Back to Journals » OncoTargets and Therapy » Volume 9
Clinicopathological significance of STAT4 in hepatocellular carcinoma and its effect on cell growth and apoptosis
Authors Li J, Liang L, Liu Y, Luo Y, Liang X, Luo D, Feng ZB, Dang Y, Yang L, Chen G
Received 7 November 2015
Accepted for publication 9 January 2016
Published 21 March 2016 Volume 2016:9 Pages 1721—1734
DOI https://doi.org/10.2147/OTT.S100040
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Jianjun Li,1,* Lu Liang,2,* Yongru Liu,2 Yihuan Luo,2 Xiaona Liang,2 Dianzhong Luo,2 Zhenbo Feng,2 Yiwu Dang,2 Lihua Yang,3 Gang Chen2
1Department of General Surgery, Western Branch, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Department of Pathology, 3Department of Medical Oncology, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China
*These authors contributed equally to this work
Background: Recent studies showed that signal transducer and activator of transcription 4 (STAT4) was downregulated in hepatocellular carcinoma (HCC) tissues. However, the role of STAT4 in HCC is still unknown. The aim of this study is to explore the association between STAT4 expression and other clinicopathological features in HCC and to test the effect of STAT4 on cell growth and apoptosis in vitro.
Methods: STAT4 was evaluated by immunohistochemistry in 171 HCC and corresponding paraneoplastic liver, 37 cirrhosis, and 33 normal liver tissues. Association between STAT4 and clinicopathological parameters was analyzed. Meta-analysis on STAT4 in cancer was performed. The effect of STAT4 small interfering RNA (siRNA) on cell growth and cell apoptosis was also detected.
Results: Positive rate of STAT4 was 29.2% (50/171) in HCC tissues, 53.2% (91/171) in paraneoplastic liver tissues, 64.9% (24/37) in cirrhosis tissues, and 72.7% (24/33) in normal liver tissues. STAT4 was upregulated in younger patients who were female, with single tumor node, early TNM stage, without portal vein tumor embolus, and α-fetoprotein (AFP)-positive tumors compared with the groups comprising older patients, males, and those with multiple tumor nodes, advanced TNM stage, with portal vein tumor embolus, and AFP negative tumors. Meta-analysis showed STAT4 was correlated with TNM stage (OR =0.50, 95% CI =0.30, 0.83, P=0.008) and age (OR =0.58, 95% CI =0.38, 0.95, P=0.032) in malignant tissues, and with AFP level (OR =1.76, 95% CI =1.06, 2.94, P=0.03) in HCC. STAT4 siRNA promoted growth and suppressed apoptosis of HepG2 cells.
Conclusion: STAT4 might play a vital role in development of HCC, via influencing cell growth and apoptosis, as a tumor suppressor.
Keywords: hepatocellular carcinoma, HCC, signal transducers and activators of transcription 4, STAT4, clinicopathological features, immunohistochemistry, meta-analysis, in vitro
Introduction
Hepatocellular carcinoma (HCC), an aggressive malignancy of liver, is the fifth most frequent form of cancer and the third cause of cancer mortality worldwide.1,2 Over 80% of cases occur in East Asia and sub-Saharan Africa, and the incidence is still increasing in Europe3 and the United States.4 Studies have shown that HCC results in 650,000 or more deaths per year worldwide, three-quarters of the deaths occur in the East Asian countries.5,6 Locally metastatic or advanced HCC patients face a dismal outcome. Diverse therapy strategies are available, including surgical resection, radiotherapy, and chemotherapy, as well as liver transplantation.7,8 Although more than 70% of patients in early stage can have a 5-year survival rate, their recurrences of HCC are inevitable,9,10 which makes long-term outcome still unsatisfactory even after curative hepatectomy.11
Signal transducers and activators of transcription (STAT) can regulate hematopoietic process and have influence on interacting tumor cells with their immune microenvironment through suppression or induction of growth factors and specific cytokines.12,13 Extensive studies have shown that STAT3 and STAT5 play a crucial role in tumor cell proliferation.12,14 Besides, STAT5 and STAT6 could upregulate genes which are important for HCC cell proliferation.12,15 However, studies focused on STAT4 are limited. A very recent study conducted in human beings by Jiang et al16 demonstrated that expression level of STAT4 mRNA in HCC tissues was downregulated. However, the clinical role and molecular mechanism of STAT4 in HCC have not been investigated extensively. Hence, we focused on studying the association between the expression of STAT4 and clinicopathological features in HCC patients in this study. A mini meta-analysis was also performed to investigate whether STAT4 was linked to the progression of different cancers. The effect of STAT4 on HCC cell growth and apoptosis was further investigated by in vitro experiments.
Materials and methods
Tissue samples
HCC tissues and corresponding paraneoplastic liver tissues (n=171), cirrhosis liver tissues (n=37), and normal liver tissues (n=33) were collected. All tissues were formalin fixed and paraffin embedded (FFPE). All patients underwent hepatectomies, and tissues of HCC patients were obtained before cancer-related therapies. Included patients were randomly selected from those who underwent the hepatic surgeries in the First Affiliated Hospital of Guangxi Medical University, the People’s Republic of China during March 2010 to December 2012. All patients were between 29 and 82 years of age, with a mean age of 52 years. Adjacent noncancerous livers were more than 2 cm away from the tumor node. Tumor size ranged from 1 to 11 cm, with a mean diameter of 6.4 cm. Of the 171 HCC patients, 104 patients were followed up; the median time of being followed up was 32.78 months (ranged from 2.68 to 68.00 months). Sixteen patients had recurrent tumor, and 88 were dead or censored in the end. The study was approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University, and written informed consents were signed by the patients and doctors to use the samples for research. Two pathologists independently conducted blind tests on all tissue samples.
Immunohistochemistry
Antibody of STAT4 was provided by Santa Cruz Biotech Company, CA, USA (mouse monoclonal antihuman STAT4 antibody, PL-68, 1:300 dilution). All stained tissues were reviewed and scored by two pathological doctors independently without any clinical information of patients. Positive cells percentage was scored as 0 (0%), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). Intensity of staining was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong).17 Final score of each case was the result of the positive score multiplied by the intensity score. Score more than 2 was considered as positive.
Meta-analysis
To compare and comprehend the clinical value of STAT4 in different malignancies, a meta-analysis was then performed. Relevant literatures were searched from the following databases: PubMed, Web of Science, EMBASE, Cochrane Library, China National Knowledge Infrastructure, Chongqing VIP, Wanfang, and Chinese Biomedical Literature Database, with the search strategy as (“STAT4” OR “STAT-4” OR “SLEB11” OR “signal transducer and activator of transcription 4”) AND (“tumor” OR “tumour” OR “cancer” OR “carcinoma” OR “sarcoma” OR “neoplasia” OR “neoplasm” OR “malignan*”). The last update time was October 31, 2015. Inclusion criteria were as follows: 1) Study tested the expression of STAT4 in malignant tissues. 2) Study assessed the relationship between STAT4 levels and clinicopathological features. 3) The written language of literatures must be English or Chinese.
The extracted information include first author name, published year, region, type of malignancy, case quantity, test method of STAT4, cutoff value, and clinicopathological features reported by each study. The quality of included literatures was assessed by Newcastle–Ottawa quality assessment scale. Fixed-effect model was used to calculate odds ratios (ORs) and their 95% confidence intervals (CIs) when heterogeneity among studies has no statistical significant (P=0.05); otherwise, random effect model was conducted. Analysis was performed with Stata 12.0 software (StataCorp LP, College Station, TX, USA).
Cell culture
HepG2 and HepB3 cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA), cultured in Dulbecco’s Modified Essential Medium (Invitrogen Corp., Grand Island, NY, USA). Fetal bovine serum (10% heat-inactivated, Invitrogen Crop), 2 mM glutamine, and gentamicin were supplied in media. Cells were grown in the environment with 37°C temperature, 5% CO2 and 100% humidity. The expression of STAT4 in human HCC cell lines HepG2 and HepB3 was detected by Western blot (data not shown). HepG2 showed higher STAT4 expression than HepB3 cell line according to the result, so HepG2 cell line was used for further in vitro experiments. All in vitro experiments were repeated three times.
Knockdown of STAT4 in HepG2 cells
HepG2 cells were seeded in a 96-well (2.5×103 cells per well) or 24-well (2.5×104 cells per well) plate, incubated at 37°C for 24 hours, then the cells were transfected with STAT4 small interfering RNA (siRNA) pool, with a final concentration of 200 nm/L, combiMAGnetofection (OZ BIOSCIENCES, Marseille cedex 9, France) was used according to manufacturer’s instruction. The STAT4 siRNA pool contains four siRNAs. The sequences are shown in Table 1. Among them, STAT4 siRNA-1537 (siSTAT4-1537), siSTAT4-2281, and siSTAT4-2314 were reported by Lv et al,18 while siSTAT4-1134 was designed according to Reynolds et al’s work.19 Four STAT4 siRNAs were compared to identify regions of similarity using BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi. Cells transfected with STAT4 scrambled siRNAs were regarded as negative control. Besides, mock and blank controls were also set up in the experiments. Samples of cell were harvested at 0, 48 and 96 hours after transfection and analyzed further.
  | Table 1 SiRNAs contained in the STAT4 siRNA pool |
Western blotting
The cells in all groups were collected after transfection for 96 hours. The process was performed as previously reported.20–22 The membrane was incubated with anti-human STAT4 rabbit polyclonal antibody (Abcam, Cambridgeshire, UK) at a dilution of 1:500 and β-actin antibody (Sigma-Aldrich, St Louis, MO, USA) at a dilution of 1:2,000 as control. Chemiluminescence detection assay was performed after incubation.
Cell viability
Cell viability was determined by fluorimetric detection of resorufin (CellTiter-Blue Cell Viability Assay, G8080, Promega, Madison, WI, USA). The manufacturer’s protocol was followed for the procedure. FL600 fluorescence plate reader (Bio-Tek, Richmond, VA, USA) was used for fluorimetry (ex: 560 nm/em: 590 nm). Results were the fluorescence value of sample/mock control ×100%.
Cell proliferation
Colorimetric tetrazolium (MTS) assay (CellTiter96 AQueous One Solution Cell Proliferation Assay G3580, Promega) was used to assess the proliferation of cells. MTS reagent (20 μL) was added to each well after transfection. The cells were incubated for 2 hours at 37°C in a humidified and 5% CO2 atmosphere. Absorbance at 490 nm was documented using a microplate reader (Scientific Multiskan MK3, Thermo Fisher Scientific Oy, Vantaa, Finland). The results were expressed as the absorbance of sample/absorbance of mock control ×100%.
Fluorescent microscopy evaluation of cell growth, apoptosis, and morphology
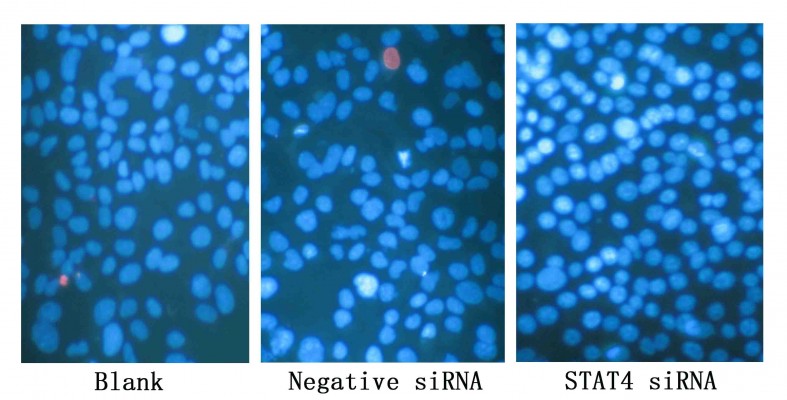
Cell growth and apoptosis were also monitored by Hoechst 33342 (Sigma-Aldrich) and propidium iodide (PI, Sigma-Aldrich) double fluorescent chromatin staining. Absolute number of different cells would be determined by this method, the brief principle was as following: necrotic cells (PI positive with debris signals), late apoptotic cells (Hoechst 33342 positive/PI positive, with red fragmentations in the cells), early apoptotic cells (Hoechst 33342 positive/PI negative with blue fragmentations in the cells), and viable cells (Hoechst 33342 positive/PI negative).23 Cells in different states (necrotic, apoptotic, and viable) were counted under the 200× magnification, and each type of cells was calculated in ten diverse fields in each well. The final results were shown by mean values and were compared with the mock control group.
Caspase-3/7 activity detection
Caspase-3/7 activity was tested using a synthetic Rhodamine-labeled caspase-3/7 substrate (Apo-ONE® Homogeneous Caspase-3/7 Assay, G7790, Promega), which was manipulated proximately after the detection of cell viability, as the instructions of the manufacturer pointed out. After incubation for 60 minutes at room temperature, the fluorescence of each well was detected (excitation: 499 nm/emission: 512 nm) by a FL600 fluorescence plate reader. Caspase-3/7 activity was expressed as fluorescence of treated sample/mock control.22
Statistical analysis
SPSS 20.0 was used for statistical analysis. Kruskal–Wallis H-test was used to test difference among three or more groups. Mann–Whitney U-test was used for the comparison between two groups. The association between STAT4 expression level and clinicopathological features was assessed by Spearman’s rank correlation. Receiver operating characteristic (ROC) curves were graphed to evaluate the significance of STAT4 in distinguishing the HCC from noncancerous tissues. Survival analysis was performed by Kaplan–Meier method, and the survival between groups was estimated with log-rank test. One-way analysis of variance (one-way ANOVA) was used in experiments in vitro. If pairwise comparison was needed, least significance difference test (LSD) was used. It was regarded as statistically significant when P<0.05, P-value was derived from two-tailed tests.
Results
STAT4 expression in different liver tissues
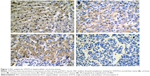
Positive rate of STAT4 expression was 29.2% (50/177) in HCC tissues, 53.2% (91/177) in the paired adjacent noncancerous tissues, 64.9% (24/37) in cirrhosis tissues, and 72.7% (24/33) in normal tissues, respectively (Table 2). Expression of STAT4 was positive in noncancerous liver tissues (Figures 1A–C and 2), while weak signal was detected in HCC tissues (Figures 1D and 2). Kruskal–Wallis H-test revealed that the expression of STAT4 was significantly different in four types of liver tissues. (H=37.635, P<0.05). Paired comparisons were performed using Mann–Whitney U-test. Significant difference was observed between the HCC group and paired noncancerous group (Z=−4.497, P<0.001). Besides, Spearman’s rank correlation analysis exhibited a positive correlation between STAT4 expression and status of liver tissues (r=0.302, P<0.001) (Table 2). Furthermore, ROC curve was analyzed to confirm the diagnostic value of STAT4 expression in HCC. The area under curve of STAT4 was 0.563 (95% CI =0.506–0.619, P=0.030), with a low clinical value of diagnosis.
Association between STAT4 and clinical features in HCC patients
Correlations of STAT4 expression and other clinicopathological features including age, sex, number of tumor nodes, clinical TNM stage, portal vein tumor embolus, AFP level, and cirrhosis were assessed in 171 HCC cases (Table 2). STAT4 positive rate was remarkably higher in younger group than that in older group when the cutoff year was 50 (Z=−2.554, P=0.011) (Table 3). Females had a higher level of STAT4 expression compared to males (Z=−2.587, P=0.010). Patients with single tumor node had a higher level of STAT4 expression than those with multiple tumor nodes (Z=−3.542, P<0.05). The expression of STAT4 was remarkably greater in early stage (I and II) than in advanced stage (III and IV) (Z=−2.971, P=0.003). When considering portal vein tumor embolus, patients without embolus had a remarkably higher level of STAT4 expression than those who had embolus (Z=−3.281, P=0.001). α-Fetoprotein (AFP)-positive patients had higher STAT4 expression compared to AFP-negative patients (Z=−2.212, P=0.027). Patients without cirrhosis had a remarkably higher expression level of STAT4 than those with cirrhosis (Z=−3.168, P=0.002).
Further analysis by Spearman’s rank correlation elucidated that the expression of STAT4 was significantly correlated with sex (r=0.198, P=0.009), number of tumor nodes (r=−0.313, P=0.000), clinical TNM stage (r=−0.228, P=0.003), portal vein tumor embolus (r=−0.290, P=0.001), AFP level (r=−0.212, P=0.026), and cirrhosis (r=0.243, P=0.001) (Table 2).
Recurrence analysis
Patients with negative STAT4 expression had a mean recurrence-free survival time of 56.811 months (95% CI =51.071–62.550), while patients with positive STAT4 expression only had 47.747 months (95% CI =50.890–61.044). However, no significant change was noted between two groups (χ2=0.496, P=0.481, Figure 3).
Meta-analysis of STAT4 in cancers
Three studies of HCC (including the current study), one study on primary hepatocarcinoma, one study on gastric adenocarcinoma, and one study on non-small-cell lung cancer14,24–27 were included in mini meta-analysis. The different expression levels of STAT4 in cancer tissues and noncancerous tissues were analyzed. Meanwhile, the association between the expression of STAT4 and clinicopathological features including metastasis, differentiation, sex, TNM stage, the number of tumor nodes, and age of patients were analyzed. In subgroup analysis, the relationship between STAT4 level and AFP, hepatitis B virus (HBV) status, and vascular invasion status of HCC patients was analyzed. The final result showed that the expression of STAT4 between cancer and noncancerous tissues had no difference (OR =1.33, 95% CI =0.33, 9.88, P=0.494) (Figure 4A). The expression of STAT4 was found discrepant in different TNM stages (I–II versus III–IV) (OR =0.50, 95% CI =0.30, 0.83, P=0.008) (Figure 4B), different age stage (OR =0.58, 95% CI =0.38, 0.95, P=0.032) (Figure 4C), and different serum AFP level in HCC patients (OR =1.76, 95% CI =1.06, 2.94, P=0.03) (Figure 4H). No evidence showed any relationship between STAT4 and other clinicopathological features (Figure 4D–H).
STAT4 siRNAs promote proliferation and inhibit apoptosis in HepG2 cells
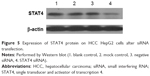
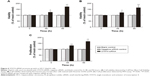
Transfected efficiency was confirmed by Western blotting (Figure 5). The viability of HepG2 cells transfected with STAT4 siRNA was increased significantly in 48 hours (P<0.05) and 96 hours (P<0.01) compared with blank control and negative siRNA control when tested using CellTiter-Blue Cell Viability Assay (Figure 6A). To verify the result of viability assay, viable cells were counted by microscopy after they were stained with Hoechst 33342 and PI (Figures 6B and 7), and cell proliferation was determined by MTS tetrazolium assay (Figure 6C). Both results showed consistent tendency with viability assay. To estimate the effect of STAT4 on cell apoptosis, human HCC cells were observed microscopically after Hoechst 33342 and PI staining. STAT4-siRNA-transfected cells showed an apoptosis resistance tendency at 96 hours incubation when compared with blank control (P<0.01) and negative siRNA control (P<0.01) (Figure 8A). Meanwhile, fluorescent caspase-3/7 assay was performed. The result showed that the activity of caspase 3/7 was reduced in STAT4 siRNA group when incubated for 96 hours, with a P-value less than 0.01 when compared with blank and negative siRNA control groups (Figure 8B).
Discussion
HCC is an aggressive solid malignancy in humans and is becoming a serious health threat with increasing morbidity and mortality.28,29 As relative genomic and molecular technology develops, some biomarkers for HCC diagnosis and treatment was discovered.30 However, sensitive biomarkers which are able to diagnose HCC in early stage and predict prognosis of HCC patients are needed. In this research, we aimed at seeking the potential diagnostic and prognostic value of STAT4 in HCC.
When exploring the diagnostic value of STAT4 in HCC by IHC, we found that STAT4 expression level was lower in HCC tissues than in noncancerous liver tissues. Consistent with our study, Wubetu et al14 and Wang et al27 found that STAT4 expression in HCC tissues was lower than that in paired adjacent noncancerous tissues. Additionally, in genome-wide association study, STAT4 single nucleotide polymorphism (SNP) rs7574865 was significantly associated with HCC,28,31 and STAT4 was expressed at low levels in HCC subjects when tested by quantitative real-time polymerase chain reaction16 and microarray.32 However, Chen et al33 found no relationship between rs7574865 and HCC. A meta-analysis indicated no statistically significant alteration regarding the expression of STAT4 in cancer tissues and noncancerous tissues. Inconsistent results suggested that more studies on STAT4 in HCC are needed in the future. Except for HCC, STAT4 was found relevant in other malignancies like colon and rectal cancer,34 breast cancer,35 and lymphoma.36
In the analysis of the association between STAT4 and clinicopathological features of patients, Wubetu et al14 and Wang et al27 found that tumor size and differentiation were linked with STAT4 expression; however, no links were found in our study. We found similar result with Wubetu et al14 that STAT4 expression in advanced TNM stage HCC patients was lower than in early TNM stage patients; the meta-analysis was also suggested that advanced TNM stage patients (one on NSCLC and two on HCC) have low STAT4 level. This study showed that the expression of STAT4 was lower in elder patients than younger patients. It was consistent with the results from the current meta-analysis, which showed a tendency of low STAT4 expression in older patients. This might be result in different metabolism rate and immunity between younger and elder patients. Meanwhile, we found positive rate of STAT4 was higher in female patients than males. It might be caused by different hormone environments in females and males, and it might be the reason that male patients are more susceptible to HCC than females.37 However, the relationship between STAT4 and age, and sex need more studies with larger sample size to clarify. STAT4 seemed to act as a cancer suppressor gene in current meta-analysis. However, STAT4 expression was surprisingly higher in AFP-positive group with statistical significance. Currently, AFP was found inefficient with low sensitivity and specificity in HCC diagnosis.38,39 But serum level of AFP was still frequently used for the diagnosis of HBV-relative HCC with considerable sensitivity and specificity in the People’s Republic of China.40 In the meta-analysis of AFP and STAT4 expression, only three studies (including the current study) with 327 cases were included.14,27 No relationship was found between AFP and STAT4 in the studies of Wubetu et al14 and Wang et al.27 The relationship between STAT4 and AFP needs to be illuminated.
To test the effect of STAT4 in HCC, the growth and apoptosis of HCC HepG2 cells transfected with STAT4 siRNA were detected. Consistent with STAT4 expression tested in HCC tissues, results in vitro implied STAT4 plays a role as cancer suppressor in HCC. Wang et al27 also found that downregulation of STAT4 could enhance proliferation of liver L02 cells, consistent with the current effect produced in HepG2 cells. However, Cheng et al41 found the proliferation and invasion of colorectal cancer (CRC) cells was inhibited when STAT4 gene was knocked down. Zhou et al42 revealed that STAT4 was regulated by micro RNA-141 (miR-141) directly, and a negative relationship was found between the expression of STAT4 and miR-141 in gastric cancer cells. STAT4 was upregulated when miR-155 was knocked down in patient-derived MyLa cell.43 The effect of Cryptotanshinone (CPT) on inhibiting breast tumor growth was studied,44 and it was found that CPT regulated cytotoxic CD4+ T cells through upregulated Janus kinase 2 (JAK2) and STAT4, and played an antitumor role in breast cancer. It is known that the JAK/STAT pathway is involved in cell growth.34 Therefore, STAT4 could play different roles according to variant cancers. Since there was little research on STAT4 in HCC cells, the mechanism of action of STAT4 in HCC is still unclear. As different HCC cell lines originate from different hosts, individual variation leads to HepG2 showing higher expression of STAT4 than HepB3, which was the result in our preliminary test via Western blotting. STAT4 has been reported to play an important role in the process of immunoreactions. STAT4 can be activated by IL-12, which is produced by antigen-presenting cells (APCs) and cells that have similar function as APCs; activated STAT4 can promote inflammatory cells like NK cells to produce IFN-γ.45 Recent studies have suggested that IFN-γ can induce cells to undergo apoptosis through the change of microenvironment46 and induced autophagy.47 As an authorized activator of STAT4, IL-12 has the potential to evolve as a hot point in the STAT4-targeted therapy. We also noticed that dasatinib, one class of chemotherapy drugs targeting STAT3 in the clinic, can inhibit the growth of cells through preventing activation of STAT3, another member of the STAT family. Analogously, STAT4 also gains the potential to be a molecular target in future cancer treatment.48–50
Our study, together with literatures mentioned above, suggested STAT4 might have a potential role in the prediction of prognosis and progress in HCC and other malignancy. However, limitations existed in this study. First, the amount of included patients was limited. All tissues tested in this study were FFPE tissues, there may be some aberrance existed as compared with fresh biopsies. Second, only a small number of six papers were included in the meta-analysis because of language limitation and because not many studies on the expression of STAT4 in malignant tissues are available to date. Third, the mechanism of STAT4 in influencing the growth and apoptosis of HCC cells has not been explored, and no in vivo study has been available at present. Therefore, we will overcome the limitations mentioned above and explore the mechanism of STAT4 in HCC in a future study. Large cohort studies with high quality are needed to validate the conclusions. Also, patients recruited from multiple ethnicities would make it more convincing. Moreover, the molecular mechanism of STAT4 in HCC needs to be explored further.
Conclusion
Above all, STAT4 protein was downregulated in HCC tissues compared with noncancerous tissues and was negatively correlated with TNM stage, number of tumor nodes, portal vein tumor embolus, and cirrhosis. Along with other previous research, our study strongly prompts that STAT4 is a tumor-suppressive gene. STAT4 may play an antioncogenic role in HCC and suppresses deterioration of HCC. Furthermore, STAT4 may be a potential targeted gene for molecular therapy in HCC. However, the potential role and the intrinsic molecular mechanism of STAT4 in HCC need further investigations.
Acknowledgment
The study was supported by the Fund of Natural Science Foundation of Guangxi, the People’s Republic of China (2015GXNSFBA139157). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Boman F, Sfeir R, Priso R, Bonnevalle M, Besson R. Advantages of intraoperative semiquantitative evaluation of myenteric nervous plexuses in patients with Hirschsprung disease. J Pediatr Surg. 2007;42(6):1089–1094. | ||
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. | ||
Capocaccia R, Sant M, Berrino F, et al. Hepatocellular carcinoma: trends of incidence and survival in Europe and the United States at the end of the 20th century. Am J Gastroenterol. 2007;102(8):1661–1670; quiz 1660, 1671. | ||
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. | ||
Yuen MF, Hou JL, Chutaputti A; Asia Pacific Working Party on Prevention of Hepatocellular Carcinoma. Hepatocellular carcinoma in the Asia Pacific region. J Gastroenterol Hepatol. 2009;24(3):346–353. | ||
Asia-Pacific Working Party on Prevention of Hepatocellular Carcinoma. Prevention of hepatocellular carcinoma in the Asia-Pacific region: consensus statements. J Gastroenterol Hepatol. 2010;25(4):657–663. | ||
Bhangoo MS, Karnani DR, Hein PN, et al. Radioembolization with Yttrium-90 microspheres for patients with unresectable hepatocellular carcinoma. J Gastrointest Oncol. 2015;6(5):469–478. | ||
Paik EK, Kim MS, Choi CW, et al. Dosimetric comparison of volumetric modulated arc therapy with robotic stereotactic radiation therapy in hepatocellular carcinoma. Radiat Oncol J. 2015;33(3):233–241. | ||
Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334(11):693–699. | ||
Roxburgh P, Evans TR. Systemic therapy of hepatocellular carcinoma: are we making progress? Adv Ther. 2008;25(11):1089–1104. | ||
Zhao X, Jiang K, Liang B, Huang X. STAT4 gene polymorphism and risk of chronic hepatitis B-induced hepatocellular carcinoma. Cell Biochem Biophys. 2015;71(1):353–357. | ||
Yu H, Jove R. The STATs of cancer – new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. | ||
Liao Y, Cai B, Li Y, et al. Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One. 2014;9(11):e111677. | ||
Wubetu GY, Utsunomiya T, Ishikawa D, et al. High STAT4 expression is a better prognostic indicator in patients with hepatocellular carcinoma after hepatectomy. Ann Surg Oncol. 2014;21(Suppl 4):S721–S728. | ||
Bruns HA, Kaplan MH. The role of constitutively active Stat6 in leukemia and lymphoma. Crit Rev Oncol Hematol. 2006;57(3):245–253. | ||
Jiang DK, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45(1):72–75. | ||
Zhang XL, Dang YW, Li P, et al. Expression of tumor necrosis factor receptor-associated factor 6 in lung cancer tissues. Asian Pac J Cancer Prev. 2014;15(24):10591–10596. | ||
Lv L, Zhang J, Huang X, Zhao Y, Zhou Z, Zhang H. Lentivirus-mediated RNA interference targeting STAT4 inhibits the proliferation of vascular smooth muscle cells. Arch Med Res. 2008;39(6):582–589. | ||
Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. | ||
Chen G, Kronenberger P, Teugels E, De Greve J. Influence of RT-qPCR primer position on EGFR interference efficacy in lung cancer cells. Biol Proced Online. 2011;13:1. | ||
Chen G, Kronenberger P, Teugels E, Umelo IA, De Greve J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Med. 2012;10:28. | ||
Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. | ||
Chen G, Noor A, Kronenberger P, Teugels E, Umelo IA, De Greve J. Synergistic effect of afatinib with su11274 in non-small cell lung cancer cells resistant to gefitinib or erlotinib. PLoS One. 2013;8(3):e59708. | ||
Wen S, Yang S, Guli K, et al. Study on the expression of ICAM-1, STAT4 in gastric cancer and the relationship between clinical signs and their expression. J Hebei Med Coll Continu Educ. 2008;25(02):15–18. | ||
Xin C, Zuo Z, Song B, Lan X, Wang C, Wang H. The expressions of STAT4, Bax and c-myc in human non-small cell lung cancer tissues and correlativity analysis. J Luzhou Med Coll. 2011;34(05):534–538. | ||
Zhou J, Zhong D, Yang Z, Hu J, Deng X. Expression of STAT1, STAT2 and STAT4 in primary hepatocellular carcinoma and its significantce. Chin J Hepatobiliary Surg. 2004;10(12):49–51. | ||
Wang G, Chen JH, Qiang Y, Wang DZ, Chen Z. Decreased STAT4 indicates poor prognosis and enhanced cell proliferation in hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3983–3993. | ||
Chen W, Wang M, Zhang Z, et al. Replication the association of 2q32.2-q32.3 and 14q32.11 with hepatocellular carcinoma. Gene. 2015;561(1):63–67. | ||
Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6(6):4505–4515. | ||
Xu M, Xie F, Qian G, et al. Peritumoral ductular reaction: a poor postoperative prognostic factor for hepatocellular carcinoma. BMC Cancer. 2014;14:65. | ||
Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of Hepatitis B virus infection. J Med Virol. Epub August 8, 2015. doi:10.1002/jmv.24350. | ||
Kim LH, Cheong HS, Namgoong S, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci of STAT4 and HLA-DQ in a Korean population. Infect Genet Evol. 2015;33:72–76. | ||
Chen K, Shi W, Xin Z, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PLoS One. 2013;8(10):e77315. | ||
Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol Carcinog. 2013;52(2):155–166. | ||
Kristensen VN, Vaske CJ, Ursini-Siegel J, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A. 2012;109(8):2802–2807. | ||
Lupov IP, Voiles L, Han L, et al. Acquired STAT4 deficiency as a consequence of cancer chemotherapy. Blood. 2011;118(23):6097–6106. | ||
Chen W, Armstrong BK, Zheng R, Zhang S, Yu X, Clements M. Cancer burden in China: a Bayesian approach. BMC Cancer. 2013;13:458. | ||
Wan HG, Xu H, Gu YM, Wang H, Xu W, Zu MH. Comparison osteopontin vs AFP for the diagnosis of HCC: a meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38(6):706–714. | ||
Baig JA, Alam JM, Mahmood SR, et al. Hepatocellular carcinoma (HCC) and diagnostic significance of A-fetoprotein (AFP). J Ayub Med Coll Abbottabad. 2009;21(1):72–75. | ||
Dong X, He H, Zhang W, Yu D, Wang X, Chen Y. Combination of serum RASSF1A methylation and AFP is a promising non-invasive biomarker for HCC patient with chronic HBV infection. Diagn Pathol. 2015;10:133. | ||
Cheng JM, Yao MR, Zhu Q, et al. Silencing of stat4 gene inhibits cell proliferation and invasion of colorectal cancer cells. J Biol Regul Homeost Agents. 2015;29(1):85–92. | ||
Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 induced by helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem. 2014;33(4):1003–1012. | ||
Litvinov IV, Cordeiro B, Fredholm S, et al. Analysis of STAT4 expression in cutaneous T-cell lymphoma (CTCL) patients and patient-derived cell lines. Cell Cycle. 2014;13(18):2975–2982. | ||
Zhou J, Xu XZ, Hu YR, Hu AR, Zhu CL, Gao GS. Cryptotanshinone induces inhibition of breast tumor growth by cytotoxic CD4+ T cells through the JAK2/STAT4/ perforin pathway. Asian Pac J Cancer Prev. 2014;15(6):2439–2445. | ||
Weaver JR, Nadler JL, Taylor-Fishwick DA. Interleukin-12 (IL-12)/STAT4 axis is an important element for beta-cell dysfunction induced by inflammatory cytokines. PLoS One. 2015;10(11):e0142735. | ||
Khalkhali-Ellis Z, Abbott DE, Bailey CM, et al. IFN-gamma regulation of vacuolar pH, cathepsin D processing and autophagy in mammary epithelial cells. J Cell Biochem. 2008;105(1):208–218. | ||
Singh SB, Ornatowski W, Vergne I, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12(12):1154–1165. | ||
Lue HW, Cole B, Rao SA, et al. Src and STAT3 inhibitors synergize to promote tumor inhibition in renal cell carcinoma. Oncotarget. Epub November 26, 2015. | ||
Chen J, Lan T, Zhang W, et al. Dasatinib enhances cisplatin sensitivity in human esophageal squamous cell carcinoma (ESCC) cells via suppression of PI3K/AKT and Stat3 pathways. Arch Biochem Biophys. 2015;575:38–45. | ||
Egloff AM, Grandis JR. Improving response rates to EGFR-targeted therapies for head and neck squamous cell carcinoma: candidate predictive biomarkers and combination treatment with Src inhibitors. J Oncol. 2009;2009:896407. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.