative serum soluble E-selectin is a risk factor for postoperative hematogenous recurrence, and it is a prognostic factor for ESCC.53
SHH
The Hedgehog signal (Hedgehog signaling) is activated in glioma, neuroblastoma, basal cell carcinoma, lung cancer, esophageal cancer, gastric cancer, pancreatic cancer, breast cancer and other tumors. Tumors are triggered by epigenetic or genetic alterations, leading to over-positive feedback or negative feedback of the Hedgehog signaling pathway.54 PI3K/AKT and MAPK signals interact with the SHH signaling pathway to promote esophageal cancer cell survival and proliferation.55 Barrett’s esophagus is a precancerous lesion of EAC. It was found that NO promoted the expression of reduced squamous cell phenotype, and Hh signaling pathway mediated the columnar cell phenotype gene expression. Inhibition of esophageal NO production or Hh signals may prevent Barrett’s esophagus.56 Ma et al57 reported that in 22 cases of primary EC, 14 patients showed increased Hh target gene expression. Hh signal activation was not related to tumor subtype, staging or differentiation. The expression of Hh signaling pathway and its target genes was fairly common in esophageal cancer.
Mori et al58 tested the expression of Hh signaling pathway in 34 human ESCC cells. The results showed that 34 patients with SHH protein expressed 100%. Cyclopamine, a specific inhibitor of Hh pathway, can significantly inhibit the differentiation, proliferation and migration of ESCC cells, suggesting that the Hh pathway may be a new research target for the treatment of esophageal cancer. Sims-Mourtada et al59 found that blocking the Hh signal enhanced the cytotoxicity of esophageal cancer cells, and the activation of the Hh pathway could promote the regeneration of tumor after chemoradiotherapy and facilitate the chemotherapy of esophageal cancer.
TSP1
TSP1 is an extracellular matrix glycoprotein that can affect cell adhesion, exercise and growth. Oshiba et al10 suggested that the positive rate of TSP1 in stage T3 ESCC was significantly higher than that in stage T1 (70.6% vs 26.9%, respectively, P<0.001). Lymph node metastasis and venous metastasis were significantly enhanced in TSP1-positive ESCC at 71.4% and 80.0%, respectively, suggesting that increased expression of TSP1 plays an important role in the growth and metastasis of ESCC. As the malignant degree increased, the expression of TSP1 increased. However, Tzeng et al60 found that low ESCC patients with Rab37 or TSP1 expression were significantly associated with poor prognosis. Multivariate Cox regression analysis showed that a low expression of both Rab37 and TSP1 was an independent prognostic factor in ESCC patients. The two conclusions are inconsistent.
In this study, the expression level of TSP1 in the normal control group was 1.8 times as high as that in the early stage cancer group, which was in agreement with the results of Tzeng et al.60 Possible reasons for this phenomenon are listed as follows. 1) The testing object was different. Particular geographic distribution of disease patterns and different areas of the population present different genetic backgrounds, health and food intake habits and diverse protein components. Previous reports on the proteomic study of esophageal cancer in high-incidence area (Henan, People’s Republic of China) and low-incidence area (Beijing, People’s Republic of China) were different.61,62 2) Different detection methods may affect results. Magnetic bead-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry technology was used by Jia et al63 is a commonly used method of proteomics analysis. However, due to the cumbersome operation and high experimental conditions, it is not conducive and highly applicable. AAH-BLG-507 protein chip used in this study showed a high sensitivity and simple operation procedures. However, the cost is relatively high.
IL-6
IL-6 is a kind of cytokine with extensive biological function, which plays an important role in regulating inflammatory response and immune response. Studies have shown that IL-6 is closely associated with tumors. 1) IL-6 activates phosphatidylinositol 3-kinase (PI3K/Akt) and JAK/STAT3 pathways to inhibit the pro-apoptotic protein TGFβ.64 On the one hand, it can also induce the expression of p21 and Bcl-2 to play an anti-apoptotic effect.65,66 2) IL-6 can directly affect the expression of angiogenic factors such as VEGF and promote angiogenesis.67 3) IL-6 may play an important role in tumor invasion and extracellular matrix differentiation by recruiting cytokines such as MMPs.68
IL-6 may be an important inducement of esophageal cancer angiogenesis and endothelial cell formation. The ESCC cells can produce IL-6 and IL-6 receptors, and the IL-6 concentration in cancer cells is significantly higher than that in normal epithelial cells.69 IL-6 mRNA is in high expression in ESCC tissues (P<0.01). IL-6 protein was significantly higher than that of nontumor tissues (P<0.01).70 The expression of IL-6 was positively correlated with distant metastasis (P=0.0003) and low therapeutic response rate (P=0.0001). Inhibition of IL-6 expression, invasion and radiation resistance of cancer cells can also restrain.71
Elevated IL-6 levels are associated with poor tumor progression and poor prognosis in patients with ESCC. Patients with IL-6-positive tumors have significantly shorter overall survival than IL-6-negative patients.72 The overall survival and progression-free survival of patients with a high IL-6 expression in ESCC receiving cisplatin was significantly poorer.73 IL-6 expression may serve as a predictor of cisplatin-based resistance to esophageal cancer.74 Elevated levels of IL-6 in ESCC patients were expected to respond poorly to preoperative chemoradiation.75 Targeted inhibition of IL-6 may be an effective strategy for the treatment of esophageal cancer.76,77
IL-8
IL-8 is a member of the chemotactic cytokine CXC family, which inhibits apoptosis and promotes tumor cell growth. IL-8 is involved in the immunosuppression of tumors through the promotion of neutrophil and marrow. Myeloid-derived suppressor cells (MDSCs) accumulate in the tumor microenvironment, leading to immune escape of tumor cells.78 There are differences in the expression of IL-8 in cancer patients, which can be used as the clinical stage, prognosis and disease monitoring indicators of esophageal cancer. Tang et al79 reported that IL-8 levels in early and advanced esophageal cancer tissues were significantly higher than those in normal esophageal tissues (P<0.05). Esophageal cancer in advanced esophageal cancer was significantly higher than that in early esophageal cancer (P<0.01). There were differences in IL-8 expression in cancer patients, which can be used as clinical staging, prognosis and disease monitoring indicators of esophageal cancer. Consistent with the findings of Chen et al,80 Kitadai et al81 reported that IL-8 was not expressed in esophageal atypical hyperplasia and mucosal carcinomas but was expressed in advanced squamous cell carcinomas. Krzystek-Korpacka et al82 found that IL-8 was associated with tumor size and spread of ESCC patients, especially lymph node metastasis. IL-8 expression increase can significantly enhance the ability of ESCC cell invasion and migration.83
In the present study, the expression of IL-6 and IL-8 in early ESCC was upregulated, which was similar to previous studies. It also suggested that the serum could be an effective test indicator for early diagnosis of ESCC. Although the expression levels of 11 proteins in the current experiment are significantly different from those in the normal control group, the diagnostic value of early ESCC needs to be further explored due to the lack of expression of related proteins in advanced EC.
ELISA was selected to verify the differential proteins, indicating that IL-6 and IL-8 were significantly upregulated in the serum of early cancer patients, which is in agreement with the results of protein chips. It confirmed the reliability of protein chip results. IL-6 and IL-8 proteins are expected to be used in early stage of ESCC diagnosis. However, tumor markers for the diagnosis of esophageal cancer only served as a screening role. Tumor markers detected by the suspicious population further confirmed by endoscopy is a more feasible strategy.
Acknowledgments
We are grateful to all the participants involved in the study who donated blood for research purposes. This study was funded by the Doctor’s Start-up Funding of Taihe Hospital (2012QD09), the Scientific Research Project of the Department of Science and Technology of Hubei (2012FFB03901), the Department of Education of Hubei (B2013107) and the 2016 Joint Diagnostic Medicine Research Project of Taihe Hospital (2016JD02).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Chen LQ, Hu CY, Ghadirian P, Duranceau A. Early detection of esophageal squamous cell carcinoma and its effects on therapy: an overview. Dis Esophagus. 1999;12(3):161–167. | ||
Mcguire S, Report WC. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7(2):418–419. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Schweigert M, Dubecz A, Stein HJ. Oesophageal cancer – an overview. Nat Rev Gastroenterol Hepatol. 2013;10(4):230–244. | ||
Hao JJ, Gong T, Zhang Y, et al. Characterization of gene rearrangements resulted from genomic structural aberrations in human esophageal squamous cell carcinoma KYSE150 cells. Gene. 2013;513(1):196–201. | ||
Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer. Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21(6):921–945. | ||
Li N, Lu Y, Li D, et al. All-trans retinoic acid suppresses the angiopoietin-Tie2 pathway and inhibits angiogenesis and metastasis in esophageal squamous cell carcinoma. PLoS One. 2017;12(4):e174555. | ||
Dong J, Zeng BH, Xu LH, Lh X, et al. Anti-CDC25B autoantibody predicts poor prognosis in patients with advanced esophageal squamous cell carcinoma. J Transl Med. 2010;8:81. | ||
Oshiba G, Kijima H, Himeno S, et al. Stromal thrombospondin-1 expression is correlated with progression of esophageal squamous cell carcinomas. Anticancer Res. 1999;19(5C):4375–4378. | ||
Cao X, Zhang L, Feng GR, et al. Preoperative Cyfra21-1 and SCC-Ag serum titers predict survival in patients with stage II esophageal squamous cell carcinoma. J Transl Med. 2012;10:197. | ||
Cheng J, Zheng G, Jin H, Gao X. Towards Tyrosine Metabolism in Esophageal Squamous Cell Carcinoma. Comb Chem High Throughput Screen. 2017;20(2):133–139. | ||
Jin H, Qiao F, Chen L, Lu C, Xu L, Gao X. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J Proteome Res. 2014;13(9):4091–4103. | ||
Cheng J, Jin H, Hou X, Lv J, Gao X, Zheng G. Disturbed tryptophan metabolism correlating to progression and metastasis of esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2017;486(3):781–787. | ||
Wasinger VC, Cordwell SJ, Cerpa-Poljak A, et al. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis. 1995;16(7):1090–1094. | ||
Liotta LA, Petricoin EF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J Clin Invest. 2006;116(1):26–30. | ||
Zhang H, Liu AY, Loriaux P, et al. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6(1):64–71. | ||
Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. | ||
Matta A, Ralhan R, Desouza LV, Siu KW. Mass spectrometry-based clinical proteomics: head-and-neck cancer biomarkers and drug-targets discovery. Mass Spectrom Rev. 2010;29(6):945–961. | ||
Wilkins MR, Sanchez JC, Gooley AA, et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19–50. | ||
Fan NJ, Gao CF, Wang XL. Tubulin beta chain, filamin A alpha isoform 1, and cytochrome b-c1 complex subunit 1 as serological diagnostic biomarkers of esophageal squamous cell carcinoma: a proteomics study. OMICS. 2013;17(4):215–223. | ||
Gao H, Zheng Z, Mao Y, et al. Identification of tumor antigens that elicit a humoral immune response in the sera of Chinese esophageal squamous cell carcinoma patients by modified serological proteome analysis. Cancer Lett. 2014;344(1):54–61. | ||
Hou G, Lou X, Sun Y, et al. Correction to “Biomarker Discovery and Verification of Esophageal Squamous Cell Carcinoma Using Integration of SWATH/MRM”. J Proteome Res. 2016;15(2):680. | ||
Liu WL, Zhang G, Wang JY, et al. Proteomics-based identification of autoantibody against CDC25B as a novel serum marker in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2008;375(3):440–445. | ||
Zhang LY, Ying WT, Mao YS, et al. Loss of clusterin both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. World J Gastroenterol. 2003;9(4):650–654. | ||
Zhao J, Fan YX, Yang Y, et al. Identification of potential plasma biomarkers for esophageal squamous cell carcinoma by a proteomic method. Int J Clin Exp Pathol. 2015;8(2):1535–1544. | ||
Elkabets M, Pazarentzos E, Juric D, et al. AXL mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27(4):533–546. | ||
Kanlikilicer P, Ozpolat B, Aslan B, et al. Therapeutic Targeting of AXL Receptor Tyrosine Kinase Inhibits Tumor Growth and Intraperitoneal Metastasis in Ovarian Cancer Models. Mol Ther Nucleic Acids. 2017;9:251–262. | ||
Hong J, Peng D, Chen Z, Sehdev V, Belkhiri A. ABL regulation by AXL promotes cisplatin resistance in esophageal cancer. Cancer Res. 2013;73(1):331–340. | ||
Hsieh MS, Yang PW, Wong LF, Lee JM. The AXL receptor tyrosine kinase is associated with adverse prognosis and distant metastasis in esophageal squamous cell carcinoma. Oncotarget. 2016;7(24):36956–36970. | ||
Paccez JD, Duncan K, Vava A, et al. Inactivation of GSK3β and activation of NF-κB pathway via Axl represents an important mediator of tumorigenesis in esophageal squamous cell carcinoma. Mol Biol Cell. 2015;26(5):821–831. | ||
Kang J, Qian PX, Pandey V, et al. Artemin is estrogen regulated and mediates antiestrogen resistance in mammary carcinoma. Oncogene. 2010;29(22):3228–3240. | ||
Zhang M, Zhang W, Wu Z, et al. Artemin is hypoxia responsive and promotes oncogenicity and increased tumor initiating capacity in hepatocellular carcinoma. Oncotarget. 2016;7(3):3267–3282. | ||
Li S, Li Z, Guo F, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. | ||
Chen YQ, Jian SH, Huang YF, et al. Expression and Clinical Significance of ARTN and GFRα3 in Esophageal Squamous Cell Carcinoma. Journal of Chongqing University of Technology (Natural Science). 2018;32(5):151–155. Chinese. | ||
Loges S, Clausen H, Reichelt U, et al. Determination of microvessel density by quantitative real-time PCR in esophageal cancer: correlation with histologic methods, angiogenic growth factor expression, and lymph node metastasis. Clin Cancer Res. 2007;13(1):76–80. | ||
Dreikhausen L, Blank S, Sisic L, et al. Association of angiogenic factors with prognosis in esophageal cancer. BMC Cancer. 2015;15:121. | ||
Zhou YZ, Fang XQ, Li H, et al. Role of serum angiopoietin-2 level in screening for esophageal squamous cell cancer and its precursors. Chin Med J. 2007;120(14):1216–1219. | ||
Tsukinoki K, Saruta J, Sasaguri K, et al. Immobilization stress induces BDNF in rat submandibular glands. J Dent Res. 2006;85(9):844–848. | ||
Vasant DH, Payton A, Mistry S, Thompson DG, Hamdy S. The val66met polymorphism of brain-derived neurotrophic factor is associated with human esophageal hypersensitivity. Neurogastroenterol Motil. 2013;25(2):162–185. | ||
Feng RT, Mei JZ, Li M, Zhao JZ, Bai H, Liu GJ. Effect of brain-derived neurotrophic factor on in vitro metastasis of esophageal carcinoma cell line ECa9706. World Chin J Digest. 2015;08:1218–1223. Chinese. | ||
Xu GH, Feng F, Zhao GH, et al. Effects of esophageal cancer on the nerve fiber growth and guidance. Zhonghua Wei Chang Wai Ke Za Zhi. 2013;16(5):474–478. | ||
Megumi K, Ishigami S, Uchikado Y, et al. Clinicopathological significance of BMP7 expression in esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19(6):2066–2071. | ||
Mclaren PJ, Barnes AP, Terrell WZ, et al. Specific gene expression profiles are associated with a pathologic complete response to neoadjuvant therapy in esophageal adenocarcinoma. Am J Surg. 2017;213(5):915–920. | ||
Zou HY, Lv GQ, Dai LH, et al. A truncated splice variant of human lysyl oxidase-like 2 promotes migration and invasion in esophageal squamous cell carcinoma. Int J Biochem Cell Biol. 2016;75:85–98. | ||
Klauzinska M, Castro NP, Rangel MC, et al. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin Cancer Biol. 2014;29:51–58. | ||
Luo D, Yin J, Yl K. Clinical Significance of PEBP4, Cripto-1 Expressions in Esophageal Carcinoma. Guang Xi Yi Xue. 2014;9:1205–1208. Chinese. | ||
Liu Q, Cui X, Yu X, et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):81. | ||
van Lint P, Libert C. Matrix metalloproteinase-8: cleavage can be decisive. Cytokine Growth Factor Rev. 2006;17(4):217–223. | ||
Korpi JT, Kervinen V, Mäklin H, et al. Collagenase-2 (matrix metalloproteinase-8) plays a protective role in tongue cancer. Br J Cancer. 2008;98(4):766–775. | ||
Decock J, Thirkettle S, Wagstaff L, Edwards DR. Matrix metalloproteinases: protective roles in cancer. J Cell Mol Med. 2011;15(6):1254–1265. | ||
Wang DM, Guo H, Chen XD, Guan XY, Liu D, Bai Y. Expression of matrix metalloproteinase 8 in esophageal squamous cell carcinoma. Di San Jun Yi Da Xue Xue Bao. 2011;33(5):519–522. Chinese. | ||
Shimada Y, Maeda M, Watanabe G, Imamura M. High serum soluble E-selectin levels are associated with postoperative haematogenic recurrence in esophageal squamous cell carcinoma patients. Oncol Rep. 2003;10(4):991–995. | ||
Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9(7):873–886. | ||
Wei L, Xu Z. Cross-signaling among phosphinositide-3 kinase, mitogen-activated protein kinase and sonic hedgehog pathways exists in esophageal cancer. Int J Cancer. 2011;129(2):275–284. | ||
Souza RF. From Reflux Esophagitis to Esophageal Adenocarcinoma. Dig Dis. 2016;34(5):483–490. | ||
Ma X, Sheng T, Zhang Y, et al. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer. 2006;118(1):139–148. | ||
Mori Y, Okumura T, Tsunoda S, Sakai Y, Shimada Y. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology. 2006;70(5):378–389. | ||
Sims-Mourtada J, Izzo JG, Apisarnthanarax S, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res. 2006;12(21):6565–6572. | ||
Tzeng HT, Tsai CH, Yen YT, et al. Dysregulation of Rab37-Mediated Cross-talk between Cancer Cells and Endothelial Cells via Thrombospondin-1 Promotes Tumor Neovasculature and Metastasis. Clin Cancer Res. 2017;23(9):2335–2345. | ||
Zhang J, Wang K, Zhang J, Liu SS, Dai L, Zhang JY. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res. 2011;10(6):2863–2872. | ||
du XL, Hu H, Lin DC, et al. Proteomic profiling of proteins dysregulted in Chinese esophageal squamous cell carcinoma. J Mol Med. 2007;85(8):863–875. | ||
Jia K, Li W, Wang F, et al. Novel circulating peptide biomarkers for esophageal squamous cell carcinoma revealed by a magnetic bead-based MALDI-TOFMS assay. Oncotarget. 2016;7(17):23569–23580. | ||
Garbers C, Hermanns HM, Schaper F, et al. Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012;23(3):85–97. | ||
Ara T, Declerck YA. Interleukin-6 in bone metastasis and cancer progression. Eur J Cancer. 2010;46(7):1223–1231. | ||
Kishimoto T. IL-6: from its discovery to clinical applications. Int Immunol. 2010;22(5):347–352. | ||
Middleton K, Jones J, Lwin Z, Coward JI. Interleukin-6: an angiogenic target in solid tumours. Crit Rev Oncol Hematol. 2014;89(1):129–139. | ||
Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38(7):904–910. | ||
Oka M, Yamamoto K, Takahashi M, et al. Relationship between serum levels of interleukin 6, various disease parameters and malnutrition in patients with esophageal squamous cell carcinoma. Cancer Res. 1996;56(12):2776–2780. | ||
Chen D, Jin L, Zhu L, Mou X, Wang S, Mao C. Expressions and correlations of let-7a and IL-6 in esophageal squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29(11):1181–1184. Chinese. | ||
Chen MF, Chen PT, Lu MS, Lin PY, Chen WC, Lee KD. IL-6 expression predicts treatment response and outcome in squamous cell carcinoma of the esophagus. Mol Cancer. 2013;12:26. | ||
Yoneda M, Fujiwara H, Furutani A, et al. Prognostic impact of tumor IL-6 expression after preoperative chemoradiotherapy in patients with advanced esophageal squamous cell carcinoma. Anticancer Res. 2013;33(6):2699–2705. | ||
Qiao Y, Zhang C, Li A, et al. IL6 derived from cancer-associated fibroblasts promotes chemoresistance via CXCR7 in esophageal squamous cell carcinoma. Oncogene. 2018;37(7):873–883. | ||
Suchi K, Fujiwara H, Okamura S, et al. Overexpression of Interleukin-6 suppresses cisplatin-induced cytotoxicity in esophageal squamous cell carcinoma cells. Anticancer Res. 2011;31(1):67–75. | ||
Makuuchi Y, Honda K, Osaka Y, et al. Soluble interleukin-6 receptor is a serum biomarker for the response of esophageal carcinoma to neoadjuvant chemoradiotherapy. Cancer Sci. 2013;104(8):1045–1051. | ||
Zhao ZF, Li JX, Ye R, Wu X, Gao LL, Niu BL. Interleukin-6 as a potential molecular target in esophageal squamous cell carcinoma. Oncol Lett. 2016;11(2):925–932. | ||
Fujiwara H, Suchi K, Okamura S, et al. Elevated serum CRP levels after induction chemoradiotherapy reflect poor treatment response in association with IL-6 in serum and local tumor site in patients with advanced esophageal cancer. J Surg Oncol. 2011;103(1):62–68. | ||
Yu J, Wang Y, Yan F, Li H, Ren X. Response to comment on “Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer”. J Immunol. 2013;190(11):5341–5342. | ||
Tang RG, Yuan XH, Long XK, He T, Chen HM, Fang WZ. Detection of peripheral serum level of IL-8 and IL-18 in patients with esophageal cancer. Jian Yan Yi Xue. 2009;24(2):117–119. Chinese. | ||
Chen HM, Rz L, Tang RG, et al. Serum levels of interleukin and acute phase reaction proteins in patients with esophageal cancer and their clinical significance. Guang Xi Yi Xue. 2016;38(7):929–931. Chinese. | ||
Kitadai Y, Onogawa S, Kuwai T, et al. Angiogenic switch occurs during the precancerous stage of human esophageal squamous cell carcinoma. Oncol Rep. 2004;11(2):315–319. | ||
Krzystek-Korpacka M, Matusiewicz M, Diakowska D, et al. Elevation of circulating interleukin-8 is related to lymph node and distant metastases in esophageal squamous cell carcinomas – implication for clinical evaluation of cancer patient. Cytokine. 2008;41(3):232–239. | ||
Ren Y, Cao B, Law S, et al. Hepatocyte growth factor promotes cancer cell migration and angiogenic factors expression: a prognostic marker of human esophageal squamous cell carcinomas. Clin Cancer Res. 2005;11(17):6190–6197. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

