Back to Journals » Journal of Inflammation Research » Volume 15
Coactosin-Like Protein in Breast Carcinoma: Friend or Foe?
Authors Wang B , Zhao L, Chen D
Received 15 February 2022
Accepted for publication 10 July 2022
Published 15 July 2022 Volume 2022:15 Pages 4013—4025
DOI https://doi.org/10.2147/JIR.S362606
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Bei Wang,1 Limiao Zhao,1 Dandan Chen2
1Institute of Integration of Traditional Chinese and Western Medicine, the Hospital Affiliated to Jiangnan University, Wuxi, People’s Republic of China; 2Department of Education, School of Humanities, Jiangnan University, Wuxi, People’s Republic of China
Correspondence: Bei Wang, Email [email protected]
Background: Coactosin-like protein (COTL1) was first identified as protein that binds 5-lipoxygenase and F-actin; its functions in tumors remain unknown. COTL1 could inhibit the proliferation of breast cancer (BRCA) in vivo and in vitro; however, online public databases including UALCAN and Kaplan–Meier plotter showed high COTL1 expression in breast cancer tissue, which was correlated with poor prognosis. Therefore, we studied the role of COTL1 expression in human breast cancer and its use in determining clinical prognosis.
Methods: We first used the UALCAN database and immunohistochemical analysis to elucidate COTL1 expression in BRCA. We then performed Kaplan–Meier plotting and immunohistochemical analysis to assess prognosis in BRCA in relation to COTL1 expression. Finally, we used the CancerSEA and LinkedOmics databases to evaluate the function of COTL1 in BRCA. The TIMER and TISIDB databases were used to evaluate the association between COTL1 expression and immune cell infiltration in BRCA.
Results: UALCAN and immunohistochemical analysis showed that COTL1 was highly expressed in breast cancer. Furthermore, high COTL1 expression was correlated with poor prognosis in BRCA. We also found that COTL1 is involved in immune response via the CancerSEA and LinkedOmics databases. The TIMER and TISIDB databases showed that high COTL1 expression was correlated with immune cell infiltration.
Conclusion: COTL1 expression was higher in breast cancer tissues than in normal tissues, and high COTL1 expression was correlated with poor prognosis and immune cell infiltration. These results provide a basis for the development of applications of COTL1 in determining the prognosis of breast cancer and its treatment.
Keywords: COTL1, breast cancer, prognosis, Immune cell infiltration, biomarker
Introduction
Coactosin-like protein (COTL1) was first identified as a binding protein of 5-lipoxygenase and F-actin;1–3 its functions in tumors remain unclear.4–7 Xia et al recently reported that COTL1 could inhibit the proliferation of breast cancer in vivo and in vitro.6 Previous studies have focused on the function of COTL1 in cell and animal models, but not in humans. Online public databases including UALCAN and Kaplan-Meier plotter show that COTL1 is highly expressed in breast cancer tissue and high COTL1 expression correlates with poor prognosis. Therefore, we aimed to determine the role of COTL1 expression in human breast cancer, and the related clinical prognosis.
Breast cancer is one of the most common malignancies among women, and is a major cause of cancer-related deaths worldwide.8 The immune system plays two roles in breast cancer. It can promote tumor formation through inflammatory pathways, and can suppress tumor formation through active immune surveillance.9 In this study, we used immunohistochemistry and public online databases including UALCAN and Kaplan-Meier plotter to evaluate COTL1 expression and related prognosis in breast cancer. In addition, we explored the correlation between COTL1 expression and immune cell infiltration in breast cancer by using the CancerSEA, LinkedOmics, and TISIDB public online databases. Our results could provide a basis for the clinical application of COTL1 in breast cancer in the future.
Materials and Methods
Immunohistochemistry
We used a tissue microarray (obtained from XinChao Biological Technology Co., Ltd), which included 170 samples of breast cancer tissues and sixty-two samples from para-carcinoma tissues. Before dewaxing, the tissue microarray was incubated for 30 minutes at 60 °C. The slides were repaired with citrate antigen retrieval solution (Beyotime, #P0083). After cooling down, non-specific binding was blocked with endogenous peroxidase blocking buffer (Beyotime, #P0100A) and immunol staining blocking buffer (Beyotime, #P0102). Primary antibody (COTL1 (1: 400, Proteintech, #17119-1-AP)) was added dropwise and incubated overnight at 4°C. The increasing agent was added dropwise and the samples were placed in a humidified incubator at room temperature for 20 min. The enzyme-labeled secondary antibody (MXB, #KIT 9902) was added dropwise and the samples were placed in a humidified incubator at room temperature for 30 min. DAB horseradish peroxidase color development kit (Beyotime, #P0203) and hematoxylin contrast (Beyotime, #C0107) were used for staining. Routine alcohol gradient dehydration, transparent xylene, and neutral resin mounting were performed. The negative control was exposed to PBS instead of the primary antibody. Staining was assessed by scanning the entire tissue specimens. An immunoreactivity score system was adopted based on the scale and the intensity of positively-stained target cells. The following standards were used: 1) the number of cells with positive staining (≤ 5% = 0; 6% - 25% = 1; 26% - 50% = 2; 51% - 75% = 3; and > 75% = 4); 2) the staining intensity (colorless = 0; pallide-flavens = 1; yellow = 2; brown = 3). We multiplied the scores of 1 and 2 in the standard, and the dyeing grade was divided into none (0 points), weak (1–4 points), medium (5–8 points), or strong (9–12 points). Immunohistochemistry staining and evaluation were performed as described previously.10 This work was approved by the affiliated hospital of Jiangnan University (#LS2021076).
UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html)11 is a comprehensive, user-friendly, and interactive web resource for analyzing COTL1 expression in breast cancer.
Kaplan-Meier Plotter Analysis
Kaplan-Meier plotter (http://kmplot.com/analysis/),12 a database that integrates gene expression data and clinical data, was used to analyze the prognostic value of COTL1 in breast cancer.
LinkedOmics
LinkedOmics (http://www.linkedomics.org/login.php),13 was used to study the associated RNA Sequence and for enrichment analysis.
CancerSEA
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp),14 was used to comprehensively analyze COTL1 function in BRCA at single-cell resolution.
Timer
TIMER (https://cistrome.shinyapps.io/timer/),15 was used to comprehensively analyze tumor immune cells and COTL1 expression in BRCA.
TISIDB
TISIDB (http://cis.hku.hk/TISIDB/index.php),16 was used to study the correlation between COTL1 expression and immune system in BRCA.
Statistical Analysis
Chi-square tests were used to analyze immunohistochemical score levels in different clinical pathological feature groups. Univariate and multivariate analyses were used to evaluate the effect of clinical variables on survival. A two-tailed p-value <0.05 was considered statistically significant.
Results
High COTL1 Expression in BRCA
To elucidate the differences in COTL1 expression in tumor and normal tissues, the COTL1 mRNA levels in breast tumors and normal tissues were analyzed using the UALCAN database. COTL1 mRNA expression was significantly higher in breast cancer tissues than in normal tissues (Figure 1A). In addition, subgroup analysis based on stage, race, gender, age, subclasses, menopause status, histologic subtype, metastasis, and TP53 mutation status indicated that COTL1 mRNA expression was significantly higher in breast cancer tissues than in normal tissues (Figure 1B–J). COTL1 expression was much higher in triple negative breast cancer compared to other subtypes. Triple negative breast cancer has a worse clinical prognosis than does non-triple negative breast cancer.17 Further, the TISIDB database showed that COTL1 expression was associated with molecular and immune subtypes (Figure 1K and L). We then studied COTL1 protein expression by using a tissue microarray, which included 170 breast cancer tissue samples and sixty-two para-carcinoma tissue samples. The result of immunohistochemical analysis was consistent with that of UALCAN analysis (Figure 1M and N). To further evaluate the correlation between COTL1 expression and clinical characteristics, COTL1 expression and clinical data were downloaded from the cancer genome atlas (TCGA). The results indicated that COTL1 expression was associated with clinical parameters such as laterality, ER, PR, and radiation therapy (Table 1). The results from our immunohistochemical analysis demonstrated that COTL1 expression was correlated with clinical parameters at T, N, and HER2. (Table 2). These contradictory findings could be due to our small sample size, or because COTL1 expression is unrelated to clinical parameters.
 |
Table 1 The Correlation of COTL1 Expression and Clinical Characteristics from TCGA (n=1104) |
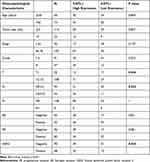 |
Table 2 The Correlation of COTL1 Expression and Clinical Characteristics in Breast Cancer Patients (n=138) |
Clinical Prognosis of COTL1 in Patients with BRCA
We determined whether COTL1 expression is correlated with prognosis in patients with breast cancer. The influence of COTL1 expression on survival rates was first evaluated using the Kaplan-Meier plotter database. The results showed that COTL1 mRNA level was positively correlated with poor prognosis (Figure 2A). We then analyzed GSE20486 and GSE12276 from the Gene Expression Omnibus (GEO) database, and found that higher COTL1 mRNA levels corresponded with poorer overall survival (OS; Figure 2B and C). To confirm the relationship between COTL1 and clinical prognosis, the overall survival analysis from our tissue microarray results demonstrated that high COTL1 expression in breast cancer tissues was associated with poor prognosis (Figure 2D). To determine whether COTL1 is an independent risk factor for overall survival in patients with breast cancer, univariate and multivariate Cox analyses were performed using the SPSS software. In these analyses, estrogen receptor (ER), and COTL1 expression were independent risk factors for OS (Table 3). Thus, COTL1 could be an independent prognostic biomarker for BRCA.
 |
Table 3 Univariate and Multivariate Analysis of the Correlation of COTL1 Expression with Overall Survival Among Breast Cancer Patients (n=139) |
COTL1 Function in BRCA
COTL1 can interact with F-actin in a calcium-independent manner and influence ALOX5 stability and activity in leukotriene synthesis as a chaperone for ALOX52. We used the CancerSEA database to investigate the new function of COTL1 in BRCA at the single-cell resolution. The results indicated that COTL1 was positively associated with differentiation, inflammation, metastasis, angiogenesis, apoptosis, and quiescence and negatively associated with DNA repair, invasion, stemness, and DNA damage (Figure 3A). According to data from Braune EB (n=369) and Jordan NV(n=70), COTL1 expression was positively correlated with angiogenesis, differentiation, inflammation and quiescence18,19 (Figure 3B and C). To elucidate the function of COTL1 in BRCA, the LinkedOmics database was used, and the results showed that COTL1 was positively and negatively correlated with the top fifty significant genes (Figure 4A). Results of gene set enrichment analysis (GSEA), including details of biological process, cellular components, and molecular function of COTL1 are shown in Figure 4B. Overrepresentation enrichment analysis (ORA) showed that COTL1 is involved in and regulates multiple immune responses (Figure 4C). Specifically, COTL1 increased interferon-gamma and interleukin-1 production and activated natural killer cells (Figure 4D). In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway showed that COTL1 is involved in multiple signal pathways such as the IL-17, TNF, and B cell receptor signaling pathways (Figure 4E). These results suggest that COTL1 plays a role in immune processes in breast cancer.
Correlations Between COTL1 and Immune Cell Infiltration in BRCA
Since COTL1 expression was associated with immunity, we investigated the correlation between COTL1 expression and immune cell infiltration in BRCA. The TIMER database was used to assess the correlation between COTL1 expression and the infiltration levels of six immune cells. COTL1 expression was strongly positively correlated with immune cell infiltration (Purity: cor = 0.415, p = 1.2e-42; B Cell: cor = 0.435, p = 2.23e-46; CD8+ T cell: cor = 0.283, p = 1.83e-71; CD4+T Cell: cor = 0.532, p = 2.08e-71; Macrophage: cor = 0.156, p = 8.84e-07; Neutrophil: cor = 0.523, p = 9.34e-68, Dendritic cell: cor = 0.622, p = 4.66–103; Figure 5). To confirm these results, we used the TISIDB database to determine the correlation between COTL1 expression and twenty-eight immune cells in BRCA. COTL1 expression in BRCA tissues was significantly correlated with immune cells including helper T cells, NK cells, NKT cells, and regulatory T cells in addition to the aforementioned six immune cell types (Figure 6A). Furthermore, TIMER was utilized to determine the correlation between COTL1 expression and immune marker genes of different immune cells including CD8+T cells, T cells, B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, NK cells, DCs, Th1, Th2, Th17, Treg, and T cell exhaustion in BRCA. COTL1 expression was strongly correlated with 94.8% immune cell gene markers (Table 4). The TISIDB database was then utilized to determine the correlation between COTL1 expression and immune inhibitors. COTL1 expression was significantly correlated with immune inhibitors. The T cell exhaustion markers: CTLA4, LAG3, PD1, and PDCD1 were positively correlated with COTL1 (Figure 6B), which was consistent with the TIMER data (Table 4). These results strongly suggest that COTL1 is closely correlated with immune cell infiltration in BRCA.
 |
Table 4 Correlation Analysis Between COTL1 and Relate Genes and Markers of Immune Cells in BRCA by TIMER |
 |
Figure 5 Correlation of COTL1 expression with tumor immune cell infiltration (purity, B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and DCs) in the TIMER database. |
Discussion
COTL1 was first identified in pancreatic cancer tissues, where it was highly expressed compared to normal tissues.20 Subsequently, Jeong et al found that COTL1 was highly expressed in small cell lung cancer (SCLC).21 COTL1 was correlated with low metastatic potential in human non-small cell lung cancer (NSCLC), which suggested that COTL1 inhibits lung cancer metastasis.4 A recent study showed that COTL1 promotes glioblastoma (GBM) cell growth in vitro and in vivo, and immunohistochemical assays showed that COTL1 is highly expressed in human GBM tissues.5 Xia et al found that COTL1 inhibited breast cancer cell growth in vitro and in vivo,6 However, multiple online databases show that COTL1 is highly expressed in breast cancer tissue compared to normal tissues. Therefore, we studied COTL1 expression and associated prognosis in human breast cancer tissues by using a tissue microarray with 170 breast cancer tissue samples and sixty-two para-carcinoma tissue samples.
Analysis using the TCGA database and immunohistochemistry showed that COTL1 expression was higher in breast cancer tissues than in normal tissues. We then determined whether COTL1 expression was correlated with prognosis in patients with breast cancer. The results from the Kaplan-Meier Plotter database were inconsistent: COTL1 mRNA level was positively correlated with poor prognosis (Figure 2A) while COTL1 protein level was negatively correlated with poor prognosis (Supplementary Figure 1). This could be because of the different data sources and processing methods of the Kaplan-Meier plotter database. Our tissue microarray data showed that high expression of COTL1 in breast cancer tissues was associated with poor prognosis (Figure 2D). Although only 139 cases were included in our study, our data strongly demonstrated that ER, and COTL1 were independent risk factors for OS (Table 3).
Breast cancer is a heterogeneous disease with unique histological, molecular, and clinical phenotypes. The expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) are considered the major determinants of breast cancer and can be used to refine molecular and prognostic taxonomy of breast cancer.22 Triple negative breast cancer (TNBC) accounts for 10–15% breast cancer cases and lacks ER and PR, and expresses low levels of HER29. TNBC involves immune activation and is associated with tumor-infiltrating lymphocytes (TILs).23 Immune checkpoint antagonists have been applied in the treatment of various tumors, specifically for CTLA-4, PD-1, and PD-L1.24 COTL1 expression is the highest in triple negative breast cancer and correlates with immune checkpoint antagonists, which suggests that COTL1 is a potential therapeutic target for breast cancer, especially triple negative breast cancer.
Xia et al found that COTL1 could inhibit the proliferation of breast cancer. In current study, it was found that the expression of COTL1 was related to the high infiltration levels of lymphocytes immune cells. Therefore, we hypothesized that COTL1 may inhibit the proliferation of breast cancer by recruiting CD8+ T lymphocytes. However, high CD8+T cell infiltration was found in breast cancer tissues with high COTL1 expression, which was associated with poor OS in BRCA. This observation appears to be contradictory assuming that the presence of CD8+ T cells improves the patient survival. However, T cell exhaustion is widely described as a mechanism that inhibits CD8+ T cells proliferation and kills tumor cells. In fact, we found that high expression of T cell exhaustion markers PD-1, CTLA4, TIM-3, GZMB, LAG-3, and PDL1 were all associated with high expression of COTL1, supporting that T cell exhaustion could suppress T cell functions in BRCA. Therefore, these findings may explain why the high level of CD8+ T cells failed to induce a survival benefit in BRCA. Xia et al proposed that COTL1 inhibited breast cancer growth in the early stage and lost its growth inhibition function in the late stage, in current study, we found COTL1 expression was higher in breast cancer tissues than in normal tissues, and high COTL1 expression was correlated with poor prognosis. Xia et al speculated that COTL1 may play a bidirectional role in tumor like TGFβ1. The mechanism by which COTL1 functions as a tumor suppressor in the early stage of breast cancer progression and as an oncogene in the late stage merits further study.
In conclusion, we used multiple online public databases and conducted immunohistochemical analysis of breast cancer tissues, and found that COTL1 is highly expressed in breast cancer and is correlated with poor prognosis. However, our study has limitations, such as small sample size and data from online public database. Future studies with larger samples can be used to verify the clinical expression of COTL1 in breast cancer and associated prognosis. Our results provide a basis for the development of clinical applications of COTL1 in breast cancer treatment.
Conclusions
We found that COTL1 expression was higher in breast cancer tissues than in normal tissues, and that high COTL1 expression was correlated with poor prognosis and immune cell infiltration. The current study provided novel insights for the future application of COTL1 in the treatment of breast cancer.
Abbreviations
COTL1, coactosin-like protein; BRCA, breast cancer; TCGA, the cancer genome atlas; GEO, gene Expression Omnibus; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, Triple negative breast cancer; SCLC, small cell lung cancer; NSCLC, non-small cell lung cancer; TILs, tumor-infiltrating lymphocytes; CTLA-4, cytotoxic T lymphocyte-associated antigen-4; PD-1, programmed cell death protein-1; PD-L1, PD ligand-1.
Ethical Approval and Consent to Participate
This work was approved by the affiliated hospital of Jiangnan University (#LS2021076).
Funding
This work was supported by the National Natural Science Foundation of China (No.31801171), China Postdoctoral Science Foundation (No. 2019M650102), Wuxi Commission of Health and Family Planning (No. 2020ZHYB04) and Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (No. HB2020039).
Disclosure
The authors declare no conflict of interest.
References
1. Provost P, Samuelsson B, Radmark O. Interaction of 5-lipoxygenase with cellular proteins. Proc Natl Acad Sci U S A. 1999;96:1881–1885. doi:10.1073/pnas.96.5.1881
2. Provost P, Doucet J, Hammarberg T, Gerisch G, Samuelsson B, Radmark O. 5-Lipoxygenase interacts with coactosin-like protein. J Biol Chem. 2001;276:16520–16527. doi:10.1074/jbc.M011205200
3. Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Radmark O. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J. 2001;359:255–263. doi:10.1042/bj3590255
4. Sun W, Guo C, Meng X, et al. Differential expression of PAI-RBP1, C1orf142, and COTL1 in non-small cell lung cancer cell lines with different tumor metastatic potential. J Investigative Med. 2012;60:689–694. doi:10.2310/JIM.0b013e31824963b6
5. Shao S, Fan Y, Zhong C, Zhu X, Zhu J. Coactosin-Like Protein (COTL1) Promotes Glioblastoma (GBM) Growth in vitro and in vivo. Cancer Manag Res. 2020;12:10909–10917. doi:10.2147/CMAR.S246030
6. Xia L, Xiao X, Liu WL, et al. Coactosin-like protein CLP/Cotl1 suppresses breast cancer growth through activation of IL-24/PERP and inhibition of non-canonical TGFbeta signaling. Oncogene. 2018;37:323–331. doi:10.1038/onc.2017.342
7. Guo S, Yang P, Jiang X, et al. Genetic and epigenetic silencing of mircoRNA-506-3p enhances COTL1 oncogene expression to foster non-small lung cancer progression. Oncotarget. 2017;8:644–657. doi:10.18632/oncotarget.13501
8. Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14–27. doi:10.1016/j.semcancer.2019.08.012
9. Emens LA. Breast cancer immunobiology driving immunotherapy: vaccines and immune checkpoint blockade. Expert Rev Anticancer Ther. 2012;12:1597–1611. doi:10.1586/era.12.147
10. Shen L, Qu X, Ma Y, et al. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-beta via EMT inhibition in colorectal cancer. Oncogenesis. 2014;3:e86. doi:10.1038/oncsis.2013.48
11. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi:10.1016/j.neo.2017.05.002
12. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi:10.1038/s41598-018-27521-y
13. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi:10.1093/nar/gkx1090
14. Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D908. doi:10.1093/nar/gky939
15. Li T, Fan J, Wang B, et al. TIMER: a Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e110. doi:10.1158/0008-5472.CAN-17-0307
16. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi:10.1093/bioinformatics/btz210
17. Li X, Yang J, Peng L, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279–287. doi:10.1007/s10549-016-4059-6
18. Braune EB, Tsoi YL, Phoon YP, et al. Loss of CSL Unlocks a Hypoxic Response and Enhanced Tumor Growth Potential in Breast Cancer Cells. Stem Cell Rep. 2016;6:643–651. doi:10.1016/j.stemcr.2016.03.004
19. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi:10.1038/nature19328
20. Nakatsura T, Senju S, Ito M, Nishimura Y, Itoh K. Cellular and humoral immune responses to a human pancreatic cancer antigen, coactosin-like protein, originally defined by the SEREX method. Eur J Immunol. 2002;32:826–836. doi:10.1002/1521-4141(200203)32:3<826::AID-IMMU826>3.0.CO;2-Y
21. Jeong HC, Kim GI, Cho SH, et al. Proteomic analysis of human small cell lung cancer tissues: up-regulation of coactosin-like protein-1. J Proteome Res. 2011;10:269–276. doi:10.1021/pr100714b
22. Rakha EA, Green AR. Molecular classification of breast cancer: what the pathologist needs to know. Pathology. 2017;49:111–119. doi:10.1016/j.pathol.2016.10.012
23. Won KA, Spruck C. Triplenegative breast cancer therapy: current and future perspectives (Review). Int J Oncol. 2020;57:1245–1261. doi:10.3892/ijo.2020.5135
24. Emens LA. Breast Cancer Immunotherapy: facts and Hopes. Clin Cancer Res. 2018;24:511–520. doi:10.1158/1078-0432.CCR-16-3001
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





