Back to Journals » OncoTargets and Therapy » Volume 13
Combination of Inositol Hexaphosphate and Inositol Inhibits Liver Metastasis of Colorectal Cancer in Mice Through the Wnt/β-Catenin Pathway
Authors Liu X, Liu C, Chen C, Sun W, Ci Y, Li Q, Song Y
Received 30 January 2020
Accepted for publication 25 March 2020
Published 16 April 2020 Volume 2020:13 Pages 3223—3235
DOI https://doi.org/10.2147/OTT.S247646
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Xiaohan Liu,1 Cuiping Liu,2 Chen Chen,1 Wenna Sun,3 Yifan Ci,1 Qianqian Li,1 Yang Song4
1School of Public Health, Qingdao University, Qingdao, Shandong, People’s Republic of China; 2School of Nursing, Qingdao University, Qingdao, Shandong, People’s Republic of China; 3Outpatient Department, Qingdao Fuwai Cardiovascular Hospital, Qingdao, Shandong, People’s Republic of China; 4Medical College, Qingdao University, Qingdao, Shandong, People’s Republic of China
Correspondence: Yang Song Email [email protected]
Introduction: Colorectal cancer, one of the most common tumors, is mainly fatal because of the occurrence of liver metastasis. Inositol hexaphosphate (IP6) and inositol (INS) were found, both, in vitro and in vivo to play an anti-tumor effect, whereas the combination of IP6 and INS was more effective than IP6 or INS alone.
Materials and Methods: The inhibitory effects of IP6, INS and the combination of IP6+INS on tumor progression and liver metastasis of colorectal cancer were investigated in an orthotopic transplantation model of colorectal cancer. The tumor-bearing mice were selected by in vivo bioluminescence imaging and were treated with IP6, INS, and IP6 combined with INS, respectively. All mice were sacrificed after 6 weeks of treatment. The cancer development and metastasis were compared among the groups. The expression of genes related to the Wnt/β-catenin in the model was analyzed.
Results: The results demonstrated that liver metastasis was inhibited after treatment with IP6, INS, and IP6+INS. Compared to that of the M_G, survival period was extended, and tumor weight was lowered in IP6_G, INS_G, and IP6+INS_G. Besides, the liver metastatic area of mice in IP6+INS_G was relatively smaller than that in M_G, IP6_G, or INS_G. The results of RNA-seq analysis showed that the expressions of Wnt10b, Tcf7, and c-Myc were significantly downregulated in IP6+INS_G compared to that in M_G (P< 0.05). Results of real-time PCR and Western blot showed that mRNA and protein expressions of β-catenin, Wnt10b, Tcf7, and c-Myc were significantly lower in IP6+INS_G compared to that in M_G (P< 0.05).
Discussion: IP6+INS was more effective in inhibiting liver metastasis of colorectal cancer than IP6 or INS alone. The better inhibition effect may be accomplished through regulating the mutation of Wnt/β-catenin signaling pathway by inhibiting Wnt10b, Tcf7, β-catenin, and c-Myc from abnormally high expression.
Keywords: colorectal cancer, liver metastasis, Wnt/β-catenin signaling, inositol hexaphosphate, inositol, mouse model
Introduction
Colorectal cancer is a common malignant tumor that threatens human health and life. The main reason for death is regarded to be poor prognosis and metastasis.1 At present, it is believed that the liver is the main target organ of hematogenous metastasis of colorectal cancer.2–4
Many efforts are being applied to prevent cancer; among these efforts is the use of natural products, which have become an important method of cancer prevention and treatment that are accessible, safe, and broadly accepted.5,6 Inositol hexaphosphate (IP6) is a natural ingredient that can be found in almost all grains and legumes.7 Both in vivo and in vitro experiments provide convincing evidence for the anticancer effect of IP6 on colorectal cancer.8–10 Inositol (INS) is the molecular skeleton of IP6, a carbohydrate and the precursor of phosphorylated compounds.11 A number of in vitro and in vivo studies have shown that IP6 and INS have independent anti-tumor effects. However, the combination of IP6 and INS has a better effect than that of IP6 or INS alone.11–13
Wnt signaling pathway is one of the most important signal pathways in cancer.14 A number of studies have provided evidence that the mutation or abnormal expression of various components in the Wnt pathway is closely related to the progression of human malignant tumors, and gene mutations in the key components of the Wnt pathway occurred in more than 90% of colorectal cancer cases.15,16 Wnt signaling pathway regulates a variety of biological and developmental processes through multiple branches, among them the classical Wnt/ β-catenin pathway is a very important branch.17,18 In vivo and in vitro studies have found that the activation of Wnt/ β-catenin pathway plays a direct role in the development of colorectal cancer.19,20 Moreover, the mutation or abnormal expression of various parts of Wnt pathway is closely related to the growth of advanced colorectal cancer.21 A study found that Wnt signaling pathway was abnormally regulated in colorectal cancer patients with liver metastasis through whole genomics.22 In vivo and in vitro studies also found that transduction of the Wnt signaling pathway would be interfered when wnt and c-myc abnormally expressed in the pathway, thus inhibited metastasis of colorectal cancer.
IP6 and INS have been found to inhibit tumor progression by regulating the abnormally expressed genes on Wnt signaling pathway in animals with colorectal cancer. IP6, extracted from rice bran, has been found to inhibit the expression of β-catenin in rats with colon cancer induced by azomethane (AOM).23 In another study of rat model with colitis induced by DSS, INS was found to inhibit the mutation of Wnt transduction by downregulating the β-catenin expression.24
In our previous studies, IP6 was also found to inhibit the colorectal cancer development by downregulating the expression of β-catenin and c-Myc in the Wnt signaling pathway in a rat model of colorectal cancer induced by 1, 2-dimethylhydrazine (DMH).25 Furthermore, another study in our laboratory established a BALB/c mouse model of colorectal cancer with liver metastasis by injecting the cell line into spleen; results showed that colorectal cancer with liver metastasis could be inhibited in this model when IP6 is combined with INS.26 This model mimics the vascular diffusion process of colorectal cancer; this process is a characteristic of colorectal cancer with liver metastasis.
Materials and Methods
Main Reagents
Phytic acid sodium salt hydrate was purchased from Macklin Biochemical (Shanghai, China). Inositol was purchased from Yuanye Biochemical (Shanghai, China). D-Luciferin Potassium Salt was purchased from Solarbio Science & Technology (Beijing, China). In addition, AIN 93M was purchased from Trophic Animal Feed High-tech (Nanjing, China) and TRIzol reagent was obtained from Tiangen Biotech (Beijing, China). Further, PrimeScript™ RT reagent Kit with gDNA Eraser and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) was purchased from Takara Biomedical Technology (Beijing, China) and reagents used in Western blot analysis were purchased from Wuhan Sanying Biotech (Wuhan, China).
Cells and Cell Culture
The CT26 mouse colorectal tumor cell line was purchased from the Shanghai Cell Bank (Chinese Academy of Sciences). We also transfected the CT26 cells with green fluorescent proteins (GFP) and a firefly luciferase (LUC)-expressing reporter, which was marked as CT26_LUC. The new cells were cultured under standard cell culture conditions in an RPMI 1640 medium, containing 10% serum at 37°C in a humidified atmosphere with 5% CO2.
Animals
A total of 120 male 6-week-old BALB/c mice purchased from Shandong Pengyue Pharmaceutical Co., Ltd. (Qingdao, China) were raised in an adequately ventilated animal room with a reversed 12-hour-light/dark-cycle. The animal room was maintained at a relative humidity around 52 ± 8% and a temperature around 24 ± 2°C, monitored by an automatic controller. The mice were supplied with AIN93M standard and regular drinking water before they were euthanized. All animal experiments in this study were conducted in strict accordance with the recommendations of the medical ethics committee of Qingdao University Medical College (reference no. QYFYWZLL 25667).
Cell Transfection
CT26 cells were cultured under standard cell culture conditions in RPMI 1640 medium containing 10% serum at 37°C in a humidified atmosphere with 5% CO2. The cells were then transfected with the lentiviral vector product with the expression of GFP and luciferase (LUC) was obtained from the previous experiment. The cells with solid and high luciferase reporter expression were selected by purocmycin treatment for 2 weeks and denominated as CT-26-luc. The CT-26-luc cells were cultured using routine protocols.
Orthotopic Mouse Model Establishment of CRC with Liver Metastasis
After adaptive feeding for one week, 100 of the 120 mice were randomly distributed into the treatment groups and were prepared to establish the mouse model of colorectal cancer with liver metastasis using the orthotopic operation approach. The other 20 mice were not operated upon. The CT-26-luc cells were detached with 0.25% Trypsin-EDTA. A total of 1 × 106 cells were washed and re-suspended in 1 mL PBS for the operation.
One hundred mice were anesthetized through intraperitoneal injection with 0.1% pentobarbital sodium (80 mg/kg of body weight) before the operation. The mice were fixed in a supine position after deep anesthesia. The left lower abdomen was disinfected. A longitudinal incision of approximately 1.5 cm was made at the operation site to expose the cecum. The cecum was gently pressed with a tweezer handle to squeeze feces from the cecum into the distal colon, and 0.2 mL of the cell suspension was injected using a 4-gauge needle under the serosa of the cecum. The contents were carefully ejected, and the injection point was gently pressed for 2 min with a cotton ball after the injection. If there was no liquid extravasation, the cecum was returned to its original position in the abdomen. The abdominal wound was closed layer by layer.
The mice were closely observed after the surgery. Pentobarbital sodium (0.1%, 80 mg/kg of body weight) and D-luciferin-potassium salt (150 mg/kg of body weight) were injected intraperitoneally about 10 min prior, and in vivo bioluminescence imaging was performed 3–4 weeks after the operation when tumors started forming in the model mice. Based on the results of in vivo bioluminescence imaging, the mice-bearing tumors were regarded as a CRC liver metastasis model.
In vivo Bioluminescence Imaging
The in vivo bioluminescence imaging method was used to detect tumor development and metastasis after the operation. D-Luciferin (150 mg/kg of body weight) and 0.1% pentobarbital sodium (80 mg/kg of body weight) were injected intraperitoneally into each mouse. The mice were fixed, and bioluminescence was detected and measured around 5 min later using the PerkinElmer IVIS Spectrum Imaging System. Living Image Software (Xenogen, PerkinElmer, and Waltham, Massachusetts, USA) was used to quantify the total flux (photons/second) uniformly sized regions of interest (ROI). Bioluminescence lasted for 30 min at about 5-min intervals. The highest number of photons/s for each mouse calculated over this time was used to determine bioluminescence.
Groups and Treatments
Three weeks after the surgery, 48 of the tumor-bearing mice were distributed randomly, according to body weight, into three treatment groups and one model group (M_G), with 12 mice per group, while another 12 mice were randomly distributed from the 20 mice that did not undergo any of the procedures described above to be the control group (C_G). The treatment groups were treated by gavage with IP6 (IP6_G, 80 mg/kg of weight), INS (INS_G, 80 mg/kg of weight), and IP6 combined with INS (IP6+INS_G, 80 mg/kg of weight, containing IP6 [80 mg/kg] and INS [80 mg/kg], v:v = 1:1), respectively. The C_G and M_G mice were treated with a volume of normal saline equal to that of each treatment group. Each mouse received 0.2 mL of treatment solution in a maximum of 5 days discontinuously in a week for about 6 weeks.
Tumor Progression and Metastasis Evaluation
All animals were sacrificed by cervical dislocation 12 hours after the last time of treatment, which lasted for 6 weeks. The number of mice surviving each day before sacrifice was recorded, and the survival percentage was calculated as follows: survival percentage (/day) = number of surviving mice each day in each group/ total number of mice in each group × 100%. The mass of each tumor and liver was measured using an electronic balance (JA5003N:Minqiao Precision instrument Co., Ltd. Shanghai, China). The number of livers with metastasis in each group was recorded, and the liver metastasis percentage was calculated as follows: liver metastasis rate = number of mice with metastasis in each group/total number of mice in each group × 100%. Liver metastasis was analyzed using ImageJ software (Rawak Software, Inc., Stuttgart, Germany). The data are presented as means ± standard deviations (SD).
RNA Extraction
Total RNA was extracted from tissues from the control, model, and treatment groups using TRIzol® Reagent, according to the manufacturer’s instructions (Invitrogen). We also removed genomic DNA with DNase I (TaKara). The RNA quality was determined using the 2100 Bioanalyser (Agilent). Concentration and purity were quantified using NanoDrop® (NanoDrop Technologies). A high-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, >10μg) was used to construct the sequencing library.
RNA-Seq Transcriptome Analysis
The TruSeqTM RNA sample preparation kit from Illumina (San Diego, CA) was used with 5 μg of total RNA. Following that, the RNA-seq transcriptome library was prepared. RNA extraction and library construction and sequencing were performed by Shanghai Mei Ji Biological Co. Ltd., differentially expressed genes between two different samples were identified, and their functions studied. The expression of transcripts was calculated according to the FRKM (the fragments per kilobase of exon per million mapped reads) method. Differential expression analysis of raw counts was performed by the DESeq2 software, using the default parameter p-adjust <0.05 & |log2FC| ≥ 1, the multiple test method BH (FDR correction with Benjamini/Hochberg), and a fold-change > 1.5. To identify the enrichment of differential expressing genes in GO terms and pathways, the functional-enrichment analyses on GO were also performed.
Real-Time PCR Analysis
A Primer Script RT Reagent Kit and a Bio-Rad c-Mycycler PCR instrument were used to reversibly transcribe total RNA (2 µg). The PCR primer sequences and sizes of the resulting PCR product are shown in Table 1. PCR was operated in a 20-µL reaction volume according to instructions. A two-step PCR cycling protocol was then performed, consisting of a denaturation step at 95°C and a combined annealing/extension step at 60°C. The protocol was useful for primers with a Tm under 60°C. As an internal reference gene, β-actin was needed to standardize the expression of the target gene. The relative levels of gene expression were enumerated using the comparative formula 2ΔΔCt. These experiments were replicated in triplicate. The PCR primer sequences and the sizes of the resulting PCR product are shown in Table 1.
 |
Table 1 RT-PCR Primer Sequences and Product Sizes |
Western Blot Analysis
A primary tumor tissue sample (50 mg, cecum tissue of the C_G) was cut into small fragments, and the total tissue protein was extracted using RIPA buffer containing phenylmethanesulfonyl fluoride (PMSF). The lytic tissue was centrifuged, and the supernatant was kept at −80°C. Protein concentration was determined using the BCA protein analysis kit. The tissue lysate of each sample was dissociated with 12% SDS-PAGE, and then the membrane protein was transferred to polyvinylidene difluoride membranes. The membranes were sealed in Tris buffer salt water-Tween (TBS-T) containing skim milk powder for 2 hours and incubated overnight with the following primary antibodies: anti-Tcf7 (1:500), anti-Wnt10b (1:1000), anti-c-Myc (1:1000), or anti-β-catenin (1:1000). After re-rinsing, the primary antibody reaction membrane was incubated with the second antibody (1:5000) for 2 hours. Vilber Fusion FX5 and BeyoECL Plus kits were used for chemiluminescent detection of binding antibodies. The bands from Fusion FX5 were measured and quantified using the Vilber FusionCapt Advance software program.
Statistical Analysis
SPSS software for Windows version 22.0 (version 22.0; IBM Corporation, NY, USA) was used to perform all statistical analyses in this study. Student’s t-test was performed to assess the differences between two groups. One-way ANOVA by the least significant difference method was performed to assess differences among groups. Data are expressed as means and standard deviations. P <0.05 was considered statistically significant.
Results
Comparison of the Results of Primary Tumor Progression
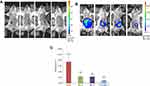
Tumor development of the 100 mice was measured by in vivo bioluminescence imaging method after the orthotopic transplantation operation. Three weeks after the operation, bioluminescence was observed around the location of the cecum in 62 mice (Figure 1A); including several that were in bad condition or had too low of a body weight. In total, 48 of the 62 tumor-bearing mice were included in the experiment and treated in groups from the day. Tumor development, observed again after 6 weeks of treatment, showed that the fluorescence area of M_G was larger than that of other groups. Fluorescence can be observed at the location around the liver. IP6+INS_G had the smallest fluorescence area (Figure 1B). The total flux at ROI of each treatment group was less than that of the M_G, whereas IP6+INS_G had the least ROI (Figure 1C, P<0.05).
The experiment ended after 6 weeks of treatment. Five mice (41.67%) in M_G survived until the end. Compared to the M_G, the survival percentages of IP6_G, INS_G, and IP6+INS_G were 75.00%, 83.33%, and 91.67%, respectively (Table 2, p<0.05). The survival of IP6+INS_G was higher than that of IP6_G or INS_G, whereas only the difference between IP6 and the combined group was significant (Figure 2A).
 |
Table 2 Deaths and Liver Metastasis in the Experiment |
The body weight of each mouse was recorded every week during the treatment period. The average weight of each group dropped after a similar increase, with no significant difference. The body weight of M_G decreased sharply after the 3rd week, whereas that of the other treatment groups slowly decreased from the 4th week during the same treatment period. The body weight of M_G and other treatment groups differed significantly from the fourth week during treatment (Figure 2B).
To further examine the effect of the treatments for inhibiting colorectal cancer in the BALB/c mouse orthotopic transplantation model, all mice in this experiment were sacrificed and dissected for the primary tumor. Each tumor was weighed, and the tumor weight was recorded. The results showed that the primary tumor of each treatment group weighed significantly less than that of M_G (p < 0.05). The primary tumor of IP6+INS_G weighed significantly less than that of IP6_G or INS_G (p < 0.05). The tumor weight between IP6_G and INS_G differed non-significantly (Figure 2C).
Comparison of the Results Regarding Liver Metastasis
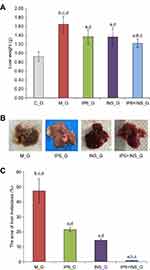
To discover the effect of different treatments on liver metastasis in this experiment, the liver weight and liver metastasis area of each group were analyzed. All mice in this experiment were sacrificed, and the liver was dissected. The number of livers with metastasis in M_G was higher than that of the other treatment groups (Table 2). Each liver was weighed. Liver weight in M_G was significantly heavier than that in other treatment groups. Moreover, liver in IP6+INS_G had the lightest weight, compared to IP6_G and INS_G, with significance (Figure 3A).
It was discovered that the liver metastasis area of each treatment group was reduced, when compared to that of M_G (Figure 3B). The liver metastasis area was further calculated and analyzed. The liver metastasis area of IP6, INS, or IP6+INS_G was significantly smaller than that of M_G. In addition, the liver metastasis area of IP6+INS_G was significantly smaller than that of IP6_G or INS_G, whereas the difference observed between IP6_G and INS_G was without statistical significance (Figure 3C).
Analysis of the RNA-Seq Transcriptome
According to the results shown above, IP6+INS resulted in a better inhibitory effect on colorectal cancer liver metastasis than IP6 or INS alone. To further investigate the inhibitory effect of IP6+INS in our experiment, RNA-seq transcriptome analysis was performed on cecal tissues from C_G and primary tumor tissues from M_G and IP6+INS_G. A total of 469 million raw reads were obtained after the RNA-seq analysis experiment, of which 99.07% were clean reads with a minimum of 98.91% in each group; 94.84% reads on average were mapped to the reference mouse genome. The outcome demonstrated that the sequencing experiment was effective.
The RNA-seq analysis indicated that the gene expression profiles of the three groups were significantly different (Figure 4A). A total of 11,692 genes were detected to be differently expressed, comparing M_G with C_G, whereas 1065 genes were differently expressed, comparing IP6+INS_G with M_G. The results of the analysis on different expressing genes of three groups two-by-two showed that 921 of the genes differing between M_G and C_G were regulated by IP6+INS_G (Figure 4B). Among the 921 genes, the expression of several colorectal cancer-related genes, such as Wnt10b, Tcf7, and c-Myc, was significantly downregulated by IP6+INS (Figure 4C). Wnt10b, Tcf7, and c-Myc play important roles in the Wnt/β-catenin signaling pathway. Furthermore, GO analysis of the 921 differently expressed genes indicated that IP6+INS was involved in several important cancer-related processes, such as “Wnt-protein binding,” “cell morphogenesis,” and “cell-cell adhesion” (Figure 4D). Taken together, the RNA-seq transcriptome analysis of C_G, M_G, and IP6+INS_G contributed in illustrating the inhibitory effect of IP6+INS on colorectal cancer with liver metastasis; moreover, it provided clues for further study on the inhibitory effect through the Wnt/β-catenin signaling pathway.
Analysis of RT-PCR
The observations of the RNA-seq analysis were confirmed by RT-PCR analysis for the main genes of Wnt/β-catenin signaling pathway, such as Wnt10b, β-catenin, Tcf7, and c-Myc. The RT-PCR analysis revealed that the mRNA expression levels of Wnt10b, β-catenin, Tcf7, and c-Myc were significantly upregulated in M_G, but significantly downregulated in the IP6+INS_G (p < 0.05) (Figure 5).
Analysis of Western Blot
Western blot analysis was performed to confirm the protein expression of Wnt10b, β-catenin, Tcf7, and c-Myc. The analysis revealed a downregulation in IP6+INS_G compared to that in M_G (p < 0.05) (Figure 6), which was consistent with the findings from the RT-PCR analysis. The expression levels of mRNA and protein collectively suggest that IP6+INS can inhibit colorectal cancer with liver metastasis through the Wnt/β-catenin pathway by inhibiting the expression of genes such as Wnt10b, β-catenin, Tcf7, and c-Myc.
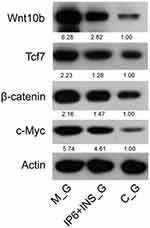 |
Figure 6 Protein expression levels of Wnt10b, Tcf7, β-catenin, and c-Myc as evaluated using Western blot analysis. Data are presented as mean ± SD, p < 0.05. |
Discussion
In this study, an orthotopic mouse model was established to mimic the approach of colorectal cancer with liver metastasis. On this basis, the inhibitory effect on colorectal tumor growth and liver metastasis in BALB/C mice of IP6, INS, and IP6+INS was observed. The results showed that the three treatment methods significantly inhibited tumor progression by reducing the tumor weight, extending the survival period, and increasing the body weight of mice. Moreover, IP6, INS, and IP6+INS were effective in controlling the liver metastasis area and liver weight, whereas the inhibitory effect on liver metastasis by IP6+INS was better than that by IP6 or INS alone. Further analysis of RNA-seq, RT-PCR, and Western-blot showed that IP6+INS might inhibit liver metastasis of colorectal cancer through Wnt/β-catenin pathway by targeting the genes such as, Wnt10b, β-catenin, Tcf7, and c-Myc.
Metastasis of colorectal cancer is a multi-step process involving epithelial-mesenchymal transformation, apoptosis inhibition, local invasion and cell migration, angiogenesis and vascular infiltration, and exudation to distant organs. For a better understanding of these metastatic processes, it is necessary to establish relevant models to describe the characteristics of tumor spontaneous metastasis. Cecum was the orthotopic site for colorectal cancer and serosa was quite important to the growth, spread, and invasive and metastatic capability in mice.27 In this study, we established an orthotopic mouse model of colorectal cancer with liver metastasis by injecting a colorectal cancer cell line under the serosa of the cecum. This approach mimics the microenvironment of human tumors and provides a realistic level of heterogeneity among tumor cells.28 Furthermore, this approach comprehensively simulates the process of spontaneous metastasis of colorectal cancer from the formation of the primary tumor to the development of liver metastasis.
Liver metastasis of colorectal cancer is a complex process involving multiple pathways and factors, in which genomic instability is an important feature of the development of colorectal cancer.29 Wnt signaling pathway is an important pathway in tumor development. There are many branches of Wnt signaling pathway, among which Wnt/β-catenin is a classic pathway. Wnt/β-catenin signal is found to be activated in many researches on cancer. Wnt/β-catenin pathway is also a key pathway in colorectal cancer development.18 Wnt, Tcf, β-catenin and c-Myc are all important roles in Wnt/ β-catenin signaling pathway. In the Wnt signaling pathway, the Wnt signal cascade reaction is activated by abnormally expressed Wnt in normal colon cells, which promotes the β-catenin to translocate in nuclear.30 β-catenin is inhibited to be phosphorylated by Wnt signal pathway in the tumor microenvironment and would accumulate in the nucleus.31 At this point, β-catenin interacts with transcription factors belonging to T cytokines/lymphoenhancer family (Tcf/Lef) and pushes the genes related to cell proliferation, migration, and embryonic development to express. Tcf is a gene located downstream of β-catenin. The combination of β-catenin and Tcf can mediate Wnt signal,32 it can also activate the transcription of oncogenes such as c-Myc and mediate cell transformation downstream the pathway.33 The activation of β-catenin/Tcf complex is considered a common feature of colorectal cancer cells. The expression of Tcf, β-catenin, and c-Myc is closely related.34 Activating β-catenin can lead to the activation of downstream genes on Wnt/β-catenin, such as Tcf and c-Myc, which may cause excessive proliferation of the tumor cells and promote the cancer to be malignant.
IP6 has broad-spectrum anticancer effect. It can inhibit the development of tumor by inhibiting the activity of tumor cells. In vitro and in vivo experiments have proved that IP6 can trigger the death of cells in many cancer cell lines, including prostate cancer,35 melanoma,36 and colon cancer.11,37 INS is a common component of almost all mammalian and plant cells. It has moderate anticancer effect and can slightly slowdown the proliferation of tumor cells in colon, breast, and lungs.38 However, INS and IP6 can work together to inhibit tumor progression with synergistic effect in vitro and in vivo.39 Our results also found that IP6 and INS could give an anti-tumor effect, either alone or in combination, whereas the combination of IP6+INS worked better in inhibition of the liver metastasis in mice with colorectal cancer.
The secondary phosphorylated forms of IP6 may achieve the better anti-tumor effect of IP6 + INS. IP6 can be hydrolyzed rapidly in body, and then be dissociated into secondary phosphorylated forms.40 It can be hydrolyzed in the intestine into INS and inorganic phosphate by phytase.41 The hydrolyzed INS and inorganic phosphate will then be transported in cells, spread to the whole body and distributed into other organs. The anticancer effect of IP6 can be achieved by not only the molecule itself, but also the combination of IP6 and its secondary phosphorylated forms.42 The number of phosphorylated forms of IP6 is increased when IP6 binds to INS in vivo. Moreover, multiple phosphorylated forms in vivo work synergistically to enhance the effect of IP6.39 Our previous study found in the rat colorectal cancer model that IP6 could inhibit the development of colorectal cancer by affecting the phosphorylation and the degradation of β-catenin.25 It is well known that β-catenin is a key factor of Wnt signaling pathway. β-catenin can be continuously destroyed and the β-catenin/Tcf complex can be degraded when Wnt is inhibited.43
Our study found that Wnt10b, β-catenin, Tcf7, and c-Myc were differentially expressed between model group and IP6+INS group. Wnt10b (belongs to the Wnt family) is a key regulator in cancer that can activate β-catenin/Tcf complex to promote tumor progression.44,45 Tcf7 is one of the members of the Tcf/lef family. Abnormally expressed high Tcf7 was found in many types of cancer, such as prostate cancer46 and renal cell carcinoma.47 It plays an important role in cancer on Wnt signaling pathway.48,49C-Myc is strongly over-expressed in most cases of colorectal cancer.50 The over-expression caused by the mutation of Wnt signaling pathway is a characteristic in almost all colorectal cancers.51 Moreover, many studies have found that Wnt10b, Tcf7, and c-Myc are associated with metastasis cancer. In vitro, it was found that knockout of wnt10b could inhibit the migration of gastric cancer cells52 and prostate cancer cells.53 Some studies have found that knockout of LncTcf7 in colorectal cancer cell lines can interfere in the mutation of Wnt/β-catenin pathway and reduce cell migration and invasion.54,55 Abnormally expressed TCF7 has also been found to promote the metastasis of nasopharyngeal carcinoma and gastric cancer.31,56 Over-expression of c-Myc is considered to be the common basis of tumorigenesis and a marker for metastasis cancer,57–60 including metastasis of colorectal cancer.61 According to our study, the mRNA and protein expressions of Wnt10b, β-catenin, Tcf7, and c-Myc were found abnormally high in mice with liver metastasis. The result is consistent with that of most studies. After treated with the combination of IP6+INS, the mRNA and protein expressions of the four genes were significantly downregulated. Therefore, we believe that the combination of IP6 and INS may have a synergistic effect in inhibiting liver metastasis of colorectal cancer instead of IP6 or INS alone by affecting the phosphorylation of β-catenin and inhibiting the abnormal expression of Wnt10b, Tcf7, and c-Myc in Wnt/β-catenin pathway.
There are still some limitations in our study. We established an orthotopic mouse model of colorectal cancer in BALB/c mice and studied on the possible mechanism of IP6+INS on inhibiting liver metastasis of colorectal cancer. The molecular mechanism of IP6 or INS alone on liver metastasis was not discussed in this study. A large number of differentially expressed genes were found by RNA-seq. We only discussed several among them in this study. We still have a lot to explore. Therefore, we are trying to design a better study to further understand the potential inhibition mechanisms on colorectal cancer of IP6, INS, and IP6+INS.
Conclusions
Using an orthotopic transplantation mouse model of colorectal cancer with liver metastasis in our experiment, we found that IP6 and INS could inhibit colorectal cancer and liver metastasis, whereas the combination of IP6 and INS was more effective than IP6 or INS alone. The inhibition of colorectal cancer with liver metastasis by IP6+INS may be accomplished via regulation of the Wnt/β-catenin signaling pathway through inhibition of the abnormal expression of Wnt10b, β-catenin, Tcf7, and c-Myc.
Acknowledgments
We thank Meng Yang and Zhao Xuemei for the assistance with the animal experiment. We also thank the teachers and students of the School of Public Health, Qingdao University Medical College, for their assistance.
Funding
This research was funded by National Natural Science Foundation of China, grant number 81973033.
Disclosure
The authors declare no conflict of interest in this study. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
1. Zarour LR, Anand S, Billingsley KG, et al. Colorectal cancer liver metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017;3(2):163–173. doi:10.1016/j.jcmgh.2017.01.006
2. Li W, Chang J, Wang S, et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget. 2015;6(27):24448–24462. doi:10.18632/oncotarget.4423
3. Lintoiu-Ursut B, Tulin A, Constantinoiu S. Recurrence after hepatic resection in colorectal cancer liver metastasis -review article. J Med Life. 2015;8:12–14.
4. Chen H, Peng H, Liu W, et al. Silencing of plasminogen activator inhibitor-1 suppresses colorectal cancer progression and liver metastasis. Surgery. 2015;158(6):1704–1713. doi:10.1016/j.surg.2015.04.053
5. Saadatdoust Z, Pandurangan AK, Ananda Sadagopan SK, et al. Dietary cocoa inhibits colitis associated cancer: a crucial involvement of the IL-6/STAT3 pathway. J Nutr Biochem. 2015;26(12):1547–1645. doi:10.1016/j.jnutbio.2015.07.024
6. Rajamanickam S, Agarwal R. Natural products and colon cancer: current status and future prospects. Drug Dev Res. 2008;69(7):460–471. doi:10.1002/ddr.20276
7. Arya M, Mishra N, Singh P, et al. In vitro and in silico molecular interaction of multiphase nanoparticles containing inositol hexaphosphate and jacalin: therapeutic potential against colon cancer cells (HCT-15). J Cell Physiol. 2019. doi:10.1002/jcp.28200
8. Pandurangan AK, Ismail S, Esa NM, et al. Inositol-6 phosphate inhibits the mTOR pathway and induces autophagy-mediated death in HT-29 colon cancer cells. Arch Med Sci. 2018;14(6):1281–1288.
9. MalGorzata K, Joanna W, Jesse K, et al. Inositol hexaphosphate inhibits proliferation and induces apoptosis of colon cancer cells by suppressing the AKT/mTOR signaling pathway. Molecules. 2017;22(10):1657.
10. Kapral M, Wawszczyk J, Sośnicki S, et al. Modulating effect of inositol hexaphosphate on arachidonic acid-dependent pathways in colon cancer cells. Prostaglandins Other Lipid Mediat. 2017;131:41–48. doi:10.1016/j.prostaglandins.2017.08.002
11. Schröterová L, Hasková P, Rudolf E, et al. Effect of phytic acid and inositol on the proliferation and apoptosis of cells derived from colorectal carcinoma. Oncol Rep. 2010;23:787–793.
12. Ivan B, Nikica D, Karlo R, et al. Efficacy of IP6 + inositol in the treatment of breast cancer patients receiving chemotherapy: prospective, randomized, pilot clinical study. J Exp Clin Cancer Res. 2010;29:12. doi:10.1186/1756-9966-29-12
13. Vucenik I, Yang GY, Shamsuddin AM. Inositol hexaphosphate and inositol inhibit DMBA-induced rat mammary cancer. Carcinogenesis. 1995;16(5):1055–1058. doi:10.1093/carcin/16.5.1055
14. Taciak B, Pruszynska I, Kiraga L, et al. Wnt signaling pathway in development and cancer. J Physiol Pharmacol. 2018;69(2):185–196.
15. Hua F, Shang S, Yang YW, et al. TRIB3 interacts With β-catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology. 2019;156(3):708–721. doi:10.1053/j.gastro.2018.10.031
16. Wang YD, Nie X, Wu RB, et al. Down regulation of human Wnt3 in gastric cancer suppresses cell proliferation and induces apoptosis. Onco Targets Ther. 2016;9:3849–3860. doi:10.2147/OTT.S101782
17. Bienz M, Clevers H. Linking colorectal cancer to wnt signaling. Cell. 2000;103(2):311–320. doi:10.1016/S0092-8674(00)00122-7
18. Sena P, Saviano M, Monni S, et al. Subcellular localization of beta-catenin and APC proteins in colorectal preneoplastic and neoplastic lesions. Cancer Lett. 2006;241(2):203–212. doi:10.1016/j.canlet.2005.10.011
19. Jardé T, Evans RJ, McQuillan KL, et al. In vivo and in vitro models for the therapeutic targeting of Wnt signaling using a Tet-OΔN89β-catenin system. Oncogene. 2013;32(7):883–893. doi:10.1038/onc.2012.103
20. Han J, Gao B, Jin X, et al. Small interfering RNA-mediated down regulation of beta-catenin inhibits invasion and migration of colon cancer cells. Med Sci Monit. 2012;18(7):BR273–BR280. doi:10.12659/MSM.883205
21. Peng H, Jingwen L, Bo-miao Z, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1
22. Liuxing F, Shifu H, Jin G, et al. Whole-exome sequencing characterized the landscape of somatic mutations and pathways in colorectal cancer liver metastasis. J Oncol. 2019;11:2684075.
23. Saad N, Esa NM, Ithnin H, et al. Suppression of β-catenin and cyclooxygenase-2 expression and cell proliferation in azoxymethane-induced colonic cancer in rats by rice bran Phytic Acid (PA). Asian Pac J Cancer Prev. 2013;14(5):3093–3099. doi:10.7314/APJCP.2013.14.5.3093
24. Bradford EM, Thompson CA, Goretsky T, et al. Myo-inositol reduces β-catenin activation in colitis. World J Gastroenterol. 2017;23(28):5115–5126. doi:10.3748/wjg.v23.i28.5115
25. Yu W, Liu C, Li X, et al. Inositol hexaphosphate suppresses colorectal cancer cell proliferation via the Akt/GSK-3β/β-catenin signaling cascade in a 1,2-dimethylhydrazine-induced rat model. Eur J Pharmacol. 2017;805:67–74. doi:10.1016/j.ejphar.2017.03.011
26. Fu M, Song Y, Wen Z, et al. Inositol hexaphosphate and inositol inhibit colorectal cancer metastasis to the liver in BALB/c mice. Nutrients. 2016;8(5):286. doi:10.3390/nu8050286
27. Hoffman RM. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs. 1999;17(4):343–359. doi:10.1023/A:1006326203858
28. Oh BY, Hong HK, Lee WY, et al. Animal models of colorectal cancer with liver metastasis. Cancer Lett. 2017;387:114–120. doi:10.1016/j.canlet.2016.01.048
29. Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–1099. doi:10.1053/j.gastro.2008.07.076
30. Basu S, Haase G, Ben-Ze’Ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Res. 2016;19:5.
31. Zhan Y, Feng J, Lu J, et al. Expression of LEF1 and TCF1 (TCF7) proteins associates with clinical progression of nasopharyngeal carcinoma. J Clin Pathol. 2019;72(6):425–430. doi:10.1136/jclinpath-2019-205698
32. Najdi R, Holcombe RF, Waterman ML. Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog. 2011;10(1):5. doi:10.4103/1477-3163.78111
33. Hao YH, Lafita-Navarro MC, Zacharias L, et al. Induction of LEF1 by MYC activates the WNT pathway and maintains cell proliferation. Cell Commun Signal. 2019;17(1):129. doi:10.1186/s12964-019-0444-1
34. Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4(5):a008052–a008052. doi:10.1101/cshperspect.a008052
35. Singh RP
36. Rizvi I, Riggs DR, Jackson BJ, et al. Inositol hexaphosphate (IP6) inhibits cellular proliferation in melanoma. J Surg Res. 2006;133(1):3–6. doi:10.1016/j.jss.2006.02.023
37. Jenab M, Thompson LU. Phytic acid in wheat bran affects colon morphology, cell differentiation and apoptosis. Carcinogenesis. 2000;21(8):1547–1552. doi:10.1093/carcin/21.8.1547
38. Vucenik I, Shamsuddin AM. Protection against cancer by dietary IP6 and inositol. Nutr Cancer. 2006;55:109–125. doi:10.1207/s15327914nc5502_1
39. Vucenik I, Shamsuddin AM. Cancer inhibition by inositol hexaphosphate (IP6) and inositol: from laboratory to clinic. J Nutr. 2003;133(11):3778S–3784S. doi:10.1093/jn/133.11.3778S
40. Ferry S. Inositol hexakisphosphate blocks tumor cell growth by activating apoptotic machinery as well as by inhibiting the Akt/NFkappaB-mediated cell survival pathway. Carcinogenesis. 2002;23(12):2031–2041. doi:10.1093/carcin/23.12.2031
41. Barker CJ, Wright J, Kirk CJ, et al. Inositol 1,2,3-trisphosphate is a product of InsP\\r, 6\\r, dephosphorylation in WRK-1 rat mammary epithelial cells and exhibits transient concentration changes during the cell cycle. Biochem Soc Trans. 1995;23(2):169S. doi:10.1042/bst023169s
42. Felix G, Bartolomé M, Ivana V, et al. Effects of exogenous inositol hexakisphosphate (InsP6) on the levels of InsP6 and of inositol trisphosphate (InsP3) in malignant cells, tissues and biological fluids. Life Sci. 2002;71(13):1535–1546. doi:10.1016/S0024-3205(02)01927-6
43. Novellasdemunt L, Antas P, Li VS. Targeting Wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;309(8):C511–C521. doi:10.1152/ajpcell.00117.2015
44. Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9(2):119–131. doi:10.1023/B:JOMG.0000037157.94207.33
45. Wend P, Wend S, Krum SA, et al. The role of WNT10B in physiology and disease. Acta physiologica. 2011;204(1):34–51. doi:10.1111/j.1748-1716.2011.02296.x
46. Siu MK, Chen W-Y, Tsai H-Y, et al. TCF7 is suppressed by the androgen receptor via microRNA-1-mediated downregulation and is involved in the development of resistance to androgen deprivation in prostate cancer. Prostate Cancer Prostatic Dis. 2017;20(2):172–178. doi:10.1038/pcan.2017.2
47. Nikuševa-Martić T, Šerman L, Zeljko M, et al. Expression of secreted frizzled-related protein 1 and 3, T-cell factor 1 and lymphoid enhancer factor 1 in clear cell renal cell carcinoma. Pathol Oncol Res. 2013;19(3):545–551. doi:10.1007/s12253-013-9615-3
48. Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4(11):a007906–a007906. doi:10.1101/cshperspect.a007906
49. Cui L, Guan Y, Qu Z, et al. WNT signaling determines tumorigenicity and function of ESC-derived retinal progenitors. J Clin Invest. 2013;123(4):1647–1661. doi:10.1172/JCI65048
50. He T. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi:10.1126/science.281.5382.1509
51. Raju K, David W. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337.
52. Wu X, Bie Q, Zhang B, et al. Wnt10B is critical for the progression of gastric cancer. Oncol Lett. 2017;13(6):4231–4237. doi:10.3892/ol.2017.5992
53. Madueke I, Hu WY, Hu D, et al. The role of WNT10B in normal prostate gland development and prostate cancer. Prostate. 2019;79(14):1692–1704.
54. Li T, Zhu J, Wang X, et al. Long non-coding RNA lncTCF7 activates the Wnt/β-catenin pathway to promote metastasis and invasion in colorectal cancer. Oncol Lett. 2017;14(6):7384–7390. doi:10.3892/ol.2017.7154
55. Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi:10.1016/j.stem.2015.03.003
56. Xu X, Liu Z, Tian F, et al. Clinical Significance of Transcription Factor 7 (TCF7) as a prognostic factor in gastric cancer. Med Sci Monit. 2019;25:3957–3963. doi:10.12659/MSM.913913
57. Trotman LC, Nowak DG, Cho H, et al. Myc drives Pten/ p53-deficient proliferation and metastasis due to Il6-secretion and Akt-suppression via Phlpp2. Cancer Disc. 2015;5(6):636. doi:10.1158/2159-8290.CD-14-1113
58. Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74(14):3764–3778. doi:10.1158/0008-5472.CAN-13-3555
59. Cho H, Herzka T, Zheng W, et al. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of pten-mutant metastasis. Cancer Disc. 2014;4(3):318–333. doi:10.5114/aoms.2018.76935
60. Liu Y, Zhou S, Shi J, et al. c-Myc transactivates GP73 and promotes metastasis of hepatocellular carcinoma cells through GP73-mediated MMP-7 trafficking in a mildly hypoxic microenvironment. Oncogenesis. 2019;8(10):58. doi:10.1038/s41389-019-0166-7
61. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, et al. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1):197. doi:10.3390/ijms18010197
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.