Back to Journals » OncoTargets and Therapy » Volume 12
CLK3 Is A Direct Target Of miR-144 And Contributes To Aggressive Progression In Hepatocellular Carcinoma
Authors Li H, Cui X, Hu Q, Chen X, Zhou P
Received 24 July 2019
Accepted for publication 30 September 2019
Published 5 November 2019 Volume 2019:12 Pages 9201—9213
DOI https://doi.org/10.2147/OTT.S224527
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Hua Li,1,* Xichun Cui,2,* Qiuyue Hu,2 Xiaolong Chen,2 Pengli Zhou3
1Department of Infectious Diseases, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 2Key Laboratory of Clinical Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 3Department of Intervention, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450014, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Pengli Zhou
Department of Intervention, The First Affiliated Hospital of Zhengzhou University, 1 Jianshe East Road, Erqi, Zhengzhou 450052, People’s Republic of China
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer with high incidence. The underlying molecular mechanisms of HCC development have been intensively studied. CLK3 (CDC Like Kinase 3) is a nuclear dual-specificity kinase and regulates gene splicing. We investigated the expression profile and functional role of CLK3 in HCC.
Methods: Immunohistochemistry (IHC) and Western blot were performed to determine CLK3 expression in HCC tissues. Bioinformatics analysis using TCGA and GEO database was conducted to evaluate the relationship between CLK3 expression and HCC prognosis. Cell proliferation was assessed by CCK8, EdU and colony formation assays, while transwell and wound-healing assays were performed to investigate the cell migration and invasion in vitro. Xenograft nude mouse model was used to test the function of CLK3 on tumor growth in vivo. Luciferase reporter assay, Western blot and RT-qPCR were conducted to verify the miRNA that directly targeted CLK3.
Results: CLK3 was markedly upregulated in HCC tissues, and the expression levels of CLK3 were closely associated with TNM stages and HCC prognosis. Functional analysis indicated that knockdown of CLK3 could suppress HCC cell growth, invasion and migration in vitro, and inhibit tumor development in vivo. Moreover, CLK3 was demonstrated as a direct target of miR-144 and miR-144 expression was inversely correlated with CLK3 expression in HCC. Enforced overexpression of miR-144 markedly inhibited the CLK3 expression while overexpression of CLK3 partially reversed the inhibitory function of miR-144 on HCC cell growth and metastasis. Mechanistically, we found that miR-144 overexpression inhibited Wnt/β-catenin signaling and the inhibition could be partly abolished by overexpression of CLK3.
Conclusion: In summary, we demonstrate tumor suppressor miR-144 suppresses hepatocellular carcinoma development and metastasis via regulating CLK3 and Wnt/β-catenin signaling, indicating that miR-144/CLK3 could be used for HCC diagnosis and treatment.
Keywords: miR-144, metastasis, HCC, CLK3, Wnt/β-catenin signaling
Introduction
Liver cancer ranks the 6th most common cancer worldwide and is the 2nd most frequent cause of cancer-related death in male and the 6th most frequent in female.1 Though the introduction of novel therapies, such as surgical resection and liver transplantation, has greatly improved the prognosis of hepatocellular carcinoma (HCC) patients, the clinical outcome for advanced HCC remains poor.2 Postoperative metastasis and recurrence remain major problems in HCC patients, which highlights the urgent need to identify new therapeutic targets and improve the prognosis of HCC patients.
CLK3 (CDC Like Kinase 3) is one member of the CDC-like kinases family,3 which has four members: CLK1/STY, CLK2, CLK3 and CLK4.4 It is been demonstrated that alternative splicing could be controlled by CLK3 through phosphorylation of serine/arginine-rich domains on direct splicing factors.5 However, the significance and specific mechanism of CLK3 in cancer development is not fully understood.
Recently, microRNAs (miRNAs) have been utilized as biomarkers for prognosis and diagnosis of tumors.6 MiRNAs are small non-coding RNAs with ~22 nucleotides that could post-transcriptionally regulate protein-coding genes by binding to their 3ʹ-untranslated regions (3-UTRs).7 Accumulating studies have shown that miRNAs are important tumor regulators. They have been emerged as novel therapeutic targets to control the occurrence and progress of cancer.6 MiR-144 has been found to participate in the development of multiple cancers, including HCC.8 However, the potential function of miR-144 in HCC has not been fully documented yet.
Here, we found that CLK3 expression was markedly enhanced in HCC tissues in comparison with that in non-tumor control tissues. CLK3 upregulation was correlated with malignancy and poor prognosis of HCC patients. We demonstrated that CLK3 silencing significantly dampened HCC cell growth, invasion and migration. Moreover, we showed that CLK3 activated Wnt/β-catenin signaling and thereby promoted HCC cell malignant phenotype. Mechanistically, we identified that CLK3 was a novel functional target of miR-144. Overexpression of miR-144 inhibited HCC cell proliferation, invasion and migration. Moreover, miR-144 also suppressed the Wnt/β-catenin signaling via targeting CLK3. In summary, our results suggest that CLK3 may be served as a promising biomarker for prognosis and potential therapeutic target for HCC.
Materials And Methods
Human Specimens
A microarray of 396 HCC and adjacent non-tumor samples were obtained from HCC patients with written informed consent who underwent surgery at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) and constructed as previously described.9 Among these patients with available frozen tumor tissues, we randomly chose 8 HCC patient tissue samples for Western blot to detect CLK3 expression. All patients provided written informed consent. The experiment was conducted in accordance with the Declaration of Helsinki and approved by the First Affiliated Hospital of Zhengzhou University Institutional Review Board.
Dataset Acquisition And Process
Expression data for CLK3 in HCC and additional cancer types were extracted from level 3 TCGA (The Cancer Genome Atlas) RNAseq data, and further validated by publicly available independent HCC microarray datasets (BLIRIJP, GSE14520, GSE36376, GSE54238, GSE64041, GSE76297, GSE76427, GSE84005 and GSE102083) from GEO (Gene Expression Omnibus) database as previously described.10 In addition, the relationship between the CLK3 expression and its clinical manifestations was validated by TCGA and publicly available independent microarray dataset (GSE14520).
Cell Culture And Vector Construction
Human HCC cell lines (SMMC-7721 and SK-Hep-1) were obtained from Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM medium (Solaibio, China) supplemented with 10% fetal bovine serum (Clark, USA), 1% penicillin/streptomycin at 37°C in an incubator containing 5% CO2. CLK3 knockdown/overexpression vectors, miR-144 mimics and inhibitors were from Hanbio Company (Shanghai, China).
Immunohistochemistry (IHC)
The IHC staining was performed to investigate CLK3 expression in HCC paraffin-embedded tissues as previously described.11 Each TMA score was determined based on the staining intensity and the percentage of positively stained cells. A score of 1–3 was categorized as CLK3 low expression subgroup, and a score of 4–5 was classified as CLK3 high expression subgroup.
Western Blotting
The Western blot was processed as previously described.12 The primary antibodies used in the present study were listed below: wnt3a (1:1000, ab19925, Abcam), β-catenin (1:1000, 51067-2-AP, Proteintech), Vimentin (1:1000, 10366-1-AP, Proteintech), E-cadherin (1:500, 20874-1-AP, Proteintech), N-cadherin (1:500, 22018-1-AP, Proteintech), Fibronectin (1:500, 15613-1-AP, Proteintech), Snail (1:500, 13099-1-AP, Proteintech) and GAPDH (1:2000, 60004-1-Ig, Proteintech). The blots were developed using the ECL substrate (Beyotime, China) and photographed using the imaging system (Bio-Rad, CA, USA).
CCK-8, Cell Invasion And Colony Formation Assay
These assays were performed as described previously and were detailed in the Supplementary Materials and Methods section.12,13
Dual Luciferase Reporter Gene Assay
The luciferase reporter vector containing WT 3ʹ-UTR of CLK3 (Wt CLK3) or mutated 3ʹ-UTR of CLK3 (Mut CLK3) were obtained from RiBoBio (Guangzhou, China) as previously described.14 5 nM of miR-144 mimics or control, together with luciferase reporter vectors pmiR-RBREPORT-h-CLK3 (Wt CLK3 or Mut CLK3) were co-transfected into HEK293T cells using MegaTran 1.0 (Origene, China). The relative luciferase activity was determined 36 h after transfection using the Dual-Glo luciferase assay kit (Promega, USA).
HCC Xenograft Model
12 male BALB/c nude mice (6-week) were randomly divided into two groups: negative control (Ctrl) or shRNA-CLK3, and subcutaneously implanted with 50 μL of 1.0 × 106 SMMC-7721 cells transfected. Tumor volume was monitored and calculated using the formula: volume = length × width2/2. Mice were euthanized and tumors were extracted and processed for immunohistochemistry analysis five weeks later. This animal experiment was approved by the Animal Ethics Committee of the First Affiliated Hospital of Zhengzhou University. In addition, Guide for the Care and Use of Laboratory Animals (8th edition) was strictly followed in the present study.
Statistical Analysis
Statistical analysis was conducted using SPSS V23.0 (IBM, Armonk, NY, USA). A two-tailed paired Student’s t-test was used to analyze the statistical significance. Differences in clinicopathological factors between the CLK3 high- or low-expression patients were analyzed by the Chi-square test. A p < 0.05 was considered statistically significant.
Results
CLK3 Expression Is Upregulated In HCC Tissues And High CLK3 Expression Indicates Poor Prognosis Of Patients With HCC
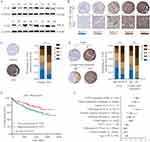
To evaluate the expression profile of CLK3 in HCC, we firstly analyzed CLK3 expression in human HCC tissues by Western blot. As shown in Figure 1A, CLK3 expression levels were significantly enhanced in HCC tissues in comparison with surrounding non-tumorous tissues (n = 8). IHC staining was performed within our TMA cohort containing 396 paired clinical HCC samples and the expression levels of CLK3 were scored between 1+ to 5+ based on the staining intensity (Figure 1B). Consistently, IHC staining further revealed the similar upregulation of CLK3 expression in HCC tissues (Figure 1C). Further analysis suggested that higher CLK3 expression in HCC was associated with several aggressive clinicopathologic features, including advanced TNM stage, lymph node metastasis and positive microvascular invasion (Figure 1D and Table 1).
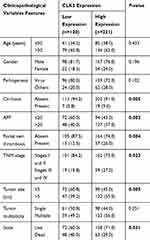 |
Table 1 Correlation Of Clinico-Pathological Features With CLK3 Expression In ZZU HCC Cohort |
Furthermore, a significant trend towards poorer overall survival (OS) and disease-free survival (DFS) for HCC patients with high CLK3 expression, compared with those with low CLK3 expression (Figure 1E). Univariate Cox analysis demonstrated that CLK3 expression, tumor size, lymph node metastasis, distant metastasis, and TNM stage were associated with the prognosis (Figure 1F). The multivariate analyses suggested a possible independent prognostic value of CLK3 in HCC patients (Table 2). Together, our data imply that high expression of CLK3 may play crucial function in HCC development.
 |
Table 2 Correlation Of Clinico-Pathological Features With CLK3 Expression In ZZU HCC Cohort |
High Expression Of CLK3 Correlates With The Clinicopathologic Characteristics And Survival Of Patients With HCC
To further explore the function of CLK3 in HCC development, expression levels of CLK3 in different cancers were analyzed by exploring TCGA cancer database. We found CLK3 mRNA levels were frequently dysregulated in most cancer types, while significantly enhanced in HCC tissues in comparison with non-tumor tissues in TCGA (Supplementary Figures 1 and 2A). In addition, we confirmed enhanced expression of CLK3 in HCC tissues in 7 additional published datasets (Figure 2A). Moreover, as shown in Figure 2B, high expression of CLK3 was positively correlated with late TNM stages in HCC patients.
The prognostic value of CLK3 mRNA expression in HCC patients was further evaluated by Kaplan-Meier survival analysis. HCC patient with high CLK3 expression had significantly worse OS than those with low CLK3 expression in GSE14520 cohort (n = 110, Figure 2C) and TCGA cohort (n = 367, Figure 2D). Furthermore, HCC patients with high CLK-3 expression exhibited a poor DFS rate (Figure 2E). Subgroup analysis revealed that late-stage (stages III and IV) patients with high CLK3 expression had shorter OS and DFS than those with low CLK3 expression in the TCGA HCC cohort, indicating that upregulated CLK3 was a potential prognostic biomarker for late-stage HCC patients (Figure 2F and G). Meanwhile, CLK3 expression was positively correlated to the expression of proliferation marker Ki67 and PCNA, indicating that CLK3 levels were positively associated with cell proliferation (Figure 2H). Furthermore, Gene Set Enrichment Analysis (GSEA) exhibited that liver cancer gene signatures of poor survival was enriched in CLK3 high expression group (Figure 2I). Together, the results further demonstrated that CLK3 played a critical role in HCC progression.
CLK3 Promotes HCC Cell Growth And Metastasis In Vitro
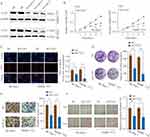
CLK3 mainly localized in the cytosol of HCC cells, as demonstrated by immunofluorescence staining (Supplementary Figure 3). We next evaluate the biological function of CLK3 in HCC. Three different shRNAs targeting CLK3 were transfected into SMMC-7721 or SK-Hep-1 cells and the knockdown efficiency was evaluated by Western blot (Figure 3A). The most efficient sh-CLK-3-3 was used for the following experiments. CCK-8 assays showed that cell proliferation was dramatically suppressed in CLK3-downregulated SK-Hep-1 or SMMC-7721 cells (Figure 3B). EdU assay showed that CLK3 knockdown markedly decreased the DNA synthesis rate in HCC cells (Figure 3C). Meanwhile, compared with negative controls, CLK3 silencing inhibited colony formation of HCC cells (Figure 3D). Moreover, transwell and wound-healing assays revealed that knockdown of CLK3 suppressed HCC cell invasion and migration, indicating an inhibitory effect on HCC metastasis (Figure 3E and F).
Since epithelial-mesenchymal transition (EMT) plays a critical role in cancer cell malignant transformation, we further tested the function of CLK3 on EMT. The results demonstrated that CLK3 knockdown enhanced the expression of E-cadherin and decreased the expression levels of N-cadherin, vimentin, and fibronectin (Supplementary Figure 2). In summary, these findings suggest that CLK3 promotes cell proliferation/invasion and regulates EMT progress in HCC.
Knockdown Of CLK3 Suppresses HCC Tumorigenesis In Vivo
Xenograft tumor model was established to further define the oncogenic role of CLK3. SK-Hep-1 cells transfected with control or sh-CLK3 were subcutaneously implanted into nude mice. The sh-CLK3 group showed significantly suppressed tumor growth (Figure 4A). Compared with the tumors from control group, tumors from sh-CLK3 group had remarkably smaller tumor size and lower tumor weight (Figure 4B-D). Moreover, IHC staining found that the tumors developed from sh-CLK3 cells displayed reduced expression of CLK3 and Ki-67 in comparison with those from control group (Figure 4E and F). Our findings indicate that CLK3 might promote HCC tumor development in vivo.
CLK3 Activates The Wnt/β-Catenin Signaling Pathway
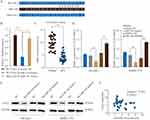
Next, Gene Set Variation Analysis (GSVA) was adopted to predict the potential signaling pathways involved. As shown in Figure 5A, GSVA results indicated that CLK3 might be associated with regulation of Wnt/β-catenin signaling pathway. Consistently, we found that CLK3 knockdown significant inhibited the expression of wnt3a and β-catenin in SK-Hep-1 or SMMC-7721 cells (Figure 5B and C). Collectively, these data suggest that CLK3 might exert its function via activating Wnt/β-catenin signaling pathway.
MiR-144 Negatively Regulates CLK3 Expression By Directly Binding To 3ʹ-UTR Of CLK3
We further performed comprehensive bioinformatics analysis using online database to predict the miRNAs that could post-transcriptionally regulate CLK3 expression. As shown in Figure 6A, miR-144 had the putative complementary binding site with 3ʹ-UTR of CLK3. Luciferase reporter assay found that miR-144 mimics markedly inhibited the luciferase activity in HEK293T cells transfected with pmiR-RB-REPORT containing WT 3ʹ-UTR of CLK3, but not mutated 3ʹ-UTR of CLK3 (Figure 6B). TCGA datasets analysis found that miR-144 expression was drastically lower in HCC tissues compared with that in normal control tissues (Figure 6C). While CLK3 mRNA and protein were significantly suppressed in SK-Hep-1 or SMMC-7721 cells transfected with miR-144 mimics, transfection of miR-144 inhibitor enhanced CLK3 expression in SK-Hep-1 or SMMC-7721 cells (Figure 6D and E). Indeed, Pearson association study demonstrated a negative correlation between miR-144 expression and CLK3 expression (Figure 6F). Thus, miR-144 might posttranscriptional regulate CLK3 expression in HCC cells.
MiR-144 Suppresses Growth And Metastasis Of HCC Cells By Repressing CLK3
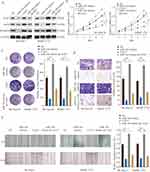
To evaluate whether CLK3 was a functional target of miR-144, we tested the inhibitory function of miR-144 could be reversed by restoration of CLK3 expression. Overexpression of CLK3 was achieved by transfection of pcDNA3.1-CLK3 into SK-Hep-1 or SMMC-7721 cells. CLK3 overexpression could partially reverse the miR-144-inhibited Wnt/β-catenin signaling pathway (Figure 7A). CCK-8 and colony formation assays suggested that while miR-144 mimics inhibited cell growth and CLK3 overexpression enhanced cell growth in HCC cells, CLK3 overexpression could partially reverse the miR-144 induced growth inhibition in HCC cells (Figure 7B and C). Furthermore, CLK3 overexpression could significantly reverse the miR-144 induced repression of migration and invasion in HCC cell lines, as demonstrated by transwell and wound-healing assays (Figure 7D and E). These results reinforced that miR-144 might inhibit growth and metastasis of HCC cells by repressing CLK3.
Discussion
Although therapeutic strategies and surgical technique have been advanced significantly in the past decades, the clinical survival status of HCC patients has not improved satisfactorily.2 Therefore, it is critical to fully understand the underlying mechanisms of HCC carcinogenesis and develop new promising therapeutic strategies for HCC treatment.
CLK3 regulates the intra-nuclear distribution of the serine/arginine-rich (SR) family of splicing factors.4 CLK3 is best known for its expression in normal testis.4 CLK3 functions in regulating the alternative splicing.5 However, the biological role of CLK3 in human cancers and its function in tumorigenesis is still not clear. In this study, we revealed that CLK3 was markedly upregulated in human HCC tissues and high level of CLK3 was closely associated with advanced TNM stages and poor survival of HCC patients (Figures 1 and 2). Functional experiments demonstrated the oncogene role of CLK3 in promoting HCC cell growth and metastasis (Figure 3). Interestingly, CLK3 was highly expressed not only in HCC but also in cholangiocarcinoma, esophageal carcinoma, gastric cancer and pancreatic cancers, suggesting that it might be an oncogene of higher generality in digestive tract tumors. As there are few reports that have explored the function of CLK3 in cancers, the oncogene role of CLK3 in tumor pathogenesis and progression remains unclear. More studies are required to dissect the precise role of CLK3 and underlying mechanisms in tumor development.
Various miRNAs participate in the pathogenesis and progression of different cancers.15 MiR-144, a well-known tumor suppressor, has been demonstrated to be involved in multiple cellular processes in several cancers. For instance, it has been shown that miR-144 inhibited cervical cancer development and metastasis by regulating VEGFA and VEGFC.16 Yin et al reported that miR-144 suppressed breast cancer progression through suppressing CEP55.17 Regarding HCC, it has been validated that miR-144 could play important function in multiple biological processes via targeting a variety of functional genes. Cao et al showed that miR-144 function as a tumor suppressor in HCC by targeting E2F3.18 Yu et al indicated that miR-144 regulated SMAD4 expression and inhibited HCC development.19 In addition, miR-144 could improve the chemoresistance of HCC by modulating Nrf2-dependent antioxidant pathway.20 Consistent with these studies, we revealed that CLK3 was a direct target of miR-144 and the expression of CLK3 and miR-144 were inversely associated with each other in clinical HCC specimens (Figure 6). In addition, CLK3 could partially abolish the inhibitory effect of miR-144 on HCC cell migration and invasion (Figure 7). The findings indicate that targeting miR-144/CLK3 axis could be utilized to develop novel therapeutic targets in HCC.
Constitutive activation of WNT signaling is the hallmark of multiple cancers and promotes cancer cell proliferation and invasion.21 Our study further demonstrated that miR-144 overexpression suppressed the Wnt/β-catenin signaling and the inhibition could be partly abolished by CLK3 overexpression (Figure 5). Indeed, several signaling pathways were reported to be regulated by miR-144. In bladder cancer, miR-144 targeted EZH2 and regulated Wnt signaling.22 MiR-144 could inhibit mTOR signaling pathway and suppress the colorectal cancer.23 These studies indicate the complicated role and mechanism of miR-144 depending on different cancer types and molecular targets.
In summary, our results show that CLK3 is overexpressed in HCC, and overexpression of CLK3 is a negative prognostic factor for HCC patients. Functional experiments reveal that CLK3 knockdown suppresses HCC tumorigenesis both in vitro and in vivo. Furthermore, CLK3 functions as an oncogene and activates Wnt/β-catenin signaling in HCC cells. Finally, we identify miR-144 as a tumor suppressor by controlling CLK3. Our findings uncover the mechanistic role of miR-144/CLK3 axis in HCC progression, providing a potential therapeutic biomarker in HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Dvm AJ. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):11. doi:10.3322/caac.21442
2. Ayuso C, Rimola J, Vilana R, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72. doi:10.1016/j.ejrad.2018.01.025
3. Bullock AN, Das S, Debreczeni JE, et al. Kinase domain insertions define distinct roles of CLK kinases in sr protein phosphorylation. Structure. 2009;17(3):352–362. doi:10.1016/j.str.2008.12.023
4. Becker W, Kentrup H, Heukelbach J, Joost HG. cDNA cloning and characterization of rat Clk3, a LAMMER kinase predominately expressed in testis. Biochim Biophys Acta. 1996;1312(1):63–67. doi:10.1016/0167-4889(96)00036-5
5. Cesana M, Guo MH, Cacchiarelli D, et al. A CLK3-HMGA2 alternative splicing axis impacts human hematopoietic stem cell molecular identity throughout development. Cell Stem Cell. 2018;22(4):575. doi:10.1016/j.stem.2018.03.012
6. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–587. doi:10.1016/j.molmed.2006.10.006
7. Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2011;65(9):3509–3512. doi:10.1158/0008-5472.CAN-05-0298
8. Liang H-W, Ye ZH, Yin SY, et al. A comprehensive insight into the clinicopathologic significance of miR-144-3p in hepatocellular carcinoma. Onco Targets Ther. 2017;10:3405–3419. doi:10.2147/OTT.S138143
9. Bao J, Yu Y, Chen J, et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9(10):1045. doi:10.1038/s41419-018-1020-0
10. Zhang J, He Y, Yu Y, et al. Upregulation of miR‐374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018;7(7):3351–3362. doi:10.1002/cam4.2018.7.issue-7
11. Xue C, He Y, Zhu W, et al. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am J Transl Res. 2018;10(12):4152–4162.
12. Xue C, He Y, Hu Q, et al. Downregulation of PIM1 regulates glycolysis and suppresses tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10(undefined):5101–5112. doi:10.2147/CMAR.S184381
13. He Y, Xue C, Yu Y, et al. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10(undefined):3857–3865. doi:10.2147/CMAR.S175681
14. Chen J, Yu Y, Chen X, et al. MiR‐139‐5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Proliferation. 2018;51(6):e12510.
15. CP B, HS S, GJ G. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi:10.1038/nrg.2016.134
16. Tao P, Wen H, Yang B,et al. miR-144 inhibits growth and metastasis of cervical cancer cells by targeting VEGFA and VEGFC. Exp Ther Med. 2018;15(1):562–568.
17. Yin Y, Cai J, Meng F, Sui C, Jiang Y. MiR-144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biol Ther. 2018;19(4):306–315. doi:10.1080/15384047.2017.1416934
18. Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumor Biol. 2014;35(11):10759–10764. doi:10.1007/s13277-014-2017-7
19. Sheng S, Xie L, Wu Y, et al. Mir-144 inhibits growth and metastasis in colon cancer by downregulating SMAD4. Bioscience Reports. 2019;39(3).
20. Zhou S, Ye W, Zhang Y, et al. miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am J Transl Res. 2016;8(7):2992.
21. Holland J D, Klaus A, Garratt A N, et al. Wnt signaling in stem and cancer stem cells.[J]. Current Opinion in Cell Biology, 2013, 25(2):254–264.
22. Guo Y, Ying L, Tian Y, et al. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280(18):4531–4538. doi:10.1111/febs.12417
23. Iwaya T, Yokobori T, Nishida N, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi:10.1093/carcin/bgs288
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.