Back to Journals » Cancer Management and Research » Volume 14
CMTM Family and Gastrointestinal Tract Cancers: A Comprehensive Review
Authors Li J, Wang X, Wang X, Liu Y, Zheng N, Xu P, Zhang X, Xue L
Received 18 January 2022
Accepted for publication 26 March 2022
Published 26 April 2022 Volume 2022:14 Pages 1551—1563
DOI https://doi.org/10.2147/CMAR.S358963
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Jie Li,1 Xiaozi Wang,2 Xiaoning Wang,1 Yan Liu,1 Na Zheng,1 Pengwei Xu,2 Xianghong Zhang,2 Liying Xue2
1Department of Hematology, Hebei General Hospital, Shijiazhuang, 050000, People’s Republic of China; 2Laboratory of Pathology, Hebei Medical University, Shijiazhuang, 050000, People’s Republic of China
Correspondence: Liying Xue, Laboratory of Pathology, Hebei Medical University, No. 361, Zhongshan Eastern Road, Shijiazhuang, 050000, People’s Republic of China, Tel +86 311 86265561, Email [email protected]
Abstract: Gastrointestinal tract cancers are a highly heterogeneous group of malignant diseases, contributing significantly to the burden of death worldwide. Chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTMs) plays important roles in cancer development and progression. Since the first member was cloned, there have been abundant studies on the relationships between the CMTM family and human cancers. It has been reported that the CMTM family has a large potential prognostic value for multiple cancers. Meanwhile, upregulated or downregulated expression of the family members was related to advanced tumor stage, metastasis, and overall survival. Studies have also reported that these proteins play critical roles in antitumor immunity. We performed a systematic review to sum up the latest advances of CMTM family’ roles in gastrointestinal tract cancers, with a primary focus on hepatocellular carcinoma and gastric carcinoma.
Keywords: CMTM family, gastrointestinal tract cancers, MARVEL, hepatocellular carcinoma, gastric carcinoma
Introduction
Gastrointestinal tract cancers are a highly heterogeneous group of malignant diseases, including esophageal carcinoma (EC), gastric carcinoma (GC), colorectal cancer (CRC), hepatocellular carcinoma (HCC), and pancreatic cancer (PC). These cancers contribute significantly to the burden of death worldwide, especially for CRC, with more than 1.9 million new cases estimated in the Global Cancer (GLOBOCAN) 2020 statistics and ranking third for incidence and second for mortality globally.1 Each gastrointestinal tract tumor has distinct biological characteristics, such as cellular proliferation, disruption of cell metabolism, invasion, metastasis, promotion of angiogenesis, unique follow-up therapy, and prognosis. Although the pathogenesis of these cancers has been extensively studied, much remains unclear and further studies related to cancer-associated molecules are needed.
The chemokine-like factor super family (CKLFSF) consists of nine members: CKLF and CKLFSF 1–8, and their encoding proteins are structurally similar to chemokines.2 CKLF1 was first cloned from a leukemia cell line stimulated by phytohemagglutinin (PHA) and reported by the Human Disease Gene Research Center of Peking University in 2001.3 Subsequently, CKLFSF 1–8 were demonstrated through analysis combining CKLF2 cDNA and protein sequence analysis with experimental verification in 2003. Owing to the presence of a MARVEL (MAL and related proteins for vesical trafficking and membrane link) domain, CKLFSF1-8 was renamed CKLF-like MARVEL transmembrane domain containing 1–8 (CMTM1-8).
Since the first member was cloned, there have been abundant studies on the relationships between the CMTM family of proteins and human cancers. It has been reported that the CMTM family has a large potential prognostic value for multiple cancers owing to its differential expression between tumor and normal tissues.4–8 Meanwhile, upregulated or downregulated expression of these family proteins was related to advanced tumor stage, tumor grade, metastasis, and overall survival. Studies have also reported that these proteins play critical roles in antitumor immunity.9,10 For example, CMTM4/6 reduces PD-L1 ubiquitination, increases protein half-life, and enhances the ability of PD-L1-expressing tumor cells to inhibit T cells. These findings indicate that some CMTM family members are potential therapeutic targets for the treatment of human cancer. Owing to the diverse structure and function of CMTM family members and obvious heterogeneity of different human cancers, CMTM1-8 has distinct effects on malignant tumors, oncogenes, and tumor suppressors. This review briefly outlines the structure and function of CMTM1-8 in human cancers and details the latest advances in the field regarding its roles in carcinogenesis and its potential clinical value, with a primary focus on gastrointestinal tract cancers.
Characteristics of CMTM Family
CMTM members are located on different chromosomes: CMTM1-4 cluster on 16q22.1, CMTM5 cluster on 14q11.2, and CMTM6-8 cluster on 3p22.3.2 The CMTM family is structurally and functionally characterized, similar to classic chemokines and transmembrane-4 superfamily (TM4SF). Among these, CMTM1 contains a C-C motif and shows higher sequence identity with chemokines, while CMTM8 has 39.3% amino acid similarity with TM4SF11,2 and the biological characteristics of CMTM2-7 are between those of chemokines and TM4SF.2,11–13 As a recently discovered gene family, the transcription of CMTM members and their quaternary structures has not been identified. Individual CMTM family members have different alternative RNA splicing forms and further make up several isoforms. We summarized the MARVEL domain of each CMTM1-8 in Figure 1 based on the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) and GeneCards, the human gene database (https://www.genecards.org/).
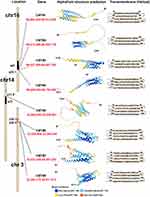 |
Figure 1 CMTM family members structure according to AlphaFold Protein Structure Database and GeneCards. |
CMTM1-8 each has a MARVEL domain, a conserved M-shaped topology: four transmembrane-helix region architectures with cytoplasmic N- and C-terminal regions (Figure 1). Thus, their functional production could be related to the cholesterol-rich membrane apposition events in various biological processes, such as the biogenesis of vesicular transport carriers or tight junction regulation.13 As MARVEL was initially identified in proteins of the myelin and lymphocyte (MAL), physins, gyrins, and occludin families, the MARVEL domain seems to be related to complex diseases, such as inflammation and schizophrenia.13 To date, CMTM1, 2, 3, and 4 are highly expressed in the testis and compartments of the bone marrow and peripheral blood cells, such as activated peripheral blood monocytes,14–16 while CMTM3, 5, 7, and 8 are broadly expressed in healthy adult and fetal tissues.8,17 Therefore, CMTM1-8 appears to be involved in various human diseases, including atopic dermatitis,18 autoimmune,11 cerebral ischemia,19–22 peripheral neuropathy,23 cardiovascular diseases,24 and spermatogenesis dysfunction.16,25 We reviewed the relevant literature, and the research advances on the CMTM family and human cancers are summarized in Table 1.
 |
Table 1 CMTM Family in Human Cancers |
Several biological processes, such as DNA methylation and microRNAs, as well as transcriptional regulation molecules, such as NF-κB, p53, and SOX10, regulate the expression of CMTM family members.26,27 DNA methylation is another important regulator of CMTM family member expression. For example, CpG methylation can inactivate the CMTM5 gene in various carcinoma cell lines, such as oral squamous cell carcinoma,28 breast carcinoma,29 and myeloid leukemia.30 Frequent CMTM7 promoter methylation was detected in ESCC and NPC cell lines with downregulated or silenced CMTM7, but not in other tumor cell lines. Most notably, methylation was not detected in several cancer cell lines (including KYSE180), in which CMTM7 was barely observable. This suggests that other genetic alterations or histone modifications may also be responsible for the downregulation of CMTM7.17
In terms of transcriptional regulation molecules, the promoter sequence of CMTM7 contains numerous regions that are highly similar to the HMG-box sequence. In line with this, SOX gene family members containing a DNA-binding domain have been reported to regulate cell differentiation and tissue formation as a highly conserved transcription factor. According to the TCGA database, a positive correlation between CMTM7 and SOX10 mRNA expression levels was detected. Additionally, knockdown of SOX10 by transfection with siRNA significantly downregulated CMTM7 expression at the mRNA and protein levels in GC cell lines.31 Thus, SOX10 may also be a potential regulator of the expression of CMTM family members. In another study, Guan et al showed that miR-10b-3p, which plays an oncogenic role, was dramatically upregulated in HCC cell lines (HepG2), and the expression of CMTM5 was significantly suppressed.32
CMTM Family Members in Digestive System Cancers
Esophageal Cancer (EC)
EC is one of the most common malignant diseases, with approximately 604,000 new cases and 544,000 deaths worldwide per year.1 Unlike other gastrointestinal tract cancers, EC has two common histologic subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), which have different etiologies and geographic variations. A recent study demonstrated that the expressions of CMTM3 and CMTM4 at the mRNA and protein levels were significantly decreased in ESCC tissues compared to those in matched non-tumor tissues.46 Meanwhile, CMTM3 expression was significantly correlated with lymph node metastasis and clinical stage in ESCC, and lower CMTM3 expression correlated with shorter survival time for ESCC patients. This indicates that the expression of CMTM3 in resected tumors may be an effective prognostic biomarker.46 CMTM5-v1 was reported to be reduced or silenced in 12 of 16 ESCC cell lines,29 while CMTM7 is frequently silenced or downregulated in eight of 18 ESCC cell lines (44.4%), such as EC109, KYSE410, and KYSE180.17 Meanwhile, promoter methylation of CMTM5 and CMTM7 was also detected in esophageal tumor tissues and cell lines.17,29 Overexpression of CMTM7 by adenovirus infection promoted the internalization of epidermal growth factor receptor (EGFR) and further downregulated p-AKT.17 CMTM7 also inhibited cell proliferation and motility in KYSE410 and KYSE180 cells by inducing G1/S cell cycle arrest by upregulating p27 and downregulating cyclin-dependent kinase 2 (CDK2) and 6 (CDK6). Interestingly, MAL proteins, which are essential components of the cellular polarized sorting system, have been reported to suppress cell invasion and tumorigenicity of cervical squamous cell cancers, suggesting that it may be a candidate tumor suppressor gene.104 It is possible that CMTM3, 5, and 7 act as novel functional critical tumor-suppressor genes in ESCC.17 At present, there are no studies on CMTM1-8 in EAC.
Gastric Carcinoma (GC)
GC remains a critical cancer worldwide and is responsible for over 1,089,103 new cases and an estimated 768,793 deaths in 2020, ranking fifth for incidence and fourth for mortality globally.1 Although GC is generally reported as an entity, it can be divided into two topographical subsites: cardia adenocarcinoma and non-cardia adenocarcinoma. The two entities differ in their risk factors, carcinogenesis, and epidemiologic patterns.47–50 Recently, an analysis using bioinformatics methods showed that the mRNA levels of CMTM1, 3, 6, 7, and 8 were upregulated in GC, but CMTM2, 4, and 5 were not significantly different between GC samples and normal tissue. Additionally, the mRNA expression of CMTM family members exhibited strong relationships with various clinical characteristics of patients, such as tumor stage, metastatic lymph node status, and H. pylori status. For example, elevated CMTM3 and 5 mRNA levels were significantly associated with poor overall survival, while upregulated mRNA expression of CMTM2, 4, and 6 was significantly associated with better overall survival. These results indicate that the CMTM family expression pattern could be a novel prognostic factor for patients with GC.35
In contrast, CMTM3 was reported as a tumor suppressor gene in GC patients with different tumor stages using immunohistochemistry (IHC). High expression of CMTM3 might correlate with favorable prognosis of patients, and CMTM3 expression significantly affected the migration and invasion of gastric cell lines (AGS and SGC-7901 cells). CMTM3 not only inhibited EGF-mediated tumorigenicity by inducing Rab5 activity, but also suppressed GC metastasis via the STAT3/Twist1/EMT pathway.5,42,43 Meanwhile, patients with diffuse-type GC and higher CMTM2 expression had a better overall survival, indicating that CMTM2 expression could predict the prognostic outcomes in diffuse-type GC, but not in intestinal-type GC.105 CMTM5-v1 was reduced or silenced in eight of 10 GC cell lines.29
CMTM6, 7, and 8 are broadly expressed in normal human epithelial cell lines. A recent study showed that GC patients with high CMTM6 expression had shorter overall survival (OS),82 and the co-expression of CMTM6 and PD-L1 may be used to judge the prognosis of GC patients.83 Although only one of 16 GC lines (6.25%) showed CMTM7 downregulated expression, the downregulation of CMTM7 was detected in GC tissues using tissue microarrays.17 Interestingly, silencing CMTM7 expression could increase the proliferation and promote tumorigenesis of GC cells, while the overexpression of SOX10-dependent CMTM7 significantly inhibited the tumor growth of GC.31 Contrary to the bioinformatics results of the Oncomine database,35 IHC results showed that the positive rate of CMTM8 protein expression was significantly decreased in GC samples compared to that in adjacent non-tumor tissues, and its expression was correlated with tumor differentiation and node metastasis stage.100 The reason for this discrepancy may be that there are some differences between mRNA and protein expression of CMTM8 regarding posttranscriptional regulation. CMTM8 overexpression significantly decreased the proliferation of the gastric cell line BGC823 and the expression of EGFR.43 In addition, multiple gene set enrichment analyses (GSEA) showed that CMTM8 could regulate the Ca2+ signaling pathway, cell adhesion molecules, and interaction of cytokines and their receptors in GC progression.106 Therefore, CMTM3, 7, and 8 tend to be regarded as tumor suppressors in the development of GC.
Hepatocellular Carcinoma (HCC)
Primary HCC is the sixth most commonly diagnosed malignancy and the third leading cause of cancer-related deaths worldwide, with 905,677 new cases and 830,180 deaths in 2020.1 The liver, one of the most common hematogenous metastasis organs of gastrointestinal malignancies, has attracted more interest from the community and research on CMTMs. Recent studies have revealed that CMTMs are closely related to HCC, and the members might represent promising targets for HCC diagnosis and treatment. Based on the Human Protein Atlas and bioinformatics analysis, a higher level of CMTM1 mRNA expression was associated with a lower survival probability in patients with HCC.34 Although CMTM2 is not a prognostic index in HCC according to data from the Human Protein Atlas, several studies have revealed that CMTM2 expression is correlated with HCC pathological grades.38 In addition, knockdown of CMTM2 promoted invasion and migration of HCC cells (Huh-7 and SMMC7721) by inducing the epithelial-mesenchymal transition (EMT) process.39 The Human Protein Atlas showed that CMTM3 expression was higher in liver tumor tissues and was associated with poor prognosis.34 However, Li et al reported that the expression of CMTM3 was lower in HCC cell lines (HepG2, 97H, Hep3B, and HCCLM3), and CMTM3 inhibited the proliferation and metastasis of HCC cells by suppressing the JAK2/STAT3 signaling pathway.40 Moreover, Zhao et al suggested that CMTM3 may participate in regulating the tumor microenvironment in an orthotopic HCC mouse model.107
A case-control study in the southern Chinese population showed a strong correlation between rs3811178 in CMTM5 and the risk of HCC.108 Compared with the paired adjacent non-tumor tissues, CMTM5 was significantly reduced in HCC tissues, as well as in Huh7, Hep3B, HepG2, and SMMC-7721 cell lines. CMTM5 overexpression significantly inhibited cell proliferation and metastasis in Huh7 cells by inhibiting PI3K/AKT signaling.65 In addition, a previous study demonstrated that CMTM5 expression could be restored in HCC treated with PXD101, a histone deacetylase inhibitor.109 Thus, CMTM5 may be a valuable therapeutic target for HCC treatment.
As for the CMTM4 expression in HCC, there are two inconsistent opinions. One study reported a lower CMTM4 protein expression level in HCC tissues,57 while another two studies showed upregulation of CMTM4 at the mRNA and protein levels.58,59 Furthermore, the upregulated CMTM4 expression in HCC facilitates escaping from antitumor T-cell immunity and contributes to an immunosuppressive tumor microenvironment.59 Similar to CMTM4, CMTM6 expression was downregulated78 or upregulated80,110 in HCC tissues. The reasons for the contradiction between these studies are still ambiguous; however, small-scale specimens, heterogeneity of tumor tissue, and observational bias are possible factors. Interestingly, immunoreactive cellular distribution showed positive CMTM6 expression in all proliferative lesions from the early stage in the piperonyl butoxide (PBO)-induced hepatocarcinogenesis mouse model.111 Meanwhile, CMTM6 had a high concordance ratio with CK8/18+ foci, a useful immunohistochemical marker for detecting hepatocellular proliferative lesions in mice.112 Individuals from Guangxi, China, with the rs164207 AA genotype of CMTM6, have a higher risk of HCC than those with the CC genotype.108 These results suggest that CMTM6 may play important roles in the early stages of carcinogenesis and could be used as a detection marker for hepatocellular proliferative lesions.111
Recent studies have shown that CMTM6 expression is upregulated in HCC tissues, and its overexpression increases cell proliferation and enhances cell invasion and migration and induces epithelial–mesenchymal transition (EMT) mechanistically by stabilizing vimentin.80 In addition, the co-expression of CMTM6/PD-L1 can regulate inflammatory cell density77 and has a close relationship with tumor recurrence.81 Given the participation of CMTM6 in PD-L1 stabilization,9,10 combined treatment with anti-CMTM6 and anti-PD-L1 may be a new method to enhance the therapeutic benefits in HCC.
The expression of CMTM7 was significantly reduced in liver cancer tissues, but was not correlated with TNM stage or metastasis. Overexpression of CMTM7 inhibited the proliferation and migration of HCC cell lines (SK-HEP-1). Thus, CMTM7 functions as a tumor suppressor in liver cancer by suppressing cell cycle progression.96
Pancreatic Cancer (PC)
PC is generally regarded as one of the most lethal malignancies, with a median overall survival of 6 months and a 5-year survival rate of less than 5%.102 The 2020 GLOBOCAN statistics showed that in 185 countries pancreatic cancer accounts for almost as many deaths (466,003) as cases (495,773) due to its poor prognosis.1 Using a pancreatic cancer tissue microarray, CMTM5 was significantly decreased in pancreatic cancer tissues and was directly correlated with the differentiation status of tumors.74 Restoration of CMTM5-v1 not only induced apoptosis of pancreatic cancer cells (MIA PaCa-2), but also had synergistic effects with TNF-A.74 Pancreatic stellate cell (PSC)-derived exosomes, miR-5703, promoted cell proliferation, and its inhibitor suppressed the function of exosomes. Furthermore, CMTM4 was downregulated by miR-5703 directly bound to the CMTM4 3ʹUTR, and CMTM4 knockdown promoted cell proliferation owing to the PAK4-activated PI3K/AKT pathway in pancreatic cancer cells.113 A recent publication reported that CMTM8 protein was co-localized with lysophosphatidic acid (LPA)-1 in pancreatic cancer cells and markedly increased after LPA treatment. However, CMTM8 mRNA abundance showed no significant change in BxPC-3 and PANC-1 cells treated with LPA, implying that LPA treatment stabilizes CMTM8 protein. Meanwhile, depletion of CMTM8 knockdown significantly inhibited the migration and invasion of pancreatic cancer cells in vivo.114 To date, with the exception of CMTM4, 5, and 8, no reports on the role of the other CMTM family members in pancreatic cancer are available. Further studies are needed to elucidate the underlying mechanisms of CMTMs in the development of pancreatic cancer.
Colorectal Cancer (CRC)
CRC ranks third in terms of incidence, but second in terms of mortality worldwide. In 2020, more than 1.9 million new CRCs were estimated to occur.1 However, after a large number of studies were reviewed, only a few studies have focused on the roles of CMTMs in CRC. CMTM3 is silenced or downregulated in colon cell lines and primary tumors, consistent with the results from the Cancer Genome Atlas (TCGA) database,44,115 while re-expression of CMTM3 can inhibit cell growth and cause apoptosis in human cancer cells.44,116 Xue et al found that CMTM4 was also frequently reduced in CRC, and CMTM4 overexpression was associated with increased overall survival rates. Overexpression of CMTM4 inhibited cell proliferation and migration in SW480 cells, and knockdown of CMTM4 showed the opposite effects in HT-29 cells.60 The phosphorylation levels of cell signaling molecules essential for CRC progression, including AKT, ERK1/2, and STAT3, were decreased by CMTM4 overexpression in SW480 cells and increased by CMTM4 knockdown in HT-29 cells.60 CMTM7 was methylated in primary esophageal carcinomas and GC, but not in any of the 11 colon tumor tissues.17 Since a protective role for PD-L1 was reported in Nature, the latest research has shown that CMTM6 is crucial in regulating the immune microenvironment in CRC, and the co-expression of CMTM6 and PD-L1 can be used to stratify the risk of progression in CRC patients.75 Meanwhile, the expression status of CMTM6 in M2 macrophages may more accurately predict the drug response to PD-1/PD-L1 inhibitors in CRC patients.85 To date, the roles of other CMTM members in CRC, except for CMTM3, 4, and 6, are largely unknown.
Conclusion
To summarize, CMTM family members have different expression profiles in human cancer samples and normal tissues and have different functions in the development and progression of gastrointestinal tract cancers. Meanwhile, they were reported to be tumor suppressors or tumor promoters owing to their relationships with the regulation of the cell cycle, EMT process, and EGFR signaling pathway (Table 2). The heterogeneity of different tumors and distinct molecular structures of each CMTM member make this family in human cancers controversial. With the exception of CMTM4/6, demonstrated to be PD-L1 regulators and may be potential immunotherapy targets, the mechanism of the other members remains partially understood. Hence, the CMTM family and their functions, mechanisms, and related signaling pathways in gastrointestinal tract cancers need to be further explored as these could provide a new viewpoint for the diagnosis or therapeutic targets of human cancers.
 |
Table 2 The Roles of CMTM Family in Gastrointestinal Tract Cancers |
Statements and Declarations
This study was reviewed and approved by The Institutional Review Board of Hebei Medical University.
Funding
This study was supported by the Natural Science Foundation of Hebei Province (No. H2021206395).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Han W, Ding P, Xu M, et al. of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics. 2003;81(6):609–617. doi:10.1016/s0888-7543(03)00095-8
3. Han W, Lou Y, Tang J, et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J. 2001;357(Pt1):127–135. doi:10.1042/0264-6021:
4. Hou X, He S, Zhang D, Yang C, Shi Y, Zhang K. Expression and clinical significance of CMTM6 in nonsmall cell lung cancer. DNA Cell Biol. 2020;1:343. doi:10.1089/dna.2020.5564
5. Su Y, Lin Y, Zhang L, et al. CMTM3 inhibits cell migration and invasion and correlates with favorable prognosis in gastric cancer. Cancer Sci. 2014;105(1):26–34. doi:10.1111/cas.12304
6. Si J, Zhang P, Tian D, et al. CMTM1_v17 is associated with chemotherapy resistance and poor prognosis in non-small cell lung cancer. World J Surg Oncol. 2017;15(1):34. doi:10.1186/s12957-016-1094-z
7. Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233–243. doi:10.1016/j.ebiom.2018.08.012
8. Zhang S, Pei X, Hu H, et al. Functional characterization of the tumor suppressor CMTM8 and its association with prognosis in bladder cancer. Tumour Biol. 2016;37(5):6217–6225. doi:10.1007/s13277-015-4508-6
9. Burr ML, Sparbier CE, Chan YC, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101–105. doi:10.1038/nature23643
10. Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106–110. doi:10.1038/nature23669
11. Duan HJ, Li XY, Liu C, Deng XL. Chemokine-like factor-like MARVEL transmembrane domain-containing family in autoimmune diseases. Chin Med J. 2020;133(8):951–958. doi:10.1097/CM9.0000000000000747
12. Wu K, Li X, Gu H, Yang Q, Liu Y, Wang L. Research Advances in CKLF-like MARVEL Transmembrane Domain-containing Family in Non-small Cell Lung Cancer. Int J Biol Sci. 2019;15(12):2576–2583. doi:10.7150/ijbs.33733
13. Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27(12):599–601. doi:10.1016/s0968-0004(02)02229-6
14. Zhong J, Wang Y, Qiu X, et al. Characterization and expression profile of CMTM3/CKLFSF3. J Biochem Mol Biol. 2006;39(5):537–545. doi:10.5483/bmbrep.2006.39.5.537
15. Song HS, Shi S, Lu XZ, et al. Intracellular CMTM2 negatively regulates human immunodeficiency virus type-1 transcription through targeting the transcription factors AP-1 and CREB. Chin Med J. 2010;123(17):2440–2445.
16. Wang L, Wu C, Zheng Y, et al. Molecular cloning and characterization of chemokine-like factor super family member 1 (CKLFSF1), a novel human gene with at least 23 alternative splicing isoforms in testis tissue. Int J Biochem Cell Biol. 2004;36(8):1492–1501. doi:10.1016/j.biocel.2003.11.017
17. Li H, Li J, Su Y, et al. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene. 2014;33(24):3109–3118. doi:10.1038/onc.2013.282
18. Yang GY, Chen X, Sun YC, Ma CL, Qian G. Chemokine-like factor 1 (CLFK1) is over-expressed in patients with atopic dermatitis. Int J Biol Sci. 2013;9(8):759–765. doi:10.7150/ijbs.6291
19. Kong LL, Wang ZY, Hu JF, Yuan YH, Li H, Chen NH. Inhibition of chemokine-like factor 1 improves blood-brain barrier dysfunction in rats following focal cerebral ischemia. Neurosci Lett. 2016;627:192–198. doi:10.1016/j.neulet.2016.06.003
20. Kong LL, Wang ZY, Hu JF, et al. Inhibition of chemokine-like factor 1 protects against focal cerebral ischemia through the promotion of energy metabolism and anti-apoptotic effect. Neurochem Int. 2014;76:91–98. doi:10.1016/j.neuint.2014.07.004
21. Kong LL, Hu JF, Zhang W, et al. Expression of chemokine-like factor 1 after focal cerebral ischemia in the rat. Neurosci Lett. 2011;505(1):14–18. doi:10.1016/j.neulet.2011.09.031
22. Kong LL, Wang ZY, Han N, et al. Neutralization of chemokine-like factor 1, a novel C-C chemokine, protects against focal cerebral ischemia by inhibiting neutrophil infiltration via MAPK pathways in rats. J Neuroinflammation. 2014;11:112. doi:10.1186/1742-2094-11-112
23. Eichel MA, Gargareta VI, D’Este E, et al. CMTM6 expressed on the adaxonal Schwann cell surface restricts axonal diameters in peripheral nerves. Nat Commun. 2020;11(1):4514. doi:10.1038/s41467-020-18172-7
24. Zhang JW, Liu TF, Chen XH, et al. Validation of aspirin response-related transcripts in patients with coronary artery disease and preliminary investigation on CMTM5 function. Gene. 2017;624:56–65. doi:10.1016/j.gene.2017.04.041
25. Zhang XW, Dun YJ, Tang X, et al. [Expression of chemokine like factor-like myelin and lymphocyte and related proteins for vesicle trafficking and membrane link transmembrane domain-containing protein 2 in rats with varicocele]. Beijing Da Xue Xue Bao Yi Xue Ban. 2016;48(4):579–583. Chinese.
26. Wu J, Li L, Wu S, Xu B. CMTM family proteins 1-8: roles in cancer biological processes and potential clinical value. Cancer Biol Med. 2020;17(3):528–542. doi:10.20892/j.issn.2095-3941.2020.0032
27. Qian C, Xu Z, Chen L, Wang Y, Yao J. Long noncoding RNA LINC01391 restrained gastric cancer aerobic glycolysis and tumorigenesis via targeting miR-12116/CMTM2 axis. J Cancer. 2020;11(21):6264–6276. doi:10.7150/jca.48365
28. Zhang H, Nan X, Li X, et al. CMTM5 exhibits tumor suppressor activity through promoter methylation in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2014;447(2):304–310. doi:10.1016/j.bbrc.2014.03.158
29. Shao L, Cui Y, Li H, et al. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res. 2007;13(19):5756–5762. doi:10.1158/1078-0432.CCR-06-3082
30. Niu J, Li H, Zhang Y, et al. Aberrant expression of CKLF-like MARVEL transmembrane member 5 (CMTM5) by promoter methylation in myeloid leukemia. Leuk Res. 2011;35(6):771–776. doi:10.1016/j.leukres.2010.11.023
31. Jin Y, Qin X, Jia G. SOX10-dependent CMTM7 expression inhibits cell proliferation and tumor growth in gastric carcinoma. Biochem Biophys Res Commun. 2018;507(1–4):91–99. doi:10.1016/j.bbrc.2018.10.172
32. Guan L, Ji D, Liang N, Li S, Sun B. Up-regulation of miR-10b-3p promotes the progression of hepatocellular carcinoma cells via targeting CMTM5. J Cell Mol Med. 2018;22(7):3434–3441. doi:10.1111/jcmm.13620
33. Wang J, Zhang G, Zhang Y, et al. CMTM1_v17 is a novel potential therapeutic target in breast cancer. Oncol Rep. 2014;32(5):1829–1836. doi:10.3892/or.2014.3429
34. Li M, Luo F, Tian X, Yin S, Zhou L, Zheng S. Chemokine-Like Factor-Like MARVEL Transmembrane Domain-Containing Family in Hepatocellular Carcinoma: latest Advances. Front Oncol. 2020;10:595973. doi:10.3389/fonc.2020.595973
35. Liang Z, Xie J, Huang L, et al. Comprehensive analysis of the prognostic value of the chemokine-like factor-like MARVEL transmembrane domain-containing family in gastric cancer. J Gastrointest Oncol. 2021;12(2):388–406. doi:10.21037/jgo-21-78
36. Delic S, Thuy A, Schulze M, et al. Systematic investigation of CMTM family genes suggests relevance to glioblastoma pathogenesis and CMTM1 and CMTM3 as priority targets. Genes Chromosomes Cancer. 2015;54(7):433–443. doi:10.1002/gcc.22255
37. Cao L, Yang C, Zhu B, et al. A novel protein CMTM1-v5 specifically induced human lymphoma cells apoptosis in vitro and in vivo. Exp Cell Res. 2019;385(1):111623. doi:10.1016/j.yexcr.2019.111623
38. Guo X, Zhang S, Tan S, et al. Downregulated CMTM2 Poses Potential Clinical Significance in Hepatocellular Carcinoma. DNA Cell Biol. 2020;39(4):683–689. doi:10.1089/dna.2019.5237
39. Zhang S, Tian R, Bei C, et al. Down-Regulated CMTM2 Promotes Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:5731–5741. doi:10.2147/OTT.S250370
40. Li W, Zhang S. CKLF-Like MARVEL Transmembrane Domain-Containing Member 3 (CMTM3) Inhibits the Proliferation and Tumorigenisis in Hepatocellular Carcinoma Cells. Oncol Res. 2017;25(2):285–293. doi:10.3727/096504016X14732523471442
41. Lu M, Huang Y, Sun W, Li P, Li L, Li L. miR-135b-5p promotes gastric cancer progression by targeting CMTM3. Int J Oncol. 2018;52(2):589–598. doi:10.3892/ijo.2017.4222
42. Yuan W, Liu B, Wang X, et al. CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 2017;386:77–86. doi:10.1016/j.canlet.2016.11.015
43. Yuan W, Li T, Mo X, et al. Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget. 2016;7(20):29507–29519. doi:10.18632/oncotarget.8789
44. Wang Y, Li J, Cui Y, et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69(12):5194–5201. doi:10.1158/0008-5472.CAN-08-3694
45. Zhou Z, Ma Z, Li Z, et al. CMTM3 Overexpression Predicts Poor Survival and Promotes Proliferation and Migration in Pancreatic Cancer. J Cancer. 2021;12(19):5797–5806. doi:10.7150/jca.57082
46. Han T, Shu T, Dong S, et al. Chemokine-like factor-like MARVEL transmembrane domain-containing 3 expression is associated with a favorable prognosis in esophageal squamous cell carcinoma. Oncol Lett. 2017;13(5):2982–2988. doi:10.3892/ol.2017.5837
47. Liu W, Li J, Zhang D, et al. Trefoil factor 1 and gastrokine 2 inhibit Helicobacter pylori-induced proliferation and inflammation in gastric cardia and distal carcinogenesis. Oncol Lett. 2020;20(6):318. doi:10.3892/ol.2020.12181
48. Xue L, Ouyang Q, Li J, et al. Different roles for p16(INK) (4a) -Rb pathway and INK4a/ARF methylation between adenocarcinomas of gastric cardia and distal stomach. J Gastroenterol Hepatol. 2014;29(7):1418–1426. doi:10.1111/jgh.12547
49. Arnold M, Laversanne M, Brown LM, Devesa SS, Bray F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: international Trends in Incidence up to 2030. Am J Gastroenterol. 2017;112(8):1247–1255. doi:10.1038/ajg.2017.155
50. Sanikini H, Muller DC, Sophiea M, et al. Anthropometric and reproductive factors and risk of esophageal and gastric cancer by subtype and subsite: results from the European prospective investigation into cancer and nutrition (EPIC) cohort. Int J Cancer. 2020;146(4):929–942. doi:10.1002/ijc.32386
51. Xie J, Yuan Y, Liu Z, et al. CMTM3 is frequently reduced in clear cell renal cell carcinoma and exhibits tumor suppressor activities. Clin Transl Oncol. 2014;16(4):402–409. doi:10.1007/s12094-013-1092-3
52. Hu F, Yuan W, Wang X, et al. CMTM3 is reduced in prostate cancer and inhibits migration, invasion and growth of LNCaP cells. Clin Transl Oncol. 2015;17(8):632–639. doi:10.1007/s12094-015-1288-9
53. Hu FZ, Yuan WQ, Wang XL, et al. [Knockdown of CMTM3 promotes migration and invasion of PC3 cell in vitro]. Beijing Da Xue Xue Bao Yi Xue Ban. 2016;48(1):594–597. Chinese.
54. Zhang H, Zhang J, Nan X, et al. CMTM3 inhibits cell growth and migration and predicts favorable survival in oral squamous cell carcinoma. Tumour Biol. 2015;36(10):7849–7858. doi:10.1007/s13277-015-3504-1
55. Abe M, Yamashita S, Mori Y, et al. High-risk oral leukoplakia is associated with aberrant promoter methylation of multiple genes. BMC Cancer. 2016;16:350. doi:10.1186/s12885-016-2371-5
56. Yuan W, Wei F, Ouyang H, et al. CMTM3 suppresses chordoma progress through EGFR/STAT3 regulated EMT and TP53 signaling pathway. Cancer Cell Int. 2021;21(1):510. doi:10.1186/s12935-021-02159-5
57. Bei C, Zhang Y, Wei R, et al. Clinical significance of CMTM4 expression in hepatocellular carcinoma. Onco Targets Ther. 2017;10:5439–5443. doi:10.2147/OTT.S149786
58. Zhou HQ, Li JH, Liu LW, Lou JM, Ren ZG. Increased CMTM4 mRNA expression predicts a poor prognosis in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2020;19(6):596–601. doi:10.1016/j.hbpd.2020.06.004
59. Chui NN, Cheu JW, Yuen VW, et al. Inhibition of CMTM4 Sensitizes Cholangiocarcinoma and Hepatocellular Carcinoma to T Cell-Mediated Antitumor Immunity Through PD-L1. Hepatol Commun. 2021. doi:10.1002/hep4.1682
60. Xue H, Li T, Wang P, et al. CMTM4 inhibits cell proliferation and migration via AKT, ERK1/2, and STAT3 pathway in colorectal cancer. Acta Biochim Biophys Sin. 2019;51(9):915–924. doi:10.1093/abbs/gmz084
61. Li T, Cheng Y, Wang P, et al. CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2015;34:122. doi:10.1186/s13046-015-0236-4
62. Zhao W, Zhao F, Yang K, et al. An immunophenotyping of renal clear cell carcinoma with characteristics and a potential therapeutic target for patients insensitive to immune checkpoint blockade. J Cell Biochem. 2019;120(8):13330–13341. doi:10.1002/jcb.28607
63. Li H, Liu YT, Chen L, et al. CMTM4 regulates epithelial-mesenchymal transition and PD-L1 expression in head and neck squamous cell carcinoma. Mol Carcinog. 2021;60(8):556–566. doi:10.1002/mc.23323
64. Plate M, Li T, Wang Y, et al. Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol Cells. 2010;29(4):355–361. doi:10.1007/s10059-010-0038-7
65. Xu G, Dang C. CMTM5 is downregulated and suppresses tumour growth in hepatocellular carcinoma through regulating PI3K-AKT signalling. Cancer Cell Int. 2017;17:113. doi:10.1186/s12935-017-0485-8
66. Wu J. CMTM5/7 are biomarkers and prognostic factors in human breast carcinoma. Cancer Biomark. 2020;29(1):89–99. doi:10.3233/CBM-191226
67. Zhou J, Lei J, Wang J, et al. Bioinformatics-Based Discovery of CKLF-Like MARVEL Transmembrane Member 5 as a Novel Biomarker for Breast Cancer. Front Cell Dev Biol. 2020;7:361. doi:10.3389/fcell.2019.00361
68. Chen Z, Cui N, Zhao JS, et al. Expressions of ZNF436, β-catenin, EGFR, and CMTM5 in breast cancer and their clinical significances. Eur J Histochem. 2021;65(1):3173. doi:10.4081/ejh.2021.3173
69. Yuan Y, Sheng Z, Liu Z, et al. CMTM5-v1 inhibits cell proliferation and migration by downregulating oncogenic EGFR signaling in prostate cancer cells. J Cancer. 2020;11(13):3762–3770. doi:10.7150/jca.42314
70. Xiao Y, Yuan Y, Zhang Y, et al. CMTM5 is reduced in prostate cancer and inhibits cancer cell growth in vitro and in vivo. Clin Transl Oncol. 2015;17(6):431–437. doi:10.1007/s12094-014-1253-z
71. Cai B, Xiao Y, Li Y, Zheng S. CMTM5 inhibits renal cancer cell growth through inducing cell-cycle arrest and apoptosis. Oncol Lett. 2017;14(2):1536–1542. doi:10.3892/ol.2017.6350
72. Shao L, Guo X, Plate M, et al. CMTM5-v1 induces apoptosis in cervical carcinoma cells. Biochem Biophys Res Commun. 2009;379(4):866–871. doi:10.1016/j.bbrc.2008.12.126
73. Li P, Liu K, Li L, et al. Reduced CMTM5 expression correlates with carcinogenesis in human epithelial ovarian cancer. Int J Gynecol Cancer. 2011;21(7):1248–1255. doi:10.1097/IGC.0b013e3182259c31
74. Guo X, Li T, Wang Y, et al. CMTM5 induces apoptosis of pancreatic cancer cells and has synergistic effects with TNF-alpha. Biochem Biophys Res Commun. 2009;387(1):139–142. doi:10.1016/j.bbrc.2009.06.148
75. Ma Y, Shi JF, Qiu HY, et al. [Pathophysiologic mechanism of CMTM5 low expression in multiple myeloma progression]. Zhonghua Xue Ye Xue Za Zhi. 2019;40(1):58–62. doi:10.3760/cma.j.issn.0253-2727.2019.01.011 Chinese.
76. Niu JH, Bao L, Zhang Y, et al. [Abnormally lower expression of cmtm5 gene in bone marrow cells from patients with multiple myeloma]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18(2):363–367. Chinese.
77. Liu LL, Zhang SW, Chao X, et al. Coexpression of CMTM6 and PD-L1 as a predictor of poor prognosis in macrotrabecular-massive hepatocellular carcinoma. Cancer Immunol Immunother. 2021;70(2):417–429. doi:10.1007/s00262-020-02691-9
78. Zhu X, Qi G, Li C, et al. Expression and Clinical Significance of CMTM6 in Hepatocellular Carcinoma. DNA Cell Biol. 2019;38(2):193–197. doi:10.1089/dna.2018.4513
79. Yugawa K, Itoh S, Yoshizumi T, et al. CMTM6 Stabilizes PD-L1 Expression and Is a New Prognostic Impact Factor in Hepatocellular Carcinoma. Hepatol Commun. 2020;5(2):334–348. doi:10.1002/hep4.1643
80. Huang X, Xiang L, Wang B, et al. CMTM6 promotes migration, invasion, and EMT by interacting with and stabilizing vimentin in hepatocellular carcinoma cells. J Transl Med. 2021;19(1):120. doi:10.1186/s12967-021-02787-5
81. Muranushi R, Araki K, Yokobori T, et al. High membrane expression of CMTM6 in hepatocellular carcinoma is associated with tumor recurrence. Cancer Sci. 2021;112(8):3314–3323. doi:10.1111/cas.15004
82. Li X, Chen L, Gu C, Sun Q, Li J. CMTM6 significantly relates to PD-L1 and predicts the prognosis of gastric cancer patients. PeerJ. 2020;8:e9536. doi:10.7717/peerj.9536
83. Zhang C, Zhao S, Wang X. Co-expression of CMTM6 and PD-L1: a novel prognostic indicator of gastric cancer. Cancer Cell Int. 2021;21(1):78. doi:10.1186/s12935-020-01734-6
84. Peng QH, Wang CH, Chen HM, et al. CMTM6 and PD-L1 coexpression is associated with an active immune microenvironment and a favorable prognosis in colorectal cancer. J Immunother Cancer. 2021;9(2):e001638. doi:10.1136/jitc-2020-001638
85. Wu X, Lan X, Hu W, et al. CMTM6 expression in M2 macrophages is a potential predictor of PD-1/PD-L1 inhibitor response in colorectal cancer. Cancer Immunol Immunother. 2021;70(11):3235–3248. doi:10.1007/s00262-021-02931-6
86. Shang X, Li J, Wang H, et al. CMTM6 is positively correlated with PD-L1 expression and immune cells infiltration in lung squamous carcinoma. Int Immunopharmacol. 2020;88:106864. doi:10.1016/j.intimp.2020.106864
87. Zugazagoitia J, Liu Y, Toki M, et al. Quantitative Assessment of CMTM6 in the Tumor Microenvironment and Association with Response to PD-1 Pathway Blockade in Advanced-Stage Non-Small Cell Lung Cancer. J Thorac Oncol. 2019;14(12):2084–2096. doi:10.1016/j.jtho.2019.09.014
88. Koh YW, Han JH, Haam S, Jung J, Lee HW. Increased CMTM6 can predict the clinical response to PD-1 inhibitors in non-small cell lung cancer patients. Oncoimmunology. 2019;8(10):e1629261. doi:10.1080/2162402X.2019.1629261
89. Wang H, Gao J, Zhang R, Li M, Peng Z, Wang H. Molecular and immune characteristics for lung adenocarcinoma patients with CMTM6 overexpression. Int Immunopharmacol. 2020;83:106478. doi:10.1016/j.intimp.2020.106478
90. Zheng Y, Wang C, Song A, et al. CMTM6 promotes cell proliferation and invasion in oral squamous cell carcinoma by interacting with NRP1. Am J Cancer Res. 2020;10(6):1691–1709.
91. Chen L, Yang QC, Li YC, et al. Targeting CMTM6 Suppresses Stem Cell-Like Properties and Enhances Antitumor Immunity in Head and Neck Squamous Cell Carcinoma. Cancer Immunol Res. 2020;8(2):179–191. doi:10.1158/2326-6066.CIR-19-0394
92. Zhang S, Yan Q, Wei S, et al. CMTM6 and PD-1/PD-L1 overexpression is associated with the clinical characteristics of malignancy in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2021;132(2):202–209. doi:10.1016/j.oooo.2021.02.019
93. Mohapatra P, Shriwas O, Mohanty S, et al. CMTM6 drives cisplatin resistance by regulating Wnt signaling through the ENO-1/AKT/GSK3β axis. JCI Insight. 2021;6(4):e143643. doi:10.1172/jci.insight.143643
94. Pang X, Wang SS, Zhang M, et al. OSCC cell-secreted exosomal CMTM6 induced M2-like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunol Immunother. 2021;70(4):1015–1029. doi:10.1007/s00262-020-02741-2
95. Ishihara S, Iwasaki T, Kohashi K, et al. The association between the expression of PD-L1 and CMTM6 in undifferentiated pleomorphic sarcoma. J Cancer Res Clin Oncol. 2021;147(7):2003–2011. doi:10.1007/s00432-021-03616-4
96. Huang ZM, Li PL, Yang P, et al. Overexpression of CMTM7 inhibits cell growth and migration in liver cancer. Kaohsiung J Med Sci. 2019;35(6):332–340. doi:10.1002/kjm2.12058
97. Liu B, Su Y, Li T, et al. CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget. 2015;6(38):41092–41107. doi:10.18632/oncotarget.5732
98. Liu Q, Su Y, Jiang GC, et al. Change of CMTM7 expression, a potential tumor suppressor, is associated with poor clinical outcome in human non-small cell lung cancer. Chin Med J. 2013;126(16):3006–3012.
99. Zhang W, Mendoza MC, Pei X, et al. Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem. 2012;287(15):11850–11858. doi:10.1074/jbc.M111.258236
100. Yan M, Zhu X, Qiao H, Zhang H, Xie W, Cai J. Downregulated CMTM8 Correlates with Poor Prognosis in Gastric Cancer Patients. DNA Cell Biol. 2021;40(6):791–797. doi:10.1089/dna.2021.0110
101. Gao D, Hu H, Wang Y, et al. CMTM8 inhibits the carcinogenesis and progression of bladder cancer. Oncol Rep. 2015;34(6):2853–2863. doi:10.3892/or.2015.4310
102. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi:10.1056/NEJMra0901557
103. Hu H, Chen JW, Xu KX, et al. [Expressions of CMTM8 and E-cadherin in primary and metastatic clear cell renal cell carcinoma]. Beijing Da Xue Xue Bao Yi Xue Ban. 2013;45(4):537–541. Chinese.
104. Hatta M, Nagai H, Okino K, et al. Down-regulation of members of glycolipid-enriched membrane raft gene family, MAL and BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004;30(1):53–58. doi:10.1111/j.1341-8076.2004.00156.x
105. Choi JH, Kim YB, Ahn JM, et al. Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp Mol Med. 2018;50(4):1–11. doi:10.1038/s12276-017-0009-6
106. Huang Y, Li F, Du Y. Effects of CKLFSF8 on expression of EGFR of BGC823 cell. J Zhengzhou Univ. 2006;41:617–619.
107. Zhao W, Xu Y, Xu J, et al. Subsets of myeloid-derived suppressor cells in hepatocellular carcinoma express chemokines and chemokine receptors differentially. Int Immunopharmacol. 2015;26(2):314–321. doi:10.1016/j.intimp.2015.04.010
108. Bei C, Tan C, Zhu X, Wang Z, Tan S. Association Between Polymorphisms in CMTM Family Genes and Hepatocellular Carcinoma in Guangxi of China. DNA Cell Biol. 2018;37(8):691–696. doi:10.1089/dna.2018.4274
109. Ma BB, Sung F, Tao Q, et al. The preclinical activity of the histone deacetylase inhibitor PXD101 (belinostat) in hepatocellular carcinoma cell lines. Invest New Drugs. 2010;28(2):107–114. doi:10.1007/s10637-009-9219-7
110. Li T, Zhong J, Chen Y, et al. Expression of chemokine-like factor 1 is upregulated during T lymphocyte activation. Life Sci. 2006;79(6):519–524. doi:10.1016/j.lfs.2006.01.042
111. Yafune A, Kawai M, Itahashi M, et al. Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model. Toxicol Lett. 2013;222(3):295–302. doi:10.1016/j.toxlet.2013.08.006
112. Kawai M, Saegusa Y, Kemmochi S, et al. Cytokeratin 8/18 is a useful immunohistochemical marker for hepatocellular proliferative lesions in mice. J Vet Med Sci. 2010;72(3):263–269. doi:10.1292/jvms.09-0329
113. Li M, Guo H, Wang Q, et al. Pancreatic stellate cells derived exosomal miR-5703 promotes pancreatic cancer by downregulating CMTM4 and activating PI3K/Akt pathway. Cancer Lett. 2020;490:20–30. doi:10.1016/j.canlet
114. Shi W, Zhang C, Ning Z, et al. CMTM8 as an LPA1-associated partner mediates lysophosphatidic acid-induced pancreatic cancer metastasis. Ann Transl Med. 2021;9(1):42. doi:10.21037/atm-20-1013
115. Li J, Chen C, Bi X, et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi:10.1016/j.gene.2017.07.082
116. Li Z, Xie J, Wu J, et al. CMTM3 inhibits human testicular cancer cell growth through inducing cell-cycle arrest and apoptosis. PLoS One. 2014;9(2):e88965. doi:10.1371/journal.pone.0088965
117. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-589. doi:10.1038/s41586-021-03819-2
118. Varadi M, Anyango S, Deshpande M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439-D444. doi:10.1093/nar/gkab1061
119. Fishilevich S, Nudel R, Rappaport N, et al. Genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017;2017:bax028. doi:10.1093/database/bax028
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
