Back to Journals » Infection and Drug Resistance » Volume 14
Co-Production of NDM-1, CTX-M-9 Family and mcr-1 in a Klebsiella pneumoniae ST4564 Strain in China
Authors Wang X, Li Q, Kang J, Zhang Z, Song Y, Yin D, Guo Q, Song J, Li X, Wang S, Duan J
Received 18 November 2020
Accepted for publication 9 January 2021
Published 5 February 2021 Volume 2021:14 Pages 449—457
DOI https://doi.org/10.2147/IDR.S292820
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Xinchun Wang,1,* Qi Li,2,* Jianbang Kang,1 Zheng Zhang,2 Yan Song,1 Donghong Yin,1 Qian Guo,1 Junli Song,1 Xiaoxia Li,1 Shuyun Wang,1 Jinju Duan1
1Department of Pharmacy, Second Hospital of Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China; 2Department of Pharmacy, School of Pharmacy, Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinju Duan
Department of Pharmacy, Second Hospital of Shanxi Medical University, No. 382, Wuyi Road, Xinghualing District, Taiyuan, Shanxi, People’s Republic of China
Tel +86 351 3365713
Email [email protected]
Purpose: To identify novel sequence types 4564 (ST4564) carbapenem-resistant Klebsiella pneumoniae (CRKP). Characterizing the feature of the clinic, resistance, and virulence of a co-producing NDM-1 and CTX-M-9 family and mcr-1 ST4564 strain.
Methods: A novel ST4564 CRKP was collected from June 2018 to July 2018. We investigated its antimicrobial susceptibility by the microdilution method. Using the modified carbapenem inactivation method (mCIM) to screen phenotype of carbapenemases. Resistance mechanisms, virulence-associated genes, multilocus sequence typing (MLST), and capsular serotypes were characterized by polymerase chain reaction (PCR) and DNA sequencing. Next-generation sequencing (NGS) was carried out to determine the genetic features of carbapenem resistance and virulence.
Results: ST4564, co-carrying NDM-1, CTX-M-9 and mcr-1, was resistant to carbapenems, cephamycin, third- or fourth-generation cephalosporins, β-lactam combination agents, quinolones and tigecycline but remained susceptible to amikacin (AMK) and colistin (COL). Through the NGS analysis with the G+C content of 56.65%, multiple resistance and virulence genomes were detected. The genes encoding the β-lactams, aminoglycosides, quinolones, macrolides, sulfonamide, polysaccharide capsule, type-I fimbriae cluster, siderophore genes, transporter and pumps, T6SS and pullulanase secretion protein. goeBURST analysis showed that ST4564 belonged to the CC1571 and it was not related to the prevalent high-risk clones.
Conclusion: We first identified the novel ST4564 CRKP. Our finding suggested that the urgent need for infection control of the new clone to prevent it from becoming a high-risk clone of CRKP.
Keywords: Klebsiella pneumoniae, carbapenem resistant, ST4564, next-generation sequencing
Introduction
Klebsiella pneumoniae (KP), a gram-negative bacterium, is an opportunistic pathogen.1 It is known to colonize the skin, respiratory and gastrointestinal tracts and could cause severe infections, including urinary tract infections, pneumonia, soft tissue infections, bacteremia, abscesses, and intra-abdominal infections. Carbapenems are considered to be an effective treatment for clinical infection caused by KP.2 To make matters worse, the numbers of carbapenem-resistance Klebsiella pneumoniae (CRKP) strains have increased steadily. The infections caused by CRKP often lead to high morbidity and high mortality.
CRKP strains are spreading as clones, especially the high-risk clones, which include but not limited to, ST11, ST15, ST16, ST23, ST25, ST101, ST147, ST307, ST258.3–5 First ST258 KP clone was reported in the United States and was disseminated around the world quickly.2 After that, ST11, ST23, ST15, and ST101 clones were continuously discovered and further investigated. These clones could cause outbreak infections and lead to unimaginable outcomes. Furthermore, the dissemination of STs has geographical differences. ST258 has become the most prevalent clone of CRKP in North America and Europe.6 ST11 was frequently found in Asia, especially China, accounting for up to 60% of CRKP clones.7
Previous studies have highlighted the complex resistance mechanisms of CRKP and the transfer of resistance genes through epidemic plasmids in highly disseminating different clones.8 These factors could increase the difficulty in treatment.2 The most main resistance mechanism in KP was the production of carbapenemases, including class A, class B, and class D. Among them, KPC-2 was the dominant carbapenemase in China.2 Other resistance mechanisms were the expression of plasmid-mediated AmpC and extended-spectrum beta-lactamases (ESBLs). CTX-M-15- or CTX-M-14-producing ST11 or ST147 are high-risk multidrug-resistant KP (MDR) clones, which have spread globally.2,9 Recently, outbreaks of different STs with different resistance mechanisms have been reported in China.9–11 In this case, the identification of new clones’ resistance mechanism was more important.
Likewise, types of capsule, fimbriae, lipopolysaccharide (LPS) and siderophores are important and well-characterized virulence factors in KP.12 Virulence factors are associated with high pathogenicity and resulted in poor outcomes and shorten the process of diseases. rmpA, rmpA2, magA were often considered as the gene markers of hypervirulent Klebsiella pneumoniae (hvKP). They could participate in the production of the capsule. In the part of studies, ST11, ST147, and ST101 clones were often related to hvKP and ST11 was the most predominant type.13,14
Due to the relationship between the epidemic STs and the features of resistance or virulence was unclear. The aim of our study was to identify the novel sequences types of CRKP and characterize the features of genetics in it.
Materials and Methods
Bacteria Identification
From June 2018 to July 2018, one CRKP strain was isolated from the Second Hospital of Shanxi Medical University. The confirmation of this CRKP isolate was conducted by the Vitek 2 instrument (BioMerieux Italia S.p.A) and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Bruker Daltonik, Bremen, Germany). This CRKP isolate was identified based on the non-susceptibility to carbapenems (imipenem, meropenem, or ertapenem), according to the Clinical and Laboratory Standards Institute (CLSI-2020) guidelines.15,16
Susceptibility to Antibacterial Agents
One CRKP strain was tested for susceptibility by the broth microdilution method using cation-adjusted Mueller-Hinton broth. The susceptibility breakpoints were interpreted following the criteria of the CLSI-2020.15 Except for tigecycline and colistin, which were interpreted based on the European Committee for Antimicrobial Susceptibility Testing (EUCAST) criteria.17 (EUCAST-2021, v 11.0, http://www.eucast.org.) Escherichia coli ATCC 25922 was used as a control for antimicrobial susceptibility testing. We tested 17 common antimicrobial agents, which were listed as folllows: meropenem (MEM), imipenem (IPM), ertapenem (ETP), ceftriaxone (CRO), ceftazidime (CAZ), cefotaxime (CTX), cefepime (FEP), aztreonam (ATM), cefoperazone/sulbactam (CSL), piperacillin/tazobactam (TZP), ciprofloxacin (CIP), levofloxacin (LEV), fosfomycin (FOS), chloramphenicol (CHL), amikacin (AMK), tigecycline (TGC) and colistin (COL).
Phenotypic Characterization
The modified carbapenem inactivation method (mCIM) was used for phenotypic detection of carbapenemases. The string test was used to detect hypermucoviscosity phenotype. When the length of the viscous string was ≧ 5 mm, the strain was defined as hypermucoviscosity Klebsiella pneumoniae (HMKP).9
PCR Amplifications and Sequencing
The presence of genes encoding CTX-M-type extended-spectrumβ-lactamases (ESBLs), plasmid-borne AmpC β-lactamases, and carbapenemases were detected by polymerase chain reaction (PCR) as previously described.18 The positive PCR amplicons were visualized by agarose gel electrophoresis and sequenced using Sanger sequencing. The DNA sequences obtained were compared with those available in the NCBI GenBank database using BLAST searches. (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiplex PCR for virulence genes including iucA, iutA, rmpA, rmpA2 and iroN was performed after phenotypic characterization. The HMKP isolates with positive results of virulence genes were defined as hvKP in our study.19–21 The capsular serotype of the KP isolate was determined by wzi gene sequencing and allele identification using Institute Pasteur, Paris, France, BIGS database. (https://bigsdb.pasteur.fr/Klebsiella).
Next-Generation Sequencing (NGS)
The genomes of resistance and virulence were fully sequenced by Ion S5 plus (Thermo fisher) with 420 flows, which obtained read lengths ranging from 100 bp to 350 bp after read filtering. De novo assembly was performed using SPAdes genome assembler. The scaffold N50 (bp) was 169157 bp; the GC content was 56.65% and the assemble percentage of the whole genome was 97%. Antimicrobial resistance and virulence genes were screened and annotated using CARD and VFDB tools with 97% coverage and an identity threshold of 97%. All draft genome sequences were deposited into the NCBI Genome database and organized under BioProject PRJNA689622 and Bio-Samples number was SAMN17183116.
Multilocus Sequence Typing (MLST)
Multilocus sequence typing (MLST) was performed for this CRKP isolate by amplifying and sequencing seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB). Alleles and sequence types (STs) are available on the MLST online database. (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). One CRKP 102 strain containing alleles that did not match an existing sequence in the MLST database was submitted in the respective database for allele and ST assignments. Clonal complexes (CC) were determined to identify the molecular epidemiological relationships with the goeBURST 1.2.1 version software (http://www.phyloviz.net/goeburst/). ST profiles sharing 100% genetic identity in at least 6 of 7 MLST loci were grouped into a clonal complex (CC), named after its presumed ancestral genotype.
Results
Bacterial and Clinical Information
The first identified ST4564 CRKP102 strain was cultured from an alveolar lavage fluid sample from a 57-year-old male with no recent travel history. He was admitted on 6 June 2018 to the department of hematopathology, due to an unknown fever and lowest level of thrombocyte. Carbapenem antibiotic treatment, meropenem, had been used as empirical therapy prior to hospitalization. Due to the lack of meropenem, he switched to imipenem for further treatment. Because of the poor effect of imipenem, linezolid was added to the treatment, and then ceftriaxone was used as de-escalation therapy after the fungus was cultured. Then, he received the combination treatment of tigecycline with ceftriaxone and cefoperazone/sulbactam, respectively, before the strain was isolated. Among these combination treatments, cefoperazone/sulbactam was used as de-escalation therapy, because of the improvement in the inflammatory response. On 10 July, the MDR CRKP was isolated, which was identified by clinical microbiologists. And he received meropenem therapy. However, this patient remained in long-term rehabilitation and has only been detected once CRKP. Furthermore, he has fever 5 times during the whole hospitalization (Figure 1). Eventually, this patient was discharged from the hospital.
Antimicrobial Sensitivity and Resistance Mechanism Results
ST4564 CRKP102 strain showed resistance to carbapenems, third- or fourth-generation cephalosporins, β-lactam combination agents and ciprofloxacin, levofloxacin but remained susceptible to amikacin, tigecycline and colistin (Table 1). CRKP102 was confirmed as a metallo-ß-lactamase-producing strain with the mCIM test. CRKP102 subsequently was only positive for the blaNDM-1, blaCTX-M-14,17 and mcr-1 genes by PCR amplification and sequencing. Other resistance genes were not detected (Table 2).
 |
Table 1 Antimicrobial Susceptibility Testing Results of Carbapenem-Resistant K. pneumoniae Isolate (Mg/L) |
 |
Table 2 Distribution of Resistance Mechanism and Virulence Genes Harbored by ST4564 CRKP Strain |
Characterization of Virulence
CRKP102 isolate was positive for the string test and was negative for the iucA, iutA, rmpA, iroN, rmpA2 gens by PCR and sequencing. By amplifying K1, K2, K47, K64 and wzi gene, none of the serotypes was detected in this study (Table 2).
Genetic Determinants of Resistance and Virulence
The next-generation sequencing analysis was performed on CRKP102 strain. It harbored multiple genes encoding resistance to β-lactams (blaCTX-M-14, blaCTX-M-17, blaNDM, blaTEM, blaLEN), aminoglycosides [aac (3)-IIb, aph (6)-Id, aph (3ʹ)-Ia, aadA6/aadA10], quinolones (qnrBs), macrolides [mphA, mrx], and sulfonamide [sul1, sul2]. From this finding, two variants of CTX-M-9 family were detected (Table 3).
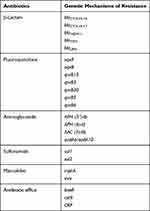 |
Table 3 The Presence of Resistance Genome in ST4564 |
Moreover, we also detected multiple virulence genes, such as: the genes associated with capsule, type-I fimbriae cluster, the siderophore genes (iutA, fepD, iroE, enterobactin genes), transporter and pumps (ABC transporter, RND efflux system (acrAB, rcsAB)), T6SS (the genes encoding pullulanase secretion protein). (Table 4)
 |
Table 4 The Presence of Virulence Genome in ST4564 |
Compared the resistance and virulence genome with the CARD and VFDB databases, CRKP102 strain had 7 blaLEN genes, 7 blaNDM genes, 5 qnrB genes, and the galF gene. These findings had a significant difference with the CARD and VFDB databases.
Molecular Typing Results
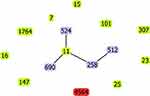
This strain had unique allelic profile (gapA_28, infB_24, mdh_21, pgi_76, phoE_146, rpoB_17, tonB_67), we submitted this ST profile in the respective database and this file was allocated 4564. This is the first time identified the ST4564. As determined by goeBURST analysis of MLST data (Figure 2), the novel sequence type 4564 was belonging to clonal cluster CC1571. Compared the allele of ST4564 with high-risk clones (ST7, ST11, ST15, ST16, ST101, ST147, ST25, ST23, ST258, ST307, ST512, ST524, ST690, ST1764), we found that ST4564 was an independent clone and was not related to any of high-risk clones.
Discussion
In this study, we have first identified the novel sequence type 4564 (ST4564) in one CRKP strain, co-carrying NDM-, CTX-M, mcr-1, and used NGS to explore its resistance and virulence characteristics. In terms of causative pathogens, the emerging of CRKP poses a huge life-threatening to public health.6 Certain STs of KP clones are more likely to cause outbreak infections.2 These clones should be timely identified, monitored and controlled. Thus, our finding enriches the available sequence types library of CRKP clones, and the molecular characteristics provide a strong microbiological basis for further studies of infections caused by CRKP clones.
Different clones have diverse resistance mechanisms. As we all know, KPC-2 enzyme is the most prevalent in China.22 NDM enzymes belong to Metallo-ß-Lactamases (MBL), it has become the epidemic resistance mechanism, since it was first found.21 NDM-1 is the main carbapenemase-producing resistance in several regions, such as India, Pakistan and Sri Lanka.8 A report from a longitudinal large-scale CRE study in China (2012–2016) showed that the isolated CRKP strains in Northwestern China produced a high level of NDM enzymes.23 Moreover, one report from our local region showed that NDM-1 enzyme was the major resistance mechanism and NDM-1 enzyme was the predominant resistance mechanism.21 To date, more than 21 different NDM variants have been observed worldwide.24,25 These various resistance mechanisms of CRKP are attributed to the diversity of geographic location.
Furthermore, ST258 and ST512 were reported in KPC-producing Klebsiella pneumoniae strains. A report showed that blaKPC is the common gene and its rapid dissemination has typically been caused by the clonal expansion of clonal complexes (CCs) 258 strains, including ST258 and ST512.26 ST15 was reported in CTX-M-15-producing clones, and ST11 was reported in KP carried different carbapenemases.12 In our study, ST4564 clone co-produced NDM-1 and CTX-M-9 variants and displayed MDR phenotype. It exhibited high MIC values against carbapenems, cephalosporins, β-lactam, quinolones and CHL, whereas was susceptible to AMK, TGC and COL. This was in consistent with an epidemiology study from China, which showed that 66.9% of the KP was resistant to CIP.27 At the same time, our results were in agreement with another study from Ohio.28 NDM-producing CRKP strains had broad hydrolytic properties, especially for most ß-lactam antibiotics, except for monobactams.8 However, MBL resistance is frequently associated with MDR, with MBL-producing isolates often co-expressing ESBLs, which inactivate monobactams.8 Our result was in line with this, CRKP102 ST4564 was a multi-drug resistance strain and was resistant to ATM. To in-depth studying the feature of its antimicrobial sensitivity, we compared the MIC values with other CRKP strains (three NDM-1-producing ST11 CRKP and one co-producing KPC-2 and CTX-M-14 ST11 CRKP clones). These strains were identified as CRKP strains from our archived clones’ collection and were not included in this study. Interestingly, except the co-producing NDM-1, CTX-M-9 variants and mcr-1 ST4564 clone had the lowest MICs of CIP and LEV, there were no obvious differences among them. However, due to the limited clones, the specific antimicrobial sensitivity of co-producing NDM-1, CTX-M-9 variants and mcr-1 ST4564 CRKP102 clone remained unclear. Further researches are still needed. Moreover, the studies about CRKP co-carrying blaNDM-1 and blaCTX-M were rare.
According to the NGS, a series of resistance genes were detected. ST4564 harbored antibiotic resistance genes to β-lactams, aminoglycosides, quinolones, macrolides, and sulfonamide. Also, we detected aad genes, multidrug resistance efflux pumps [acrA, acrB] genes, which were related to MDR phenotype.12 This was consistent with our microbiological results. In our study, ST4564 harbored two variants of blaCTX-M, which could confer resistance to third-generation cephalosporins as reported. Moreover, blaCTX-M also could confer resistance to monobactams, on the contrary, our result was in disagreement with it. We also detected oqxAB gene, another report from China demonstrated that oqxAB gene was detected in all the KP strains.28 oqxAB only can confer low-level resistance to quinolones; thus, the strain harbored oqxAB genes will still show susceptibility to quinolones.29 Our result also supported it. The ST4564 clone was susceptible to quinolones. In addition to the aforementioned resistance genes, surprisingly, the mcr-1 gene was first detected in our hospital. Regrettably, the NGS did not detect the mcr-1 gene and other resistance genes associated with colistin resistance. At the same time, CRKP102 was susceptible to colistin. It could be demonstrated that resistance phenotypes might involve complex resistance mechanisms and it still remains a challenge for genotypic detection and, consequently, phenotypic prediction.22 Consequently, more accurate sequencing technologies were needed. Furthermore, the acquisition of resistance profiles could make bacteria resistant to antibiotics frequently used within the medical therapies, thereby increasing the difficulty of clinical treatments.30 Due to these multiple resistance profiles, we speculate that novel ST4564 is associated with multi-drug resistance. In light of the dire situation, early monitoring of CRKP infection is urgent.
Apart from resistance characteristics, we found multiple virulence factors to evaluate the pathogenicity of ST4564. It was positive for the string test while negative for the PCR test of typical virulence genes. Interestingly, ST4564 was not detected rmpA or rmpA2 genes, which were related to hypermucoviscous.12 The presence in ST4564 strain of type-I fimbriae, encoded by the fimD and fimH, seems to be linked to down-regulation in biofilm cells, while these genes did not affect biofilm formation.31 This finding could state that the ST4564 clone was positive for string test without type III fimbriae because type-III fimbriae genes were expressed during the CRKP colonization.32 Another feature of the virulence factor was capsular serotypes. In ST4564, we found rcsAB. By regulating the capsule synthesis A and B genes (rcsA, rcsB), capsule production could be enhanced.31 To date, nearly 78 KP capsular serotypes were found. Among them, K1 and K2 capsular serotypes were most notorious and they were always associated with hvKP.6 However, none of the capsular serotypes was detected in ST4564. From other researches, the CRKP clones of the same capsular type with different STs were reported, such as: K1 was reported in ST23 clones, K2 was reported in ST11 clones, K149 was reported in ST307 clones; K47/K64 with ST11.6,12 Thus, virulence factors did not always singularly determine the hvKP clone, which required further investigation and identification. In addition, it is well known that antibiotic resistance genes and virulence genes could undergo the horizontal gene transfer in highly disseminating different clones.8,16,33 Thus, it is necessary to further investigate the epidemic resistance and virulence plasmids and other possible mobile genetic elements.
Up to now, more than 5000 sequence types have been discovered. To our knowledge, the dissemination of CRKP is mostly clonal, and the type of clones is geographically specific.6 ST258, ST11, ST521, and ST101 are detected worldwide and considered as the main high-risk and epidemic clones.2,13 ST11 is the predominant type in China.9 Compared the loci of housekeeping genes of ST4564 with ST11 and ST258, we found that the new clone was not related to any of them. Previous studies showed that KPC-producing KP strains were belonging to the CC258, which was the dominant clonal complex.22 In our study, ST4564 was belonging to CC1571. This was not associated with the high epidemic clonal complexes, such as CC258.
There were several limitations in this study. First, this study was explored at a single hospital. Because of that, our finding suggested that the infection control measures for CRKP infections in this region are urgently needed. Second, the total number of CRKP strain was small. The genetic characteristic of new clones was still unknown. Thus, the timely infection controls for STs, especially for new ST, were more important. Third, unfortunately, due to the NGS data were based on short-read sequencing, the specific information of plasmid content and whether resistance genes are chromosomally located or not was unclear. Thus, further experiments such as conjugation and transformation test need to be performed.
In conclusion, our study was the first research in-depth conducted MLST study of CRKP clinical clones in our local hospital. Our study showed that analysing the genetic features of a novel sequence type, ST4564, in relation to their resistance and pathogenicity may be helpful for outbreak surveillance in hospital settings. Our findings suggest the urgent need for infection control of the new clone to prevent it from becoming a high-risk clone of CRKP. Furthermore, co-producing NDM-1, CTX-M-9 variants and mcr-1 ST4564 clone should take more attention to its resistance.
Ethical Approval
This study was reviewed and approved by the research ethics committee of the Second Hospital of Shanxi Medical University (2019 YX-181). The data of patients’ clinical variables were collected from their medical records and did not contain name, address, or other personal information. The patients’ written informed consent was exempt. This study was also in line with the guidelines outlined in the Declaration of Helsinki.
Acknowledgments
We thank the Department of Pharmacy, Second Hospital of Shanxi Medical University, for supporting this research. We also thank the Institute Pasteur MLST (Paris, France) for determining alleles and profiles at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html.
Funding
This work was supported by the Shanxi Province Natural Science Foundation (grant number 201803D31124). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chung The HKA, Pham Thanh D, et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med. 2015;7(3):227–239. doi:10.15252/emmm.201404767
2. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
3. Cejas D, Elena A, Guevara Nunez D, et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: emergence of hypermucoviscous ST25 and high-risk clone ST307. J Glob Antimicrob Resist. 2019;18:238–242. doi:10.1016/j.jgar.2019.06.005
4. Espinal P, Nucleo E, Caltagirone M, et al. Genomics of Klebsiella pneumoniae ST16 producing NDM-1, CTX-M-15, and OXA-232. Clin Microbiol Infect. 2019;25(3):385e381–385 e385. doi:10.1016/j.cmi.2018.11.004
5. Mathers AJ, Peirano G, Pitout JDD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi:10.1128/CMR.00116-14
6. Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
7. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. E Bio Medicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
8. Beatriz. S-G. Pérez-Gracia maría teresa. enterobacteriaceaepresent and future of Carbapenem-resistant (CRE) infections. Antibiotics. 2019;83:3. doi:10.3390/antibiotics8030122
9. D DN G, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
10. Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous klebsiella pneumoniae st11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7. doi:10.3389/fcimb.2017.00182
11. Hernández-García M, Pérez-Viso B, León-Sampedro R, et al. Outbreak of NDM-1+CTX-M-15+DHA-1-producing Klebsiella pneumoniae high-risk clone in Spain owing to an undetectable colonised patient from Pakistan. Int J Antimicrob Agents. 2019;54(2):233–239. doi:10.1016/j.ijantimicag.2019.05.021
12. Fursova NK, Astashkin EI, Gabrielyan NI, et al. Emergence of five genetic lines ST395(NDM-1), ST13(OXA-48), ST3346(OXA-48), ST39(CTX-M-14), and Novel ST3551(OXA-48) of multidrug-resistant clinical klebsiella pneumoniae in Russia. Microb Drug Resist. 2020;26(8):924–933. doi:10.1089/mdr.2019.0289
13. Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JW. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.00542.
14. Wang X, Xu X, Li Z, et al. An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb Drug Resist. 2014;20(2):144–149. doi:10.1089/mdr.2013.0100
15. CaLS I. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
16. Tsakris A, Poulou A, Pournaras S, et al. A simple phenotypic method for the differentiation of metallo-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrobial Chemother. 2010;65(8):1664–1671. doi:10.1093/jac/dkq210
17. EUCAST Clinical Breakpoint Tables. European Committee on Antimicrobial Susceptibility Testing (EUCAST). Available from: http://www.eucast.org.
18. Politi L, Gartzonika K, Spanakis N, et al. Emergence of NDM-1-producing Klebsiella pneumoniae in Greece: evidence of a widespread clonal outbreak. J Antimicrobial Chemother. 2019;74(8):2197–2202. doi:10.1093/jac/dkz176
19. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
20. Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infection. 2015;71(5):553–560. doi:10.1016/j.jinf.2015.07.010
21. Qi L, Jiaying Z, Jianbang K, et al. Emergence of NDM-5-producing carbapenem-resistant klebsiella pneumoniae and sim-producing hypervirulent klebsiella pneumoniae isolated from aseptic body fluid in a large tertiary hospital, 2017–2018: genetic Traits of blaNDM-Like and blaSIM-like genes as determined by NGS. Infect Drug Resist. 2020;13:3075–3089. doi:10.2147/IDR.S261117
22. Mendes RE, Jones RN, Woosley LN, Cattoir V, Castanheira M. Application of next-generation sequencing for characterization of surveillance and clinical trial isolates: analysis of the distribution of β-lactamase resistance genes and lineage background in the United States. Open Forum Infectious Diseases. 2019;6(Supplement_1):S69–S78. doi:10.1093/ofid/ofz004
23. Wang QWX, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
24. Liu Y, Wan LG, Deng Q, Cao XW, Yu Y, Xu QF. First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol Infect. 2014;143(2):376–384. doi:10.1017/S0950268814000995
25. Kong Z, Cai R, Cheng C, et al. First reported nosocomial outbreak of NDM-5-producing Klebsiella pneumoniae in a neonatal unit In China. Infect Drug Resist. 2019;12:3557–3566. doi:10.2147/IDR.S218945
26. Fasciana T, Gentile B, Aquilina M, et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect Dis. 2019;19:1. doi:10.1186/s12879-019-4565-3
27. Perez F, Rudin SD, Marshall SH, et al. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob Agents Chemother. 2013;57(9):4602–4603. doi:10.1128/AAC.00725-13
28. Yuan J, Xu X, Guo Q, et al. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrobial Chemother. 2012;67(7):1655–1659. doi:10.1093/jac/dks086
29. J ZH L, Ning J, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. 2019;8:44. doi:10.1186/s13756-019-0489-3
30. Zhou K, Lokate M, Deurenberg RH, et al. Characterization of a CTX-M-15 producing klebsiella pneumoniae outbreak strain assigned to a novel sequence type (1427). Front Microbiol. 2015;6. doi:10.3389/fmicb.2015.01250
31. Paczosa MKMJ. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
32. Gona F, Bongiorno D, Aprile A, et al. Emergence of two novel sequence types (3366 and 3367) NDM-1- and OXA-48-co-producing K. pneumoniae in Italy. European J Clin Microbiol Infectious Diseases. 2019;38(9):1687–1691. doi:10.1007/s10096-019-03597-w
33. Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17:1. doi:10.1186/s12866-017-1012-8
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.